Abstract
A novel, rapid, and noninvasive test (ODK0501) to detect Streptococcus pneumoniae antigen was evaluated in a Japanese multicenter study. ODK0501 uses polyclonal antibodies to detect C polysaccharide of S. pneumoniae from sputum samples by an immunochromatographic assay. The utility of ODK0501 was evaluated for 161 adult patients with lower respiratory tract infection between March 2006 and March 2007. Bacterial culture and identification, real-time PCR, and ODK0501 assays were performed on sputum samples, and the Binax Now Streptococcus pneumoniae antigen test was performed using urine samples obtained from the same patients. The performances of all tests were compared based on the results of bacterial culture and identification. The sensitivity and specificity of ODK0501 were 89.1% (49/55 samples) and 95.3% (101/106 samples), respectively. We then compared the Binax Now Streptococcus pneumoniae antigen test with ODK0501 using samples from 142 patients. The sensitivities of ODK0501 and the Binax Now S. pneumoniae antigen test were 90.0% (45/50 samples) and 62.0% (31/50 samples), respectively (P = 0.002). The relative quantity of S. pneumoniae in expectorated sputum was calculated using real-time PCR and indicated that the possibility of false-positive results for ODK0501 due to indigenous S. pneumoniae was low. The positive and negative concordance rates of ODK0501 and Binax Now were 96.8% (30/31 samples) and 21.1% (4/19 samples), respectively. Binax Now was less capable of detecting S. pneumoniae antigen among patients with underlying chronic obstructive pulmonary disease. In conclusion, ODK0501 is noninvasive, rapid, and an accurate tool for diagnosing respiratory infection caused by S. pneumoniae.
Streptococcus pneumoniae is a frequent cause of community-acquired bacterial pneumonia and lower respiratory tract infection (10, 11, 19, 21) and a major cause of significant morbidity and mortality in Japan and the rest of the world (5).
Since pneumococcal infections are common and can be severe, appropriate initial selection of antimicrobial agents is crucial to an optimal outcome. Rapid and precise identification of the causative agents of infectious diseases is critical but challenging. Pathogen-oriented, prompt selection and application of antimicrobial agents improve prognosis, reduce medical costs, and prevent the development of drug-resistant bacteria due to their inappropriate use. The “gold standard,” bacterial culture to identify causative microorganisms, requires several days to yield a result and is thus unhelpful for the initial selection of appropriate antibacterial drugs. Although Gleckman et al. previously reported that classic Gram staining of sputum, which is simple and inexpensive, is useful for the diagnosis of bacterial infections (7), Garcia-Vazquez et al. and Reed et al. found that it is ineffective for rapid diagnoses (6, 18). In addition, staining results rely on several factors such as the quality of the sputum samples and the skill of laboratory personnel in processing samples (7). Thus, Gram staining is not recommended for all patients with community-acquired pneumonia, according to the most recent “Infectious Diseases Society of America/American Thoracic Society Consensus Guidelines on the Management of Community-Acquired Pneumonia in Adults” (12).
Urinary antigen detection is an alternative rapid diagnostic technique for detecting antigens of S. pneumoniae and Legionella pneumophila as respiratory pathogens. The Binax Now Streptococcus pneumoniae urinary antigen test (Binax, Inc., Portland, ME) detects cell wall antigens secreted in urine using an immunochromatographic method to separate capsular polysaccharide (C-ps) of S. pneumoniae. The test is noninvasive and rapid (the entire assay can be completed in about 15 min), and the specificity and sensitivity of detecting pneumococcal infections in adult patients are >90% and 50% to 80%, respectively, which allows early diagnosis (3, 8, 16). Moreover, even when S. pneumoniae cannot be identified by bacterial culture tests after the start of antibiotic therapy, the assay can still detect this organism. However, Binax Now has significant problems, including false-positive results due to indigenous S. pneumoniae in children (17) and antigens that can be detected even 1 to 3 months after treatment (13, 15).
ODK0501 (Otsuka Pharmaceutical Co., Ltd., Tokyo, Japan) is a diagnostic kit that uses polyclonal antibodies to detect antipneumococcal C-ps, a cell wall antigen of S. pneumoniae (4). This immunochromatographic method is rapid and noninvasive and analyzes sputum derived directly from a local infection site. Here, we evaluated the ability ODK0501 to detect S. pneumoniae among patients with lower respiratory tract infection at several Japanese medical institutions and compared its performance with that of the Binax Now assay. The goal of this study is to prove the usefulness of ODK0501 and its performance in clinical settings.
MATERIALS AND METHODS
Study settings.
This case-control study proceeded between March 2006 and March 2007 at the following 14 medical institutions in Japan: the Nagasaki University School of Medicine, Saitama Cardiovascular and Respiratory Center, National Hospital Organization Ureshino Medical Center, Isahaya Health Insurance General Hospital, Hokusho Central Hospital, Japanese Red Cross Nagasaki Genbaku Isahaya Hospital, Kurashiki Central Hospital, Sasebo City General Hospital, Toranomon Hospital, Nakahama Clinic, Ohmichi Internal and Respiratory Medicine, Kitakyushu Municipal Yahata Hospital, University of Tokushima School of Medicine, and Shinrakuen Hospital. This study was approved by the institutional review board of each medical institute, and informed consent was obtained from each subject.
All adult inpatients or outpatients (age, ≥15 years) with signs of lower respiratory infections including pneumonia and who had undergone sputum exploration were eligible for inclusion in this study. Prior antibiotic use was not a criterion for exclusion. Patients with a provisional diagnosis of lower respiratory infections were assessed by each study investigator to confirm the diagnosis. Clinical information such as existing underlying diseases, prior antibiotic use, number of days from onset until the day when assays were performed, white blood cell (WBC) counts, and C-reactive protein (CRP) levels were recorded when the patients registered and were retrospectively analyzed.
Sample collection and microbiological investigations.
Single expectorated sputum and urine samples were collected from patients who had provided written, informed consent to participate in this study either at a hospital visit or at the time of a presumptive diagnosis of lower respiratory infection. All expectorated sputum samples were conventionally Gram stained and transferred to the microbiology laboratory at Hokusho Central Hospital for expert evaluation. Sputum samples were also cultured at 37°C for 24 h on blood and chocolate agar at each institution. Presumptive colonies of bacteria were picked up and identified by biochemical testing. Some portions of the expectorated sputum samples were placed in different containers at the time of sampling and stored at −20°C until assay with real-time PCR to detect and quantify S. pneumoniae organisms.
Samples were immediately evaluated using ODK0501 and Binax Now or were otherwise stored at 4°C until the next available day to perform these tests (all assays were performed within 48 h after collection of samples).
Soluble Legionella pneumophila serogroup 1 antigen in urine was investigated using another immunochromatographic assay, and antibodies against Mycoplasma pneumoniae, Chlamydophila psittaci, and Chlamydophila pneumoniae were examined using complement fixation tests.
Measurement and interpretation of results obtained with ODK0501 and Binax Now.
Each of the participating institutions followed the procedures indicated below.
Sputum samples collected on swabs from containers were shaken in tubes containing sample extract and left for 5 min. Swab tips were removed from the tubes while squeezing, a filter was placed onto the tube, and the extract was dropped onto the ODK0501 instrument. Approximately 20 min later, a positive test result was indicated by the presence of one red test line and one control line (two lines), whereas a negative result was indicated by the appearance of only the control line.
Results obtained using the Binax Now assay were measured and interpreted at the each of the institutions in accordance with the instructions provided by the manufacturer (3).
Criteria for etiological diagnosis.
We evaluated only qualified samples defined according to standard criteria (presence of >25 WBC and <25 squamous cells per low-power magnification field [×100]). The following criteria were used to classify a case of lower respiratory infection as being of known etiology: (i) for M. pneumoniae C. psittaci, and C. pneumoniae, at least a fourfold increase in antibody, as determined by the complement fixation test; (ii) Legionella antigen detected in urine; and (iii) for S. pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, Staphylococcus aureus, and other bacteria, including gram-negative enterobacteria, isolation of the pathogen of the predominant organism from a qualified sputum sample. Patients who did not fulfill the etiological diagnostic criteria described above were considered to have lower respiratory infection of unknown etiology.
Real-time PCR for pneumococcal surface protein A (PspA) gene quantitation in expectorated sputum samples.
We extracted DNA from expectorated sputum samples using the QIAamp DNA minikit (Qiagen, Valencia, CA), and the relative amount of S. pneumoniae was quantified by real-time PCR using a primer and TaqMan probe established for the region of the PspA gene (2, 22) of S. pneumoniae (Table 1). Real-time PCR proceeded using an ABI7700 or ABI7900 apparatus (Applied Biosystems, Foster City, CA) at Kitazato-Otsuka Biomedical Assay Laboratory, Kanagawa, Japan, and sequences were analyzed using Sequence Detector V.1.7a, V.2.2.2 SDS (Applied Biosystems, Foster City, CA). RNase-free water and the extracted DNA from S. pneumoniae ATCC 6303 were used for negative and positive controls, respectively. After an initial denaturation step at 95°C for 15 min, PCR products were amplified by 50 cycles of 15 s at 94°C and 1 min at 60°C. Copy numbers of the PspA gene were calculated and compared among ODK0501- and culture-positive and -negative sputum samples.
TABLE 1.
Primers used for real-time PCR for quantification of Streptococcus pneumoniae
| Primer or probe | Sequencea |
|---|---|
| Primers | |
| Forward | 5′-CAAGTCTAGCCAGCGTCGCTAT-3′ |
| Reverse | 5′-GGGAGATTCTTCTGCTCTTACAAAAG-3′ |
| 5′-GGGAGATTCTTCTGCTCTTACCAAAG-3′ | |
| 5′-GGGAGATTCTTCTGCTCTTACAACAG-3′ | |
| TaqMan probes | 5′-FAM-CTGAGACGCAACAAAACCAGCCCC-TAMRA-3′ |
| 5′-FAM-CTGAGACGTAACAAAACCAGCCCC-TAMRA-3′ | |
| 5′-FAM-CGAAGACGCAACAAAACCAGCCCC-TAMRA-3′ |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
Statistical analysis.
The sensitivity and specificity based on the results of the sputum cultures of ODK0501 and Binax Now were compared using Fisher's exact probability test (two tailed, with the level of significant difference at a P value of <0.05). Characteristics of patients with pneumococcal infection diagnosed by both culture and ODK0501 as being positive were also compared with Binax Now-positive or -negative arms. We compared the proportions of qualitative variables using Fisher's exact test and applied the Mann-Whitney U test for quantitative variables. Relationships between the results obtained from real-time PCR for relative amounts of S. pneumoniae among the ODK0501-tested population were also assessed using the Mann-Whitney U test.
RESULTS
Composition of patients and etiological agents identified from patients with lower respiratory tract infection.
Of the 246 patients exhibiting signs and symptoms compatible with lower respiratory infection, 85 were excluded due to poor sputum quality (presence of <25 WBC and/or >25 squamous cells per low-power magnification field [×100]).
We thus investigated samples from 161 inpatients and outpatients (109 males [age range, 22 to 92 years; median age, 71.0 years] and 52 females [age range, 15 to 96 years; median age, 65.5 years]) with lower respiratory tract infection. Table 2 shows the clinical statuses of the patients. Most had pneumonia (79.5%), followed by acute exacerbation of chronic obstructive pulmonary disease (COPD) (AECOPD) (7.5%), acute bronchitis (6.2%), bronchiectasis (5.0%), and chronic bronchitis (1.2%), and only one had a lung abscess. We examined samples from 161 and 142 patients using ODK0501 and Binax Now, respectively. Table 2 also shows etiological agents. According to conventional microbiological criteria, etiological agents were identified in 106 patients (65.8%). The following organisms were detected and determined to be causative agents: S. pneumoniae in 55 samples (34.2%), Haemophilus influenzae in 25 samples (15.5%), M. pneumoniae in 5 samples (3.1%), Staphylococcus aureus in 8 samples (5.0%), Pseudomonas aeruginosa in 7 samples (4.3%), Moraxella catarrhalis in 5 samples (3.1%), and Klebsiella pneumoniae in 2 samples (1.2%). Four patients with mixed S. pneumoniae and H. influenzae infection were in the pneumonia and AECOPD groups. Two other patients in the pneumonia group had mixed infection due to S. pneumoniae and C. pneumoniae and due to M. pneumoniae and P. aeruginosa. The microbial etiology remained unknown after conventional microbiological culture and antibody test for 55 (34.2%) of the 161 patients.
TABLE 2.
Patient condition group and ability of ODK0501 to detect Streptococcus pneumoniae
| Patient condition group and organism | No. of patients (%) | No. of ODK0501-positive patients/total no. of patients (%) |
|---|---|---|
| Pneumonia | 128 (79.5) | |
| Streptococcus pneumoniae | 37/43 (86.0) | |
| Haemophilus influenzae | 1/16 (6.3) | |
| S. pneumoniae and H. influenzae | 3/3 (100.0) | |
| Staphylococcus aureus | 0/7 | |
| Moraxella catarrhalis | 0/3 | |
| Pseudomonas aeruginosa | 0/3 | |
| Klebsiella pneumoniae | 0/2 | |
| Streptococcus milleri group | 0/1 | |
| Mycoplasma pneumoniae | 0/4 | |
| Aspergillus spp. | 0/1 | |
| Legionella pneumophila | 0/1 | |
| Chlamydophila psittaci | 0/1 | |
| S. pneumoniae and C. pneumoniae | 1/1 (100.0) | |
| M. pneumoniae and P. aeruginosa | 0/1 | |
| Pathogen not identified | 3/41 (7.3) | |
| Acute exacerbation of COPD | 12 (7.5) | |
| S. pneumoniae | 4/4 (100.0) | |
| H. influenzae | 0/2 | |
| S. pneumoniae and H. influenzae | 1/1 (100.0) | |
| M. catarrhalis | 0/2 | |
| Pathogen not identified | 0/3 | |
| Acute bronchitis | 10 (6.2) | |
| H. influenzae | 0/2 | |
| S. pneumoniae | 1/1 (100.0) | |
| S. aureus | 0/1 | |
| Pathogen not identified | 0/6 | |
| Bronchiectasis | 8 (5.0) | |
| P. aeruginosa | 0/3 | |
| S. pneumoniae | 1/1 (100.0) | |
| Pathogen not identified | 0/4 | |
| Chronic bronchitis | 2 (1.2) | |
| S. pneumoniae | 1/1 (100.0) | |
| Pathogen not identified | 1/1 (100.0) | |
| Lung abscess | 1 (0.6) | |
| H. influenzae | 0/1 | |
| Total | 161 | 54/161 |
Overall sensitivity and specificity of ODK0501.
The ODK0501 results were positive for 54 of 161 patients (33.5%). Table 2 shows the distribution of ODK0501 positivity in each patient group. ODK0501 was positive for 41 (87.2%) of 47 patients with pneumococcal pneumonia, including four patients with mixed infections and all patients with AECOPD (n = 4), acute bronchitis (n = 1), bronchiectasis (n = 1), and chronic bronchitis (n = 1) caused by S. pneumoniae. The test sensitivity was 89.1% (49/55), the specificity was 95.3% (101/106), and the positive and negative predictive values were 90.7% and 94.4%, respectively, based on the results of the sputum cultures.
Comparison of sensitivity and specificity of ODK0501 with those of Binax Now.
We compared the sensitivity and specificity of ODK0501 with those of Binax Now against sputum culture results from 142 of the 161 patients (the Binax Now assay was not performed for 19 of 161 patients).
The sensitivity of the ODK0501 test was 90.0% (45/50 patients), the specificity was 95.7% (88/92), and the positive and negative predictive values were 91.8% and 94.6%, respectively, based on sputum cultures. The sensitivity of the Binax Now test was 62.0% (31/50 patients), the specificity was 96.7% (89/92), and the positive and negative predictive values were 91.2% and 82.4%, respectively, based on the sputum cultures. Specificities did not significantly differ between the two tests, but the sensitivity of ODK0501 for detecting S. pneumoniae was significantly higher (P = 0.002).
Quantification of the PspA gene in sputum by real-time PCR.
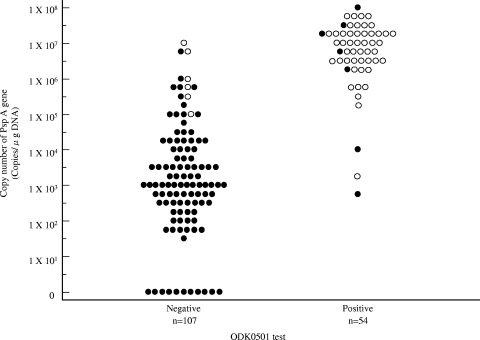
We applied real-time PCR to detect and quantify the relative amounts of S. pneumoniae organisms in expectorated sputum samples from 161 patients with lower respiratory tract infection. Figure 1 shows that PspA gene copy numbers were higher among the ODK0501-positive and culture-positive population. The population that was both ODK0501 negative and culture negative exhibited lower copy numbers of the PspA gene in sputum. The copy numbers of the PspA gene significantly differed among ODK0501-positive and -negative populations (P < 0.001).
FIG. 1.
Distributions of S. pneumoniae PspA gene copy numbers in sputum among ODK0501- and culture-positive and -negative populations. Vertical axis, copy number of the PspA gene calculated from real-time PCR. •, S. pneumoniae culture negative; ○, S. pneumoniae culture positive. ODK0501-positive and culture-positive populations contain more copies of the S. pneumoniae PspA gene. Copy numbers significantly differed between ODK0501-positive and -negative populations (P < 0.001).
False-positive and false-negative ODK0501 results among patients with lower respiratory infection.
Table 3 summarizes the clinical and laboratory data for the five S. pneumoniae culture-negative patients with lower respiratory infection and apparent false-positive ODK0501 results. Binax Now was positive and the PspA gene copy number was >6 × 106 for two patients (patients 2 and 4).
TABLE 3.
Clinical and laboratory data for patients with false-positive results by ODK0501a
| Patient | Final diagnosis | Gender, age (yr) | Underlying disease | Prior antibiotic therapy | Sputum finding | Binax Now S. pneumoniae antigen test result | No. of copies/μg DNAb | Time from symptom onset until assay performance (days) |
|---|---|---|---|---|---|---|---|---|
| 1 | Pneumonia | M, 43 | Bronchial asthma | None | ND | − | 2 × 106 | 4 |
| 2 | Pneumonia | M, 79 | COPD | SBT-ABPC, CAM | Aspergillus spp. | + | 6 × 106 | 1 |
| 3 | Pneumonia | M, 62 | Bronchiectasis | None | Haemophilus influenzae | − | 7 × 102 | 2 |
| 4 | Pneumonia | F, 90 | None | CTM-HE | ND | + | 3 × 107 | 6 |
| 5 | Chronic bronchitis | F, 50 | None | None | Mixed oral flora | − | 1 × 104 | 3 |
M, male; F, female; SBT-ABPC, sulbactam-ampicillin; CAM, clarithromycin; CTM-HE, cefotiam-hexetil; ND, not detectable.
Determined by S. pneumoniae real-time PCR.
Table 4 summarizes the clinical and laboratory data for six patients with lower respiratory infection who generated false-negative results by ODK0501 but who were positive by S. pneumoniae cultures. All of these patients had pneumonia, and Binax Now was positive for only one of them (patient 3). All of these patients had >1 × 105 copies of the PspA gene.
TABLE 4.
Clinical and laboratory data for patients with false-negative results with ODK0501a
| Patient | Final diagnosis | Gender, age (yr) | Underlying disease | Prior antibiotic therapy | Result of Gram stain for S. pneumoniae | Binax Now S. pneumoniae antigen test result | No. of copies/μg DNAb | Time from symptom onset until assay performance (days) |
|---|---|---|---|---|---|---|---|---|
| 1 | Pneumonia | M, 28 | None | None | + | − | 6 × 106 | 1 |
| 2 | Pneumonia | M, 65 | COPD | None | − | − | 1 × 106 | 3 |
| 3 | Pneumonia | M, 33 | None | CAM, CAZ | + | + | 9 × 106 | 5 |
| 4 | Pneumonia | F, 33 | None | None | − | − | 5 × 105 | 2 |
| 5 | Pneumonia | M, 42 | Interstitial pneumonia | SBTPC | + | Not tested | 4 × 105 | 2 |
| 6 | Pneumonia | M, 64 | Collagen disease | CTRX | − | − | 1 × 105 | 6 |
CAM, clarithromycin; CAZ, ceftazidime; SBTPC, sultamicillin; CTRX, ceftriaxone.
Determined by S. pneumoniae real-time PCR.
Comparison between ODK0501 and Binax Now performances among patients with pneumococcal infections.
We compared the performances of ODK0501 and Binax Now among 50 patients with pneumococcal infection. Five of 55 pneumococcal patients were excluded due to the absence of Binax Now data. The positive and negative concordance rates of ODK0501 and Binax Now were 96.8% (30/31 patients) and 21.1% (4/19), respectively.
Fifteen patients were ODK0501 positive and Binax Now negative. Table 5 summarizes the clinical and laboratory findings for the ODK0501-positive population among pneumococcal infections proven by culture test. Age, gender, final diagnosis, underlying disease, prior antibiotic use, WBC count, CRP level, copy numbers of the PspA gene, and days from onset until assay did not significantly differ between Binax Now-positive and -negative populations. However, the ODK0501-positive and Binax Now-negative population contained statistically more patients with COPD than the population that was positive by both tests (P = 0.016). Only patient 3 was ODK0501 negative and Binax Now positive (Table 4).
TABLE 5.
Comparison of clinical and laboratory data between the ODK0501-positive and Binax Now-positive arm and the ODK0501-positive and Binax Now-negative arm
| Characteristic | Value for group
|
P value | |
|---|---|---|---|
| ODK0501 positive and Binax Now positive (n = 30) | ODK0501 positive and Binax Now negative (n = 15) | ||
| Median age (yr) (range) | 67 (22-86) | 72 (34-84) | 0.718 |
| No. of male patients | 21 (70.0) | 14 (93.3) | 0.129 |
| No. of patients (%) with final diagnosis of: | |||
| Pneumonia | 28 (93.3) | 12 (80.0) | 0.315 |
| Bronchiectasis | 1 (3.3) | 0 (0.0) | 1 |
| AECOPD | 1 (3.3) | 1 (6.7) | 1 |
| Acute bronchitis | 0 (0.0) | 1 (6.7) | 1 |
| Chronic bronchitis | 0 (0.0) | 1 (6.7) | 1 |
| No. of patients (%) with underlying disease includinga: | 18 (60.0) | 10 (66.7) | 0.752 |
| COPD | 5 (16.7) | 8 (53.3) | 0.016 |
| Diabetes | 3 (10.0) | 1 (6.7) | 1 |
| Chronic lung disease | 0 (0.0) | 0 (0.0) | 1 |
| Chronic heart disease | 1 (3.3) | 0 (0.0) | 1 |
| Chronic liver disease | 1 (3.3) | 0 (0.0) | 1 |
| Chronic renal insufficiency | 0 (0.0) | 0 (0.0) | 1 |
| Cancer | 2 (6.7) | 1 (6.7) | 1 |
| Dementia | 0 (0.0) | 0 (0.0) | 1 |
| No. of patients (%) on prior antibiotic therapy | 5 (16.7) | 0 (0.0) | 0.153 |
| Avg CRP level (mg/dl) (range) | 14 (0.3-31.5) | 11 (2.0-24.0) | 0.448 |
| Avg WBC count (mm3) (range) | 12,224 (2,800-31,700) | 13,723 (6,800-48,800) | 0.754 |
| No. of copies/μg DNA (range)b | 1.6 × 107 (2 × 103-9 × 107) | 1.8 × 107 (2 × 105-7 × 107) | 0.431 |
| Time from onset of symptoms until assay (days) (range) | 4 (0-10) | 2 (0-9) | 0.420 |
One or more of these conditions.
Determined by S. pneumoniae real-time PCR.
DISCUSSION
Although early diagnosis and antimicrobial drug administration comprise the basic principles of treating pneumococcal as well as other infections, the causative agents may not be correctly identified for many patients at the start of therapy.
Binax Now is commercially available and widely used in clinical settings, but it possesses some disadvantages, including false-positive results due to indigenous S. pneumoniae (17), lasting positivity even 1 to 3 months after treatment (13, 15), and no indication for anuric patients. Furthermore, the test has a variable and relatively low sensitivity of 50 to 80% (3, 8, 16). The novel ODK0501 tests sputum samples derived directly from inflammatory sites using polyclonal antibodies to detect antipneumococcal C-ps (20), a common cell wall antigen of S. pneumoniae (4). Thus, all types of S. pneumoniae can be detected regardless of capsule serotype.
The present clinical study evaluated the applicability of ODK0501 and compared it with that of Binax Now in adult patients (age, ≥15 years). The most frequently found isolate in the present study was S. pneumoniae (34.7%) followed by H. influenzae among 161 patients with lower respiratory tract infection. These findings indicated that the frequency of detection of major isolated bacteria from patients with lower respiratory tract infection was similar to those reported previously for other studies, and thus, our patient population was valid for analysis.
ODK0501 yielded favorable results, including a sensitivity and specificity of 89.1% and 95.3%, respectively, based on sputum culture results from 161 patients. Even when a total of 120 cases with superior quality of expectorated sputum (presence of >25 WBC and <10 squamous cells per low-power magnification field [×100]) were analyzed (data not shown), the result showed higher sensitivity (92.9%) and almost equal specificity (93.6%). The sensitivity (87.2%) and specificity (95.1%) were also favorable when evaluated among 128 pneumonia cases only.
The sensitivity of ODK0501 was significantly high (P = 0.002) and the specificity was >95% compared with those of Binax Now. The negative predictive value of Binax Now was also lower than that of ODK0501. These data indicated that ODK0501 is superior to Binax Now. However, because S. pneumoniae can be indigenous to the nasal cavity and nasopharyngeal and intraoral regions of some elderly individuals, infants, and children (1, 9), sputum or nasopharyngeal samples obtained from such individuals might be S. pneumoniae positive by the ODK0501 test. The question will be raised whether the higher sensitivity of ODK0501 suggests that indigenous, colonizing S. pneumoniae in the respiratory tract detected by this method will be considered to be the cause of infection and lead to overdiagnosis. To answer this question, we measured amounts of S. pneumoniae in expectorated sputum. PspA was originally considered to be a potential antigen and virulence factor of S. pneumoniae (14) and was recently applied for epidemiological analysis of S. pneumoniae infection (2). Since a single copy of the PspA gene exists per S. pneumoniae cell and the copy number in sputum relatively correlated to the numbers of S. pneumoniae cells (data not shown), we evaluated PspA gene copy numbers in sputum samples using real-time PCR. The PCR data were then substituted as relative amounts of S. pneumoniae in sputum samples. However, because the amounts of sputum samples varied, values were corrected using human DNA concentrations in the sputum, and the number of copies/μg DNA (calculated based on human DNA concentrations in sputum) was used as the unit of measurement. The result for quantitative cultures using an adjusted bacterial suspension of 9.45 × 105 CFU/ml as the measurement sample was nearly equivalent to 4.3 × 105 copies/μg DNA, indicating a nearly 1:1 relationship (data not shown). Most ODK0501-negative patients also had negative culture results as well as fewer S. pneumoniae cells. Hence, ODK0501-positive patients also had positive culture results and more S. pneumoniae cells according to real-time PCR. These data suggested that ODK0501 does not detect small amounts of S. pneumoniae, which translates as a low false-positive rate in the presence of indigenous S. pneumoniae. Real-time PCR data also supported the finding that the analytical sensitivity of ODK0501, which was approximately 104 CFU/ml in the development phase (4), was appropriate to detect S. pneumoniae as the cause of lower respiratory infection.
Analysis of the clinical characteristics of five ODK0501 false-positive patients revealed no specific factors that could account for this result. However, Binax Now was positive for two patients in whom we detected 6 × 106 copies of the PspA gene, suggesting that culture did not detect S. pneumoniae for reasons that remain unknown. The severities of pneumonia among the five patients scored as defined by the Japanese Respiratory Society guidelines for the management of community-acquired pneumonia in adults did not differ (data not shown).
Six patients were false negative by ODK0501, and their clinical backgrounds also did not offer any apparent explanations. Only one patient was Binax Now positive, but four were Binax Now negative, indicating that Binax Now does not offer any advantages as a rapid diagnostic tool for pneumococcal infection.
Among the 50 patients with S. pneumoniae infection diagnosed by culture, 15 (30.0%) were Binax Now negative and ODK0501 positive, indicating that ODK0501 possesses more power to detect S. pneumoniae antigen than Binax Now. We investigated which clinical factors were related to the higher sensitivity of ODK0501 in diagnosing pneumococcal infections. We compared the clinical backgrounds of the ODK0501-positive and Binax Now-positive arm and the ODK0501-positive and Binax Now-negative arm. We found that only underlying COPD statistically differed among all tested factors, although the number of COPD cases was only 13. In these COPD patients, age, prior antibiotic use, WBC count, CRP level, copy numbers of the PspA gene, and days from onset until assay were compared, and no significant differences between Binax-positive and -negative populations were found (data not shown). Although whether the detection rate of Binax Now decreases in COPD patients remains unknown, ODK0501 was more sensitive in the present study.
The relationships between the severity of pneumonia and the sensitivity of ODK0501 and Binax Now are also important. We compared the performances of ODK0501 and Binax Now with the severity of pneumonia scored according to the definitions of the Japanese Respiratory Society guidelines for the management of community-acquired pneumonia in adults among patients with community-acquired pneumonia and found no significant differences between the detection rates of pneumococcal pneumonia at any level of severity between the two assays (data not shown).
In conclusion, since ODK0501 evaluates sputum samples from local sites of infection, the results reflect the real-time status of infection due to S. pneumoniae. Our study indicated that ODK0501 was statistically more sensitive than the Binax Now Streptococcus pneumoniae urinary antigen test, suggesting that ODK0501 will significantly contribute to the early, rapid, and noninvasive diagnosis of pneumococcal infection in adults.
Acknowledgments
We have no conflicts of interest.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Abe, S., K. Ishihara, and K. Okuda. 2001. Prevalence of potential respiratory pathogens in the mouths of elderly patients and effects of professional oral care. Arch. Gerontol. Geriatr. 3245-55. [DOI] [PubMed] [Google Scholar]
- 2.Brandileone, M. C., A. L. Andrade, E. M. Teles, R. C. Zanella, T. I. Yara, J. L. Di Fabio, and S. K. Hollingshead. 2004. Typing of pneumococcal surface protein A (PspA) in Streptococcus pneumoniae isolated during epidemiological surveillance in Brazil. Vaccine 223890-3896. [DOI] [PubMed] [Google Scholar]
- 3.Dominguez, J., N. Gali, S. Blanco, P. Pedroso, C. Prat, L. Matas, and V. Ausina. 2001. Detection of Streptococcus pneumoniae antigen by a rapid immunochromatographic assay in urine samples. Chest 119243-249. [DOI] [PubMed] [Google Scholar]
- 4.Ehara, N., K. Fukushima, H. Kakeya, H. Mukae, S. Akamatsu, A. Kageyama, A. Saito, and S. Kohno. 2008. A novel method for rapid detection of Streptococcus pneumoniae antigen in sputum and its application in adult respiratory tract infections. J. Med. Microbiol. 57820-826. [DOI] [PubMed] [Google Scholar]
- 5.Fine, M. J., M. A. Smith, C. A. Carson, S. S. Mutha, S. S. Sankey, L. A. Weissfeld, and W. N. Kapoor. 1996. Prognosis and outcomes of patients with community-acquired pneumonia. JAMA 275134-141. [PubMed] [Google Scholar]
- 6.Garcia-Vazquez, E., M. A. Marcos, J. Mensa, A. de-Roux, J. Puig, C. Font, G. Francisco, and A. Torres. 2004. Assessment of the usefulness of sputum culture for diagnosis of community-acquired pneumonia using the PORT predictive scoring system. Arch. Intern. Med. 1641807-1811. [DOI] [PubMed] [Google Scholar]
- 7.Gleckman, R., J. DeVita, D. Hibert, C. Pelletier, and R. Martin. 1988. Sputum gram stain assessment in community-acquired bacteremic pneumonia. J. Clin. Microbiol. 26846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutiérrez, F., M. Masiá, J. C. Rodriguez, A. Ayelo, B. Soldán, L. Cebrián, C. Mirete, G. Royo, and A. M. Hidalgo. 2003. Evaluation of the immunochromatographic Binax NOW assay for detection of Streptococcus pneumonia urinary antigen in a prospective study of community-acquired pneumonia in Spain. Clin. Infect. Dis. 36286-292. [DOI] [PubMed] [Google Scholar]
- 9.Hamer, D. H., J. Egas, B. Estrella, W. B. MacLeod, J. K. Griffiths, and F. Semoertegui. 2002. Assessment of the Binax NOW Streptococcus pneumonia urinary antigen test in children with nasopharyngeal pneumococcal carriage. Clin. Infect. Dis. 341025-1028. [DOI] [PubMed] [Google Scholar]
- 10.Ishida, T., T. Hashimoto, M. Arita, Y. Tojo, H. Tachibana, and M. Jinnai. 2004. A 3-year prospective study of a urinary antigen-detection test for Streptococcus pneumoniae in community-acquired pneumonia: utility and clinical impact on the reported etiology. J. Infect. Chemother. 10359-363. [DOI] [PubMed] [Google Scholar]
- 11.Llor, C., J. M. Cots, and A. Herreras. 2006. Bacterial etiology of chronic bronchitis exacerbations treated by primary care physicians. Arch. Bronconeumol. 42388-393. [DOI] [PubMed] [Google Scholar]
- 12.Mandell, L. A., R. G. Wunderink, A. Anzueto, J. G. Bartlett, G. D. Campbell, N. C. Dean, S. F. Dowell, T. M. File, Jr., D. M. Musher, M. S. Niederman, A. Torres, and C. G. Whitney. 2007. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 44S27-S72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcos, M. A., M. T. Jiménez-de-Anta, J. P. de-la-Bellacasa, J. González, E. Martinez, E. Garcia, J. Mensa, A. de-Roux, and A. Torres. 2003. Rapid urinary antigen test for diagnosis of pneumococcal community-acquired pneumonia in adults. Eur. Respir. J. 21209-214. [DOI] [PubMed] [Google Scholar]
- 14.McDaniel, L. S., G. Scott, J. F. Kearney, and D. E. Briles. 1984. Monoclonal antibodies against protease-sensitive pneumococcal antigens can protect mice from fatal infection with Streptococcus pneumoniae. J. Exp. Med. 160386-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murdoch, D. R., R. T. Laing, and J. M. Cook. 2003. The NOW S. pneumoniae urinary antigen test positivity rate 6 weeks after pneumonia onset and among patients with COPD. Clin. Infect. Dis. 37153-154. [DOI] [PubMed] [Google Scholar]
- 16.Murdoch, D. R., R. T. Laing, G. D. Mills, N. C. Karalus, G. I. Town, S. Mirrett, and L. B. Reller. 2001. Evaluation of a rapid immunochromatographic test for detection of Streptococcus pneumoniae antigen in urine samples from adults with community-acquired pneumonia. J. Clin. Microbiol. 393495-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro, D., L. García-Maset, C. Gimeno, A. Escribano, and J. García-de-Lomas. 2004. Performance of the Binax NOW Streptococcus pneumoniae urinary antigen assay for diagnosis of pneumonia in children with underlying pulmonary diseases in the absence of acute pneumococcal infection. J. Clin. Microbiol. 424853-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reed, W. W., G. S. Byrd, R. H. Gates, Jr., R. S. Howard, and M. J. Weaver. 1996. Sputum Gram's stain in community-acquired pneumococcal pneumonia: a meta-analysis. West. J. Med. 165197-204. [PMC free article] [PubMed] [Google Scholar]
- 19.Saito, A., S. Kohno, T. Matsushima, A. Watanabe, K. Oizumi, K. Yamaguchi, and H. Oda. 2006. Prospective multicenter study of the causative organisms of community-acquired pneumonia in adults in Japan. J. Infect. Chemother. 1263-69. [DOI] [PubMed] [Google Scholar]
- 20.Skov-Sørensen, U. B. 1995. Pneumococcal polysaccharide antigens: capsules and C-polysaccharide. Dan. Med. Bul. 4247-53. [PubMed] [Google Scholar]
- 21.Thomas, M. F., Jr. 2003. Community-acquired pneumonia. Lancet 3621991-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumonia, as revealed by sequence analysis. J. Bacteriol. 174601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]