Abstract
To further characterize the humoral immune response of pigs to porcine reproductive and respiratory syndrome virus (PRRSV), direct enzyme-linked immunosorbent assays (ELISA) were used to study the kinetics of antibody responses directed against PRRSV nonstructural proteins in pigs experimentally exposed to the virus. The highest immunoreactivities were against nsp1, nsp2, and nsp7. Using the recombinant nsp7 as an antigen, we validated a dual ELISA for the simultaneous detection and differentiation of serum antibodies against type I and type II PRRSV. Receiver operating characteristic analysis based on 1,334 known-positive and 1,357 known-negative samples showed good specificity (98.3% to type I and 99.3% to type II) and sensitivity (97.4% for type I and 99.8% for type II). To differentiate type I and type II PRRSV, 470 sera originating from experimentally inoculated pigs were tested, and positive sera were correctly differentiated in 469 of 470 samples. The capability of the nsp7 dual ELISA to detect serum antibody responses from pigs infected with various genetically different field strains was determined. The nsp7 dual ELISA possessed 97.6% agreement with the Idexx HerdChek PRRS 2XR ELISA. In further testing of Idexx ELISA suspected false-positive samples, the nsp7 dual ELISA resolved 98% of the samples as negative. Taken together, these results indicate that the nsp7 dual ELISA can be used as a differential test for PRRSV serology with high levels of sensitivity and specificity. This ELISA offers an additional tool for routine or follow-up diagnostics, as well as having substantial value in epidemiological surveys and outbreak investigations.
Porcine reproductive and respiratory syndrome (PRRS) continues to be one of the most devastating diseases of swine throughout the world. The etiological agent, PRRS virus (PRRSV), is classified in the genus Arterivirus, family Arteriviridae, order Nidovirales. Nucleotide sequence comparisons show that PRRSV can be divided into distinct European (type I) and North American (type II) genotypes, which share only about 63% nucleotide identity at the genomic level (1, 23). PRRSV is a small enveloped virus containing a positive-sense, single-stranded RNA genome about 15 kb in length, which contains nine known open reading frames (ORFs). The replicase-associated genes, ORF1a and ORF1b, are located on the 5′ end of the genome and encode the polyproteins pp1a and pp1ab. pp1a is predicted to be cleaved at eight sites to form nine nonstructural proteins (nsp): nsp1α, nsp1β, and nsp2 to nsp8 (6, 31). Proteolytic cleavage of the ORF1b portion of pp1ab generates products nsp9 through nsp12 (34). The products derived from pp1a possess proteolytic activities and are responsible for processing the other nsp cleavage products, whereas nsp9 to nsp12 are involved in virus transcription and replication (11, 31, 34). The 3′ end of the genome encodes four membrane-associated glycoproteins (GP2, GP3, GP4, and GP5; encoded by subgenomic [sg] mRNAs 2a and 3 to 5), two nonglycosylated membrane proteins (E and M; encoded by sg mRNAs 2b and 6), and a nucleocapsid (N; encoded by sg mRNA 7) (2, 17, 18, 19, 20, 21, 31, 32, 35, 36).
Serological testing to determine the PRRS status of herds and individual animals is a cost-effective tool in management strategies for monitoring and controlling PRRS. A large body of information shows that N is the most immunogenic protein and an ideal target for the serological detection of infected pigs (3, 5, 9, 29). Currently, the Idexx HerdChek PRRS 2XR enzyme-linked immunosorbent assay (ELISA), based on PRRSV N as the antigen, is widely used for the detection of antibodies produced in response to infection with North American type II or European-like type I PRRSV. However, individual unexpected positive Idexx ELISA results in otherwise seronegative herds have caused great concern, which requires the use of alternative antigens as more accurate indicators of infection. Previous studies from our laboratory and others showed that certain nsps, such as nsp2, are highly immunogenic (7, 13, 24, 25). The purpose of this study was to evaluate the humoral immune response of PRRSV-infected pigs to each of the nsps encoded by the ORF1a region of the viral genome. The kinetics of the appearance of a specific antibody response to each of the nsps was investigated in pigs experimentally infected by PRRSV. A highly immunogenic nsp, nsp7, was further evaluated to determine the feasibility for serology diagnostic-assay development.
MATERIALS AND METHODS
Viruses and cells.
MARC-145 cells were cultured in minimal Eagle's medium (Gibco BRL Life Technologies) with 10% fetal bovine serum and antibiotics (100 units/ml penicillin, 20 μg/ml streptomycin). The cells were maintained at 37°C in a humidified 5% CO2 incubator. PRRSV strains SD01-08 (type I) and VR2332 (type II) were propagated on MARC-145 cells using a method described previously (15).
Antigen production.
Recombinant proteins were generated using SD01-08 (type I) and VR2332 (type II) isolates. Based on the study of equine arteritis virus, the PRRSV ORF1a-encoded pp1a is predicted to be cleaved into eight products, nsp1 to nsp8 (31). nsp3 and nsp5 possess several predicted, nonimmunogenic hydrophobic domains, so they were not considered further. nsp6 is predicted to contain only 16 amino acids. A synthetic peptide made from these 16 amino acids was tested against sera from experimentally infected pigs. When used in an ELISA format, there was no detectable antibody response. Therefore, only PRRSV nsp1, nsp2, nsp4, nsp7, and nsp8 were considered in this study. These nsp regions from VR2332 were expressed as recombinant proteins in the pET-24b vector (Novagen) based on predicted cleavage sites (Table 1). Since nsp2 was expressed at low levels due to a C-terminal hydrophobic region, a C-terminally truncated portion was produced (13). Primers for amplifying each of the nsps are listed in Table 1. The nsp7 encoding regions amplified from SD01-08 were cloned in the pET-28a(+) vector (Novagen). Recombinant proteins were expressed and purified as we described previously (9, 13). Purified fusion proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting as described previously (13).
TABLE 1.
Primers for PRRSV ELISA antigen expression
| Proteina | Nucleotide location in the genomeb (amino acid location in pp1a) | Predicted recombinant-nsp mass (kDa) | Primer sequencesc |
|---|---|---|---|
| NA nsp1 | 191-1339 (1-383) | 42,950 | F: 5′ CGC GGA TCC TCT GGG ATA CTT GAT CGG TG |
| R: 5′ CCG CTC GAG GCC GTA CCA CTT GTG AC | |||
| NA nsp2P | 1340-3495 (384-1101)11 | 78,106 | F: 5′ CGC GGA TCC GCT GGA AAG AGA GCA AG |
| R: 5′ CCG CTC GAG TCG AGT ATC ATT TTT GGG AGG AAC | |||
| NA nsp4 | 5618-6229 (1810-2013) | 21,043 | F: 5′ CGC GGA TCC GGT GCT TTC AGA ACT CGA AAG CC |
| R: 5′ CCG CTC GAG TTC CAG TTC AGG TTT GGC AGC | |||
| NA nsp7 | 6788-7564 (2200-2458) | 28,620 | F: 5′ CGC GGA TCC TCT CTG ACT GGT GCC CTC GCT ATG |
| R: 5′ CCG CTC GAG TTC CCA TTG AAC TCT TCC AT | |||
| EU nsp7 | 6417-7223 (2066-2334) | 29,460 | F: 5′ GGAATTCTC CCT GAC GGC TGC TCT AGC TTG |
| R: 5′ GTG CTC GAG TTT CAA GGC AGT TGT CAG GCT TGG | |||
| NA nsp8 | 7565-7696 (2459-2502) | 4,872 | F: 5′ CGC GGA TCC GCT GCA AAG CTT TCC GTG G |
| R: 5′ CCG CTC GAG GTT TAA ACA CTG CTC CTT AGT C |
NA, North American genotype (type II); EU, European genotype (type I).
The numbers correspond to positions within the genome of type II PRRSV, VR2332 (GenBank accession number U87392) or type I PRRSV, SD01-08 (GenBank accession number DQ489311).
Restriction enzyme sites are boldface and italicized. F, forward; R, reverse.
Serum samples.
For type I PRRSV, a panel of serum samples (n = 320) from 32 pigs experimentally inoculated with one of four different type I PRRSV isolates, SD01-07, SD01-08, SD02-11, or SD03-15 (16), was used. They were collected at 7-day intervals for up to 85 days postinoculation. For type II PRRSV, serial serum samples (n = 1,014) were obtained from 109 pigs experimentally infected with type II PRRSV strain VR2332. They were collected at 7-day intervals for the first 2 weeks and then at 14-day intervals for up to 202 days postinoculation. In addition, 1,357 known-PRRSV-negative samples were obtained from negative control experimental pigs.
All of these serum samples, including 320 samples from type I PRRSV-infected animals, 1,014 samples from type II PRRSV-infected animals, and 1,357 samples from negative control animals, were used for validation of the nsp7-based ELISA. Among these 1,014 samples from type II PRRSV-infected animals, 510 serum samples were used for determining the kinetics of serological responses against pp1a proteins. To determine the ability of the nsp7-based ELISA to differentiate type I and type II PRRSV, a total of 470 known-positive samples were tested with 215 samples from the type I virus-infected pigs and 255 samples from the type II virus-infected pigs.
In addition to samples of known status, the nsp7-based ELISA was evaluated using field samples, i.e., 1,107 serum samples collected from 2007 to 2008 from 30 different farms in 10 different states (Minnesota, Colorado, South Dakota, Wisconsin, Illinois, Wyoming, Iowa, Kentucky, Nebraska, and Missouri). These samples were also assayed in the Idexx PRRS ELISA at the South Dakota Animal Disease Research and Diagnostic Laboratory (SD ADRDL). In addition, 100 Idexx ELISA suspected false-positive samples were also obtained from the SD ADRDL and tested in the nsp7-based ELISA.
PRRSV nsp antigen-based ELISA.
The nsp antigen-based ELISA was performed using Immulon 2 HB 96-well microtiter plates (Thermo Labsystems, Franklin, MA). A single lot of internal quality control serum samples, generated from experimentally infected pigs, was used to establish the standards for high positive (optical density [OD], ∼1.9 to 2.1), low positive (OD, ∼0.6 to 0.7), and negative (OD, <0.2). The optimal dilution of the recombinant protein was experimentally determined so that the control serum sample generated an OD as the established standard. The recombinant protein was diluted in 15 mM sodium carbonate-35 mM sodium bicarbonate (ACB), pH 8.8. The plates were coated with 100 μl (∼2 μg/ml) of the diluted protein in lanes 1, 3, 5, 7, 9, and 11. Lanes 2, 4, 6, 8, 10, and 12 were coated with 100 μl of ACB as a background control. For the nsp7-based ELISA, lanes 1, 4, 7, and 10 were coated with type I PRRSV nsp7 antigen, lanes 2, 5, 8, and 11 were coated with type II PRRSV nsp7 antigen, and lanes 3, 6, 9, and 12 were coated with ACB as a background control. The plates were incubated for 1 h at 37°C and then blocked with 10% (wt/vol) powdered dry milk in phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST) at 4°C overnight. The following day, the plates were washed with 300 μl of PBST. Test and control sera were diluted 1:50 with PBST containing 5% milk in PBST, and 100 μl of the dilution was added to the well. The plates were incubated for 1 h at 37°C and then washed, and 100 μl of goat anti-swine horseradish peroxidase conjugate (KPL, Gaithersburg, MD) was added to all wells. The plates were incubated for 1 h at 37°C and washed, and then 100 μl of 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) peroxidase substrate (KPL, Gaithersburg, MD) was added to all of the wells. Color development was observed until the positive control reached a standard OD and was then stopped by the addition of 100 μl of ABTS stop solution (KPL, Gaithersburg, MA). Color development was quantified by reading it at 405 nm with an EL800 microplate reader (BioTek Instruments Inc., Winooski, VT) controlled by XChek Software (Idexx Laboratories). The raw plate data were copied to an Excel spreadsheet to calculate the sample-to-positive (S/P) ratios using the following formula: S/P = (OD of sample − OD of buffer)/(OD of positive control − OD of buffer). Statistical analysis was performed using GraphPad InStat version 3.06 (GraphPad Software, San Diego, CA). The correlation of determination between mean S/P ratios was analyzed using Pearson R correlation analysis assuming Gaussian distribution of the data.
Validation of nsp7-based ELISA. (i) Cutoff determination, diagnostic sensitivity, and diagnostic specificity.
To accurately assess the diagnostic sensitivity and diagnostic specificity of the nsp7 ELISA, 2,691 serum samples from individual animals with established PRRSV status were analyzed using the nsp7 dual ELISA and the Idexx ELISA. The negative-testing (noninfected) validation population was composed of samples from individual animals of negative control groups. The positive-testing (infected) validation population was composed of samples from experimentally infected animals (see “Serum samples” above). Receiver operating characteristic (ROC) analysis methodology assessment was performed using GRAPH ROC software version 2.0 (14; http://members.tripod.com/refstat/GraphROC.htm).
(ii) Measurement of repeatability.
The repeatability of the nsp7 dual ELISA was assessed by running the same lot of internal quality control sera. The within-plate precision was calculated from 40 replicates on one plate, within-run precision was calculated using one serum on 10 plates in one run, and between-run precision was calculated from at least one serum in 10 different runs. Means, standard deviations, percent coefficient of variation (%CV) values, and Levey-Jennings control charts were calculated using Control Chart Pro Plus software version 7.12.24 (ChemSW).
(iii) Calculation of r.
For each positive sample, a reactivity ratio (r) value, representing the log10 of the ratio obtained by dividing the S/P ratio observed in the type I nsp7 ELISA by the S/P ratio observed in the type II nsp7 ELISA, was calculated. Thus, r values of >0 represent positives in the type I nsp7 ELISA, and r values of <0 represent positives in the type II nsp7 ELISA.
Immunofluorescence assay (IFA).
MARC-145 cells were grown in cultures for 3 to 4 days to confluence on 96-well cell culture plates (BD Biosciences, San Jose, CA). Every other lane was infected with PRRSV (5 × 103 50% tissue culture infective doses/ml), and the plates were incubated for an additional 18 to 24 h. The plates were then fixed with 300 μl of 50% (vol/vol) acetone/methanol per well for 20 min at −20°C, air dried, and frozen with a desiccant at −20°C until they were used. Serum samples to be assayed were diluted 1:20 and 1:40 with PBS, and 100 μl of each dilution was transferred to paired wells of PRRSV-infected and uninfected MARC-145 cells. The plates were incubated at 37°C for 1 h and then washed three times with 300 μl of PBS. Then, 30 μl of fluorescein isothiocyanate-labeled goat anti-swine immunoglobulin G (41.7 μg/ml; KPL) was added to each well. The plates were incubated at 37°C for 1 h and washed with PBS three times. The cells were examined for specific fluorescence with an inverted microscope and a UV light source (Nikon Eclipse TS100).
RESULTS
Antigen production.
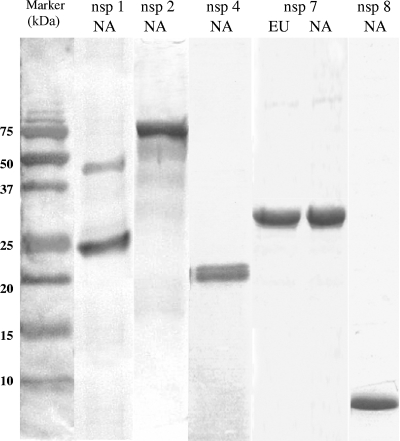
DNA fragments corresponding to all or portions of nsp1, nsp2, nsp4, nsp7, and nsp8 from type II PRRSV VR2332 and nsp7 from type I PRRSV SD01-08 were cloned and expressed in Escherichia coli. nsp4, nsp7, and nsp8 were expressed at high levels and could be purified in soluble forms. In contrast, recombinant nsp1 and nsp2 formed inclusion bodies, and a protein-refolding step was performed. The purities of the recombinant proteins were evaluated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie blue staining. As shown in Fig. 1, all of the His-tagged recombinant proteins migrated according to their predicted sizes, listed in Table 1. The identity of each protein was confirmed by Western blot analysis with monoclonal anti-His antibody and the specific swine antiserum (data not shown).
FIG. 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of recombinant PRRSV nsp preparations, followed by Coomassie blue staining. The left lane shows the protein molecular mass standard; the remaining lanes represent nsp1, nsp2, nsp4, nsp7, and nsp8 preparations, as indicated. NA, North American genotype (type II); EU, European genotype (type I). Note that nsp1 is further cleaved into nsp1α and nsp1β subunits (6, 13). Intact nsp1 and 26-kDa nsp1β eluted from the immobilized metal affinity column are shown in the second lane from the left.
Determining the kinetics of serological responses against pp1a proteins.
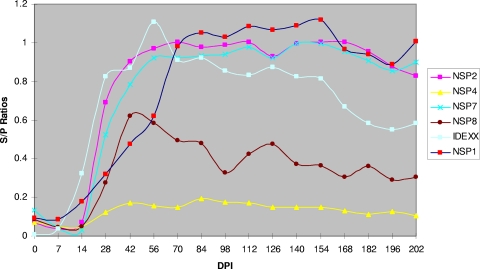
Testing of the serological responses to nsp1, nsp2, nsp4, nsp7, and nsp8 was conducted using 510 serum samples that were collected from 30 pigs experimentally infected with the type II isolate VR2332. As shown in Fig. 2, nsp4 reacted weakly with the swine immune sera. Nsp8 had a lower antibody response than nsp1, nsp2, and nsp7, and the titer dropped after 98 days postinoculation. nsp1, nsp2, and nsp7 reacted strongly with pig immune sera. The antibodies specific to these proteins were detected as early as 14 days postinoculation, and the responses lasted more than 202 days postinoculation. Interestingly, there was a decrease of antibody titer to N protein over time after 126 days postinoculation, while the antibody titers of nsp1, nsp2, and nsp7 remained relatively high. We further performed detailed comparisons of these three antigens, including their sensitivities to early seroconversion and correlation with N protein, which is the antigen used in the Idexx HerdChek PRRS 2XR ELISA. At 7 days postinoculation, all samples tested negative (S/P < 0.2) by all antigens. At 14 days postinoculation, approximately 65% of the samples were detected as seropositive (S/P > 0.5) by all ELISAs; at 21 days postinoculation, all samples were identified as positive (S/P > 0.5) irrespective of the antigen used for detection (Table 2). The correlation between the PRRSV nsp ELISA and the Idexx ELISA was evaluated using Pearson R correlation analysis. During the first 126 days postinfection, the ELISA results from nsp2 and nsp7 correlated well with those of the Idexx ELISA (R = 0.91 between nsp2 and Idexx; R = 0.84 between nsp7 and Idexx). In contrast, statistical correlation of the nsp1 ELISA to the Idexx ELISA was 0.57, which is lower than those of nsp2 and nsp7.
FIG. 2.
Kinetics of antibody response to PRRSV nsps. Pigs were experimentally infected with type II PRRSV, VR2332. The serum samples were from 0 to 202 days postinoculation (DPI) as indicated. For nsp4 and nsp8, serum samples from 10 pigs were tested; for nsp1, nsp2, and nsp7, serum samples from 30 pigs were tested.
TABLE 2.
Comparison of seroconversion detected by PRRSV nsp ELISA and Idexx ELISA
| ELISA method | No. of seropositivea animals detected/total no. of experimental animals
|
||
|---|---|---|---|
| 7b | 14 | 21 | |
| nsp1 | 0/30 | 22/30 | 30/30 |
| nsp2 | 0/30 | 21/30 | 30/30 |
| nsp7 | 0/30 | 23/30 | 30/30 |
| Idexx | 0/30 | 23/30 | 30/30 |
An S/P value of >0.5 is considered seropositive for nsp ELISAs, while an S/P value of >0.4 is considered seropositive for Idexx ELISA.
Days postinfection.
Cutoff determination, diagnostic sensitivity, and diagnostic specificity of nsp7-based ELISA.
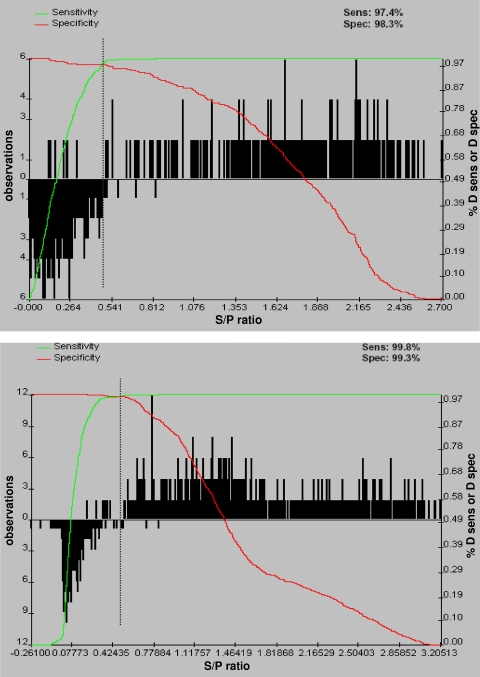
An nsp7-based ELISA was chosen to be further evaluated as a serology diagnostic assay for detection and differentiation of type I and type II PRRSV. The robustness and repeatability of the nsp7-based ELISA were assessed to determine its potential for diagnostic application. Recombinant nsp7 antigens were prepared from the type I virus, SD01-08, and the type II virus, VR2332. Serum samples from a known-positive population (type I and type II PRRSV infected) of 1,334 animals and 1,357 serum samples from a known-negative population (non-PRRSV infected) were analyzed with the nsp7-based ELISA and the Idexx ELISA. GRAPH ROC software was used for ROC analysis of nsp7-based ELISAs to determine an optimized cutoff that maximized both the diagnostic specificity and the diagnostic sensitivity of the assays (Table 3). An optimized cutoff that maximized the efficiency of the assay was calculated at an S/P of 0.51 for the type I nsp7 ELISA and an S/P of 0.52 for the type II nsp7 ELISA (Fig. 3). A diagnostic sensitivity of 97.4% (95% confidence interval, 94.4% to 99.1%) and a corresponding diagnostic specificity of 98.3% (95% confidence interval, 94.1% to 98.7%) were calculated for the type I nsp7 ELISA, while a diagnostic sensitivity of 99.8% (95% confidence interval, 99.4% to 100%) and a corresponding diagnostic specificity of 99.3% (95% confidence interval, 97.1% to 99.5%) were calculated for the type II nsp7 ELISA. When the cutoff (S/P = 0.4) determined by Idexx was used, the diagnostic sensitivity was 97.4% (95% confidence interval, 86% to 99.9%) and the diagnostic specificity of the Idexx ELISA was 99.6% (95% confidence interval, 97.8% to 99.9%). These results indicate that the nsp7-based ELISA is very comparable to the Idexx ELISA.
TABLE 3.
Comparison of sensitivity and specificity of nsp7 dual ELISA and Idexx ELISA for detection of antibodies against types I and II PRRSV
| Characteristic | Value (95% CI) for ELISA
|
||
|---|---|---|---|
| Idexx | Type I nsp7 | Type II nsp7 | |
| Optimized cutoff (S/P) | 0.4 | 0.51 | 0.52 |
| Diagnostic sensitivity (%) | 97.4 (86-99.9) | 97.4 (94.4-99.1) | 99.8 (99.4-100) |
| Diagnostic specificity (%) | 99.6 (97.8-99.9) | 98.3 (94.1-98.7) | 99.3 (97.1-99.5) |
FIG. 3.
Two-graph ROC plot of the PRRSV nsp7-based ELISA. The graphs were calculated using 965 (type I nsp7) and 1,726 (type II nsp7) individual animal serum samples and GraphROC software. The downward-pointing histogram on the left side of the figure represents the uninfected animals, and the upward-pointing histogram on the right side of the figure represents the PRRSV-infected animals. The green line represents the diagnostic sensitivity (D sens) of the assay as the cutoff S/P ratio is moving from 0 to 2.7. The red line represents the diagnostic specificity (D spec) of the assay as the cutoff S/P ratio is moving from 0 to 2.7. The black dotted vertical line represents the optimized cutoff value of 0.51 (type I) (A) and 0.52 (type II) (B), which corresponds to the maximum diagnostic sensitivity and specificity.
Repeatability of the nsp7-based ELISA.
The precision levels of the Idexx ELISA and the nsp7-based ELISA were compared using internal-control sera. The %CV was calculated using the protocol described previously (9). The Idexx ELISA within-plate %CV was 7.1, the %CV between plates in one run was 11.9, and the %CV between runs was 14.8. The nsp7-based ELISA appears to have variability similar to that of the Idexx ELISA. The type I nsp7 ELISA within-plate %CV was 6.5, the %CV between plates in one run was 11.9, and the %CV between runs was 17.1, while the type II nsp7 ELISA within-plate %CV was 2.3, the %CV between plates in one run was 5.4, and the %CV between runs was 9.5 (Table 4). These results suggest that the nsp7-based ELISA is highly repeatable in diagnostic applications.
TABLE 4.
Comparison of assay repeatability between Idexx and nsp7 dual ELISA
| ELISA | Repeatability results (%CV)a
|
||
|---|---|---|---|
| Within plate | Within run | Between runs | |
| Idexx | 7.1 | 11.9 | 14.8 |
| Type I nsp7 | 6.5 | 11.9 | 17.1 |
| Type II nsp7 | 2.3 | 5.4 | 9.5 |
The values listed are %CVs of high-level positive internal control serum.
Application of nsp7-based ELISA for the differentiation of type I and type II PRRSV.
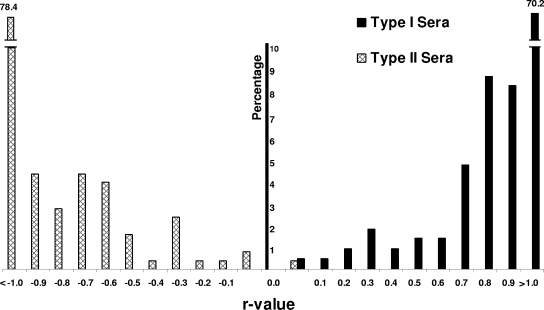
To determine if the nsp7-based ELISA can be used to differentiate the serum antibodies produced in response to infection with type I and type II viruses, a total of 470 known-positive samples were tested. The results showed that all of the samples from type I virus-infected pigs tested positive using the type I nsp7 ELISA (S/P > 0.51), and no specific antibody responses were detected in serum samples from type I PRRSV-infected animals in the type II nsp7 ELISA (S/P < 0.52) (Fig. 4). Similarly, 254 out of 255 samples from type II virus-infected pigs tested positive using the type II nsp7 ELISA (S/P > 0.52), and only one sample from type II PRRSV-infected animals tested positive in the type I nsp7 ELISA (S/P > 0.51) (Fig. 4). These results indicate that the nsp7-based ELISAs are specific for identifying the antibody response within the genotype and are capable of differentiating antibody responses to type I and type II PRRSV infections.
FIG. 4.
Differentiation of type I and type II PRRSV using the nsp7 dual ELISA. The distributions of individual samples with S/P values above the cutoff in the type I and type II nsp7 ELISAs are shown according to the calculated r values. The percentage of serum samples compared to the total number of positive sera in each test is shown on the vertical axis. For each positive sample, an r value, representing the log10 of the ratio obtained by dividing the S/P ratio observed in the type I nsp7 ELISA by the S/P ratio observed in the type II nsp7 ELISA, was calculated. Thus, r values of >0 represent positives in the type I nsp7 ELISA, and r values of <0 represent positives in the type II nsp7 ELISA.
Comparison of the nsp7-based ELISA with the Idexx ELISA for the detection of pigs infected with field viruses.
We used a broad spectrum of field serum samples (1,107 samples) submitted to the SD ADRDL to determine if the nsp7-based ELISAs could be applicable for detecting the serum antibody responses of pigs infected with various genetically different field strains. Since the sources of field serum samples were unknown (i.e., whether pigs were infected by type I or type II PRRSV), we used antigens from both type I and type II PRRSV and designated this test the nsp7 dual ELISA. When comparing the nsp7 dual ELISA with the Idexx ELISA, 490 out of 502 (97.6%) Idexx-positive samples were tested as positive by the nsp7 dual ELISA, and 590 out of 605 (97.5%) Idexx-negative samples were tested as negative by the nsp7 dual ELISA. We further investigated the application of the nsp7-based ELISA in samples with unexpected positive Idexx results. An unexpected positive result was defined as Idexx positive but negative when tested by IFA with no evidence of exposure to PRRSV. One hundred samples with suspected false-positive Idexx ELISA results were obtained from the SD ADRDL, and these samples were verified by IFA as seronegative. The nsp7 dual-ELISA results showed that 98 samples (98%) tested as negative (Table 5).
TABLE 5.
Evaluation of field sera and samples with Idexx ELISA unexpected positive results using the nsp7 dual ELISA
| Serum group | Results | Total no. of samples | No. Idexx positive | No. IFA positivea | No. nsp7 dual-ELISA positive |
|---|---|---|---|---|---|
| 1 | Idexx positiveb | 502 | 502 | NA | 490 |
| 2 | Idexx negativec | 605 | 0 | NA | 15 |
| 3 | Unexpected Idexx ELISA positived | 100 | 100 | 0 | 2 |
NA, not applicable. IFA was not performed for these samples.
97.6% tested positive in nsp7 dual ELISA.
97.5% tested negative in nsp7 dual ELISA.
98% showed negative in nsp7 dual ELISA.
DISCUSSION
The current study aimed to determine the humoral immune response to the PRRSV nsps and to develop new tools for identification of PRRSV-infected animals. Previous studies of the humoral immune response to PRRSV focused mainly on detection of antibodies to viral structural proteins, especially N (9, 22, 26, 29). Several studies showed that certain nsps, such as nsp1 and nsp2, are highly immunogenic (4, 7, 13, 24, 25). Antibody responses to linear epitopes in nsp2 have been reported to appear within 1 to 4 weeks of infection in type I and type II PRRSV strains (4, 24, 25). Johnson et al. (13) observed robust and rapid cross-reactive antibody responses induced by nsp1 and nsp2 to vaccine and field isolates and substantially higher levels of immunoreactivity related to conformational epitopes. In this study, our data demonstrated that nsp7 is also highly immunogenic. Analysis of the kinetics of antibody response showed that response to nsp7 is comparable to antibody responses to nsp1 and nsp2, as well as antigens used in the commercial Idexx ELISA. As indicated by Johnson et al. (13), nsps are available from the earliest time of infection for presentation to the immune system in the context of major histocompatibility complex class I antigen presentation pathways. As cytolytic infection also releases viral proteins into interstitial spaces, it is hypothesized that a pronounced antibody response, equivalent to the immune response to structural proteins, would be generated to nsps. One intriguing feature of the antibody response to nsp antigens was the sustained antibody titers over a 202-day period of infection, while the antibody response to Idexx antigen, N protein, showed a gradual decay in titers after 126 days postinoculation. The mechanism for sustained levels of nsp antigen may reflect the long-term retention and presentation of nsp to the immune system.
To select an antigen for diagnostic-test development, we compared the correlation between the PRRSV nsp ELISA and the Idexx ELISA. Our results showed that nsp2- and nsp7-based ELISAs had higher correlation with the Idexx ELISA. We further compared the amino acid sequences of nsp2 and nsp7. Our previous studies showed that the PRRSV nsp2 region is highly variable within and between genotypes, with 70.6% to 91.6% amino acid identity within type I PRRSV and 74.9% to 95.6% amino acid identity within type II PRRSV, but only 33.8% identity between type I and type II genotypes (7, 8, 23, 28). The central region of nsp2 contains hypervariable domains with insertions and deletions (10, 12, 27, 30, 33), and most identified B-cell epitopes are located in these regions (4, 24). In contrast, nsp7 is relatively conserved within each genotype and is divergent between genotypes. Amino acid sequence comparisons showed that nsp7 shares 96.7% to 97.4% amino acid identity within type I PRRSV and 84.9% to 100% amino acid identity within type II PRRSV, but only about 45% identity between type I and type II genotypes (7, 8, 23, 28). These results suggest that the nsp7-based ELISA could be able to detect genotype-specific anti-nsp7 antibody responses.
Further validation of the nsp7-based ELISA showed good sensitivity and specificity of the assay as determined by ROC analysis. The two-graph ROC plots of both type I and type II nsp7 ELISAs display the histograms of the uninfected and PRRSV-infected populations and demonstrate minimal overlap of the two populations (Fig. 3). The overlap between the two populations was attributed to eight samples from the type I PRRSV-infected population and nine samples from the type II PRRSV-infected population that had values below the established cutoff. Closer examination of these 17 samples revealed that all demonstrated strong background on the negative control well of the ELISA plate, which suggests that the serum may contain other nonspecific components that interacted with the secondary antibody. In addition, eight of these samples were hemolyzed, which indicates that the serum collection and processing steps were not completed under optimal conditions. There were four samples from the negative population that demonstrated positive results on the type I PRRSV ELISA and three samples from the negative population showing positive results on the type II PRRSV ELISA. The Idexx ELISA S/P values of these seven samples ranged from 0.2 to 0.3. This observation may support the practice by some veterinarians of using follow-up testing for any samples having an S/P value greater than 0.20. We suspected that these samples might be from a herd that had a history of PRRSV infection, since the nsp7 ELISA was able to detect an antibody response up to 202 days postinoculation.
Serology is a standard diagnostic and surveillance method for determining if pigs have been exposed to PRRSV. Currently, the Idexx PRRS ELISA is the most widely used serological assay for determining the serostatus of swine herds. However, positive Idexx ELISA results in otherwise seronegative herds cause concern for producers, which necessitates a variety of follow-up assays to verify that the result is either positive or negative. This indicates that there is still a need for a reliable assay to identify the serological status of single reactors compared to herd reactors. While there is no standard protocol to verify false-positive serological results for PRRSV, most diagnostic laboratories use the IFA and/or virus neutralization assays. However, the results from both of these assays are affected by antigenic variation, and they may not detect a serological response against antigenically diverse PRRSV isolates, such as the European-like PRRSV strains, known as North American type I isolates. The appearance of the type I PRRSV isolates in the United States also complicates the diagnosis of PRRSV, as there is presently no standard serological assay that clearly differentiates between type I and type II strains of PRRSV (7, 28). The movement of the swine industry toward strategies to eliminate or eradicate PRRSV will require an adequate serological diagnostic assay that can detect acutely and persistently infected pigs, detect various strains of PRRSV, and have the capacity to differentiate between type I and type II PRRSV isolates. The results generated in this study suggest that PRRSV nsp7 could be a potential new antigen for use in ELISA-based diagnostic assays. In particular, using a target other than the N protein, any false positives specifically associated with the N antigen would be avoided.
In summary, our results showed that nsp1, nsp2, and nsp7 induced high levels of antibody response during the course of PRRSV infection. Among these three proteins, nsp7 is the most suitable for diagnostic development, with the following characteristics: (i) nsp7 is expressed as a soluble recombinant protein in bacterial culture, which is convenient for ELISA antigen preparation, especially when applied to diagnostic tests dealing with massive numbers of diagnostic samples; (ii) the PRRSV nsp7 protein coding region is more homologous among different strains within the genotype than those of the other two immunogenic proteins, nsp1 and nsp2; (iii) it is able to detect antibody responses later than 126 days postinoculation. The nsp7-based ELISAs showed good sensitivity and specificity for identification and differentiation of type I and type II PRRSV. Furthermore, the nsp7 dual ELISA resolved 98% of samples with suspected false-positive Idexx ELISA results. Therefore, nsp7-based ELISA has the potential to serve as an alternative or follow-up test for the Idexx ELISA.
Acknowledgments
This project was supported by National Pork Board grant 05-155; National Research Initiative of the USDA Cooperative State Research, Education and Extension Service grant 2004-35605-14197; and the South Dakota Center for Infectious Disease Research and Vaccinology 2010 program and the SD ADRDL.
We thank John Schwartz, Rachael Breen, Haixia Liu, and Jingjing Bao for excellent technical assistance.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Allende, R., T. L. Lewis, Z. Lu, G. F. Rock, A. Kutish, A. Ali, A. R. Doster, and F. A. Osorio. 1999. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J. Gen. Virol. 80307-315. [DOI] [PubMed] [Google Scholar]
- 2.Bautista, E. M., S. M. Goyal, I. J. Soon, H. S. Joo, and J. E. Collins. 1996. Structural polypeptides of the American (VR-2332) strain of porcine reproductive and respiratory syndrome virus. Arch. Virol. 1411357-1365. [DOI] [PubMed] [Google Scholar]
- 3.Dea, S., L. Wilson, D. Therrien, and E. Cornaglia. 2000. Competitive ELISA for detection of antibodies to porcine reproductive and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Investig. 4117-126. [Google Scholar]
- 4.de Lima, M., A. K. Pattnaik, E. F. Flores, and F. A. Osorio. 2006. Serologic marker candidates identified among B-cell linear epitopes of Nsp2 and structural proteins of a North American strain of porcine reproductive and respiratory syndrome virus. Virology 353410-421. [DOI] [PubMed] [Google Scholar]
- 5.Denac, H., C. Moser, J. D. Tratschin, and M. A. Hofmann. 1997. An indirect ELISA for the detection of antibodies against porcine reproductive and respiratory syndrome virus using recombinant nucleocapsid protein as antigen. J. Virol. Methods 65169-181. [DOI] [PubMed] [Google Scholar]
- 6.den Boon, J. A., K. S. Faaberg, A. L. Meulenberg, P. G. Wassenaar, A. Plagemann, E. Gorbalenya, and E. A. Snijder. 1995. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papain-like cysteine proteases. J. Virol. 694500-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, Y., K. Dal-Young, S. Ropp, P. Steen, J. Christopher-Hennings, E. A. Nelson, and R. R. R. Rowland. 2004. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 100229-235. [DOI] [PubMed] [Google Scholar]
- 8.Fang, Y., P. Schneider, W. P. Zhang, K. S. Faaberg, E. A. Nelson, and R. R. R. Rowland. 2007. Diversity and evolution of a newly emerged North American Type 1 porcine arterivirus: analysis of isolates collected between 1999 and 2004. Arch. Virol. 1521009-1017. [DOI] [PubMed] [Google Scholar]
- 9.Ferrin, N. H., Y. Fang, C. R. Johnson, M. P. Murtaugh, D. D. Polson, M. Torremorell, M. L. Gramer, and E. A. Nelson. 2004. Validation of a blocking enzyme-linked immunosorbent assay for detection of antibodies against porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 11503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, Z. Q., X. Guo, and H. C. Yang. 2004. Genomic characterization of two Chinese isolates of porcine respiratory and reproductive syndrome virus. Arch. Virol. 1491341-1351. [DOI] [PubMed] [Google Scholar]
- 11.Gorbalenya, A. E., V. M. Blinov, A. P. Donchenko, and E. V. Koonin. 1989. An NTP binding motif is the most conserved sequence in a highly diverged monophyletic group of proteins involved in positive strand RNA viral replication. J. Mol. Evol. 28256-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han, J., G. Liu, Y. Wang, and K. S. Faaberg. 2007. Identification of nonessential regions of the nsp2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture. J. Virol. 819878-9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, C. R., Y. Wanquin, and M. P. Murtaugh. 2007. Cross-reactive antibody responses to nsp1 and nsp2 of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 881184-1195. [DOI] [PubMed] [Google Scholar]
- 14.Kairisto, V., and A. Poola. 1995. Software for illustrative presentation of basic clinical characteristics of laboratory tests—GraphRoc for Windows. Scand. J. Clin. Lab. Investig. Suppl. 22243-60. [DOI] [PubMed] [Google Scholar]
- 15.Kim, H. S., J. Kwang, K. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133477-483. [DOI] [PubMed] [Google Scholar]
- 16.Lawson, S., Y. Fang, R. R. R. Rowland, C. Christopher-Hennings, and E. A. Nelson. 2005. Experimental infection of pigs with European-like (Type 1) PRRSV isolates of US origin, abstr. 99. 86th Annu. Meet. Conf. Res. Workers Anim. Dis., 4 to 6 December 2005, St. Louis, MO.
- 17.Mardassi, H., B. Massive, and S. Dea. 1996. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 22198-112. [DOI] [PubMed] [Google Scholar]
- 18.Meng, X. J., P. S. Paul, P. G. Halbur, and M. A. Lum. 1995. Phylogenetic analyses of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the USA and Europe. Arch. Virol. 140745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meulenberg, J. J. M., A. Petersen-den Besten, E. P. de Kluyver, R. J. M. Moormann, W. M. M. Schaaper, and G. Wensvoort. 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 206155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meulenberg, J. J. M., and A. Petersen-den Besten. 1996. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF2 is incorporated in virus particles. Virology 22544-51. [DOI] [PubMed] [Google Scholar]
- 21.Mounir, S., H. Mardassi, and S. Dea. 1995. Identification and characterization of the porcine reproductive and respiratory syndrome virus ORFs 7, 5, and 4 products. Adv. Exp. Med. Biol. 80317-320. [DOI] [PubMed] [Google Scholar]
- 22.Mulupuri, P., J. J. Zimmerman, J. Hermann, C. R. Johnson, J. P. Cano, W. Yu, S. A. Dee, and M. P. Murtaugh. 2008. Antigen-specific B-cell responses to porcine reproductive and respiratory syndrome virus infection J. Virol. 82358-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelsen, C. J., M. P. Murtaugh, and K. S. Faaberg. 1999. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 73270-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oleksiewicz, M. B., A. Bøtner, P. Toft, P. Normann, and T. Storgarrd. 2001. Epitope mapping porcine reproductive and respiratory syndrome virus by phage display: the nsp2 fragment of the replicase polyprotein contains a cluster of B-cell epitopes. J. Virol. 753277-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oleksiewicz, M. B., A. Bøtner, and P. Normann. 2001. Semen from boars infected with porcine reproductive and respiratory syndrome virus (PRRSV) contains antibodies against structural as well as non-structural viral proteins. Vet. Microbiol. 81109-125. [DOI] [PubMed] [Google Scholar]
- 26.Oleksiewicz, M. B., A. Bøtner, and P. Normann. 2002. Porcine B-cells recognize epitopes that are conserved between the structural proteins of American- and European-type porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 831407-1418. [DOI] [PubMed] [Google Scholar]
- 27.Ran, Z. G., X. Y. Chen, X. Guo, X. N. Ge, K. J. Yoon, and H. C. Yang. 2008. Recovery of viable porcine reproductive and respiratory syndrome virus from an infectious clone containing a partial deletion within the Nsp2-encoding region. Arch. Virol. 153899-907. [DOI] [PubMed] [Google Scholar]
- 28.Ropp, S. L., C. E. Wees, Y. Fang, E. A. Nelson, K. D. Rossow, M. Bien, B. Arndt, S. Preszler, P. Steen, J. Christopher-Hennings, J. E. Collins, D. A. Benfield, and K. S. Faaberg. 2004. Characterization of emerging European-like porcine reproductive and respiratory syndrome virus isolates in the United States. J. Virol. 783684-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seuberlich, T., J. D. Tratschin, B. Thur, and M. A. Hofmann. 2002. Nucleocapsid protein-based enzyme-linked immunosorbent assay for detection and differentiation of antibodies against European and North American porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 91183-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen, S., J. Kwang, W. Liu, and D. X. Liu. 2000. Determination of the complete nucleotide sequence of a vaccine strain of porcine reproductive and respiratory syndrome virus and identification of the Nsp2 gene with a unique insertion. Arch. Virol. 145871-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snijder, E. J., and J. M. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79961-979. [DOI] [PubMed] [Google Scholar]
- 32.Snijder, E. J., and W. J. Spaan. 2007. Arteriviruses, p. 1337-1355. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 33.Tian, K., X. Yu, T. Zhao, Y. Feng, Z. Cao, C. Wang, et al. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE 2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Dinten, L. C., A. L. Wassenaar, A. E. Gorbalenya, W. J. Spaan, and E. A. Snijder. 1996. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 706625-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, W. H., Y. Fang, R. Farwell, M. Steffen-Bien, R. R. R. Rowland, J. Christopher-Hennings, and E. A. Nelson. 2001. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF 2b. Virology 287183-191. [DOI] [PubMed] [Google Scholar]
- 36.Wu, W. H., Y. Fang, R. R. R. Rowland, S. R. Lawson, J. Christopher-Hennings, K. J. Yoon, and E. A. Nelson. 2005. The 2b protein as a minor structural component of PRRSV. Virus Res. 114177-181. [DOI] [PMC free article] [PubMed] [Google Scholar]