Abstract
Improved diagnostic reagents and testing are currently needed for the serological detection of human herpesvirus 8 (HHV-8) infections. We evaluated the luciferase immunoprecipitation systems (LIPS) for profiling antibody responses to a panel of HHV-8 proteins for diagnosis of Kaposi sarcoma (KS)-infected individuals. Using a pilot serum set, LIPS detected robust antibody responses to several known antigens, and a screen of 14 additional HHV-8 proteins identified v-cyclin as a potentially new diagnostic antigen. In evaluating a training-serum set, a four-antigen panel (K8.1, v-cyclin, ORF65, and a LANA fragment) was found to provide sufficient information for diagnosis. Analysis of a validation serum set using the combined results from these four separate antigen tests showed 100% sensitivity and 100% specificity. Furthermore, a LIPS format using a mixture of the four antigens, which simplifies data collection and analysis, closely matched the diagnostic performance of the combined separate tests (R = 0.95). This four-antigen mixture format analyzed with the validation serum set also showed 100% sensitivity and 100% specificity but was not statistically different from two separate enzyme-linked immunosorbent assays (94% sensitivity and 100% specificity) using baculovirus-produced LANA and bacterially produced K8.1. Heat map analysis of KS patient antibody titers revealed marked heterogeneity in humoral responses to this four-antigen panel. Overall, the LIPS assay showed 97% sensitivity, and positive anti-v-cyclin antibodies were detected in approximately 75% of the KS sera. These results suggest that LIPS screening using an antigen mixture is a sensitive and high-throughput method for serological screening of HHV-8 infection in individuals with KS.
Kaposi sarcoma (KS) is an opportunistic disease in human immunodeficiency virus (HIV) patients and the most common cancer associated with AIDS worldwide (12). Identified a decade ago as the causative agent of KS, human herpesvirus 8 (HHV-8), also known as Kaposi's sarcoma-associated herpesvirus, has an approximately 165-kb genome encoding about 90 gene products (21). Many of these gene products allow the virus to evade the human immune system (8). KS primarily affects AIDS patients, but it can also occur in non-HIV-infected individuals and presents as classical, endemic, and posttransplant forms. HHV-8 is also associated with two other rare B-cell lymphoproliferative disorders, primary effusion lymphoma and multicentric Castleman disease, which are primarily found in HIV-infected or other immunosupressed patients.
Currently, there is a need for sensitive and specific testing to identify HHV-8-infected individuals, especially among potential blood donors (14). Low viral loads in blood limit the sensitivity and thus the usefulness of PCR-based approaches (20). Alternatively, a variety of serological tests, including immunofluorescence assays (23), Western blotting (26), and enzyme-linked immunosorbent assays (ELISAs) (11, 15, 18), have been employed to detect antibodies to HHV-8 proteins and to diagnose infection. Considerable progress has been made in employing defined recombinant HHV-8 antigens, including LANA, K8.1, ORF65, for testing. The most sensitive ELISAs require separate determinations of two or three HHV-8 antigens and typically rely on diagnostic algorithms to achieve 90 to 95% sensitivity and 90 to 95% specificity at best (15, 18). Furthermore, one major problem plaguing the assessment of the performance of any given HHV-8 serological test is the lack of gold standard reference serum samples (19). Typically KS patients are the only true positives available, which may cause the sensitivity of the assay to be overestimated, because KS patients generally have much higher antibody titers than asymptomatic HHV-8-infected individuals (13).
Recently, Renilla luciferase (Ruc)-antigen fusions, produced in Cos1 cells, were used in a simple immunoprecipitation assay called the luciferase immunoprecipitation systems (LIPS) to quantitatively measure antibody responses to cancer-associated autoantigens (2), autoantigens associated with autoimmune diseases (3, 4), and a variety of infectious agents, including hepatitis C virus (1), HIV (1), human T-cell leukemia virus type I (6), herpes simplex virus types 1 and 2 (5), and filarial infections (7, 24). LIPS is based on fusing protein antigens to a light-emitting enzyme reporter, Ruc, and expressing these fusions in mammalian cells. The Ruc-labeled antigen extracts are then used in immunoprecipitation assays with serum samples and protein A/G beads. Following washing, light production is measured, yielding highly quantitative antibody titers. In this study, we used LIPS to evaluate known antigens and potential HHV-8 ORFs for the serological diagnosis of KS. Following the evaluation of pilot and training-serum sets, a four-antigen panel (K8.1, v-cyclin, ORF65, and LANA fragment) was selected. This four-antigen panel, evaluated separately or as a mixture with a validation serum set, showed 100% sensitivity and 100% specificity. These results suggest that a LIPS antigen mixture is an efficient high-throughput method for serological screening of HHV-8 infection in individuals with KS.
MATERIALS AND METHODS
Patient sera.
Sera were obtained from patients or volunteers under institutional review board-approved protocols at the Clinical Center, NIH, and at Georgetown University. In the initial pilot set, 6 colon cancer, 15 KS, and 1 HIV-positive serum samples were analyzed. Subsequent analysis of the single HIV-positive sample revealed that it was strongly positive by both ELISA and LIPS for anti-HHV-8 antibodies, and for simplicity, it was designated a KS sample, making “16” KS samples. The training (n = 83 blinded sera) and validation (n = 71) serum sample sets were provided as coded samples for both LIPS and ELISAs, and the code was broken only after titers were established and categorization of the HHV-8 infection status had been done. The training set consisted of 39 KS samples, while the validation set contained 34 KS samples. In the training and validation cohorts, the HHV-8-negative samples represented healthy, HIV-negative volunteers obtained from the NIH Blood Bank and were expected to be at low risk for HHV8 positivity. One volunteer blood donor sample in each of the training and validation sets was also found to be HHV-8 positive by both ELISA and LIPS, and these samples were included as KS/HHV-8 positive in the figures.
HHV-8 ELISA.
Anti-HHV-8 antibodies were determined using two different ELISAs employing baculovirus-produced LANA (ORF73) and bacterially produced K8.1 antigen as described previously (15).
Generation of Ruc-antigen fusion constructs.
pREN2, a mammalian Ruc expression vector, was used to generate all plasmids (2). HHV-8 clones were amplified by PCR-specific linker-primer adapters using HHV-8 genomic DNA and a LANA cDNA template. Using gene-specific primers in PCR amplifications, the full-length coding sequences, as well as subfragments, were generated for fusions at the C terminus of Ruc. For each construct, including deletion mutants, a stop codon was included at the end of the coding sequence. The primer adapter sequences used to clone each protein or protein fragment are available on request. DNA sequencing was used to confirm the integrity of all of the DNA constructs. Plasmid DNA was then prepared from the different pREN2 expression vectors using a Qiagen Midi preparation kit.
LIPS analysis.
Following the transfection of mammalian expression vectors, crude protein extracts were obtained as described previously (3). The LIPS assay was performed in a 96-well plate format at room temperature as described previously (3). First, a “master plate” was constructed by diluting patient sera 1:10 in assay buffer A (20 mM Tris, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100) in a 96-well polypropylene microtiter plate. For evaluating antibody titers by LIPS, 40 μl of buffer A, 10 μl of diluted human sera (1 μl equivalent), and Ruc-antigen Cos1 cell extract, diluted in buffer A were added to each well of a polypropylene plate and incubated for 1 h at room temperature. The inputs for K8.1-Δ1, K8.1-Δ2, K8.1-Δ4, v-cyclin, LANA-Δ2, LANA-Δ3, and ORF65 Ruc antigen lysates were 10 × 106, 10 × 106, 6 × 106, 10 × 106, 4 × 106, 10 × 106, and 10 × 106 light units (LU), respectively. Next, 7 μl of a 30% suspension of Ultralink protein A/G beads (Pierce Biotechnology, Rockford, IL) in phosphate-buffered saline was added to the bottom of each well of a 96-well filter HTS plate (Millipore, Bedford, MA). To this filter plate, the 100-μl antigen-antibody reaction mixture was transferred and incubated for 1 h at room temperature on a rotary shaker. The washing steps of the retained protein A/G beads were performed on a BioMek FX (3) work station (Beckman Coulter, Fullerton, CA) using an integrated vacuum manifold. After the final wash, the LU were measured in a Berthold LB 960 Centro microplate luminometer (Berthold Technologies, Bad Wilbad, Germany) using coelenterazine substrate mix (Promega, Madison,WI). All LU data were obtained from the average of at least two independent experiments. All LU data for the training and validation cohorts were obtained from the average of two separate experiments and corrected for background by subtracting the LU values of beads incubated with Cos1 cell extract, but without sera. In addition to the individual tests, the combined summed values of the separate antigen tests were also calculated. The data presented (see Fig. 4) are log10-transformed values and are coded using a color palette ranging from red to green, indicating high and low titers, respectively.
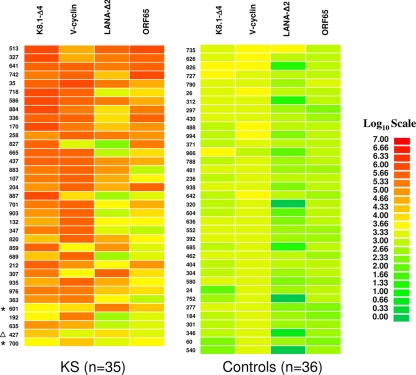
FIG. 4.
Heat map representation of patient antibody profiles to the four HHV8 antigens. The titer values for each serum were log10 transformed, and then the titer levels were color coded as indicated by the log10 scale on the right, in which signal intensities range from red to green, indicating high and low titers, respectively. The samples were rank ordered from highest to lowest based on the sum of the antibody titers to the four-antigen panel. The samples on the left are from KS-infected sera. The asterisks mark the samples that were negative or indeterminate for anti-K8.1 and anti-LANA antibodies by ELISA. A volunteer sample (non-KS) from the validation cohort that was found HHV-8 positive by both ELISA and LIPS is denoted by the triangle.
For the antigen mixture tests, the assay was modified slightly. In these tests, the Ruc-antigen extracts were harvested in lysis buffer without glycerol and used immediately upon collection. Antigen mixtures were added to each well and processed as described above using similar inputs of LU for each antigen.
Data analysis.
The GraphPad Prism software (San Diego, CA) was used for statistical analysis. Results for qualitative antibody titers between the controls and KS individuals are reported as the mean ± standard deviation (SD). Mann-Whitney U tests were used to compare the antibody titers among the groups. For the calculation of sensitivity and specificity, a cutoff limit of approximately 9,000 LU was used, which was derived from the combined value of the mean of the six control samples plus 5 SD for each antigen from the pilot set.
RESULTS
Patient antibody responses to known HHV-8 antigens.
Evaluation of the utility of LIPS in HHV-8 diagnosis began by screening serum samples from 16 KS and 6 control patients for antibodies against known HHV-8 antigens, including LANA, ORF65, and K8.1. Because of the small pilot set, the discriminatory potential of each of these HHV-8 antibody tests was evaluated by a variety of statistical and diagnostic methodologies, including receiver operator characteristics (ROC), area under the curve, Mann-Whitney U tests, and using sensitivity and specificity calculations derived from the mean plus 5 SD of the control samples (Table 1).
TABLE 1.
LIPS performances of top 10 HHV-8 antigens for KS diagnosis
| Antigen | AUCa | P valueb | Sensitivityc | Specificityc |
|---|---|---|---|---|
| K8.1-Δ4 | 0.96 | 0.0011 | 13/16 | 0/6 |
| K8.1-Δ1 | 0.92 | 0.0028 | 12/16 | 0/6 |
| LANA-Δ3 | 0.91 | 0.0052 | 13/16 | 0/6 |
| v-Cyclin | 0.89 | 0.0068 | 12/16 | 0/6 |
| ORF65 | 0.84 | 0.0105 | 10/16 | 0/6 |
| LANA-Δ2 | 0.82 | 0.0212 | 10/16 | 0/6 |
| K8.1-Δ2 | 0.81 | 0.0271 | 12/16 | 0/6 |
| ORF52 | 0.58 | 0.5669 | 3/16 | 1/6 |
| ORF47 | 0.55 | 0.7667 | 1/16 | 0/6 |
| LANA-Δ1 | 0.52 | 0.8530 | 1/16 | 0/6 |
ROC as determined by the area under the curve (AUC).
The P value was calculated using the Mann-Whitney U test.
For LIPS, the cutoff limit for calculating the sensitivity and specificity for each antigen was derived from the value of the mean plus 5 SD of the six control samples.
In the case of LANA, we generated three different protein fragments spanning amino acids 6 to 286, 274 to 1089, and 863 to 1089, designated LANA-Δ1, LANA-Δ2, and LANA-Δ3, respectively. Using the N-terminal LANA-Δ1 fragment, only one of the 16 KS sera showed positive results by the cutoff compared to the controls (Table 1). In contrast, LANA-Δ2, representing the central region of LANA, detected positive antibody responses in 10 of 16 KS sera and none of the 6 controls (Table 1). The LANA-Δ3 fragment, representing the C terminus of LANA, performed slightly better than LANA-Δ2 and reacted with 13 of the 16 KS samples (Table 1). Several other larger LANA constructs did not increase the diagnostic sensitivity of the LIPS test over the values obtained with the LANA-Δ2 and LANA-Δ3 fragments (data not shown).
Analysis of full-length ORF65 by LIPS correctly identified 10 of 16 KS samples and showed no immunoreactivity in the controls (Table 1). Initial tests of K8.1 utilized two different K8.1 exon fragments. Both of these protein fragments, K8.1-Δ1 (containing 123 amino acid residues) and K8.1-Δ2 (containing 89 amino acids), identified the same 12 KS samples as positive (Table 1 and data not shown). Subsequent testing of K8.1-Δ4, containing the complete coding sequence of the K8.1 protein, demonstrated diagnostic superiority in comparison to the two smaller K8.1 fragments and detected 13 of 16 KS samples (Table 1).
Identification of v-cyclin as a new informative HHV-8 antigen.
To identify potentially new HHV-8 antigens, the pilot serum set was screened with a panel of 14 different HHV-8 fusion proteins, i.e., v-cyclin, vFLIP/ORF71/K13, v-BCL2/ORF16, vIL-6/K2, v-GPCR/Orf74, ORF48, ORF52, Kaposin/K12, ORF57, vIRF-1/ORF K9, vIRF-3/LANA2, ORF45, ORF47, and MCP-1. Most of these HHV-8 proteins showed weak or nonexistent antibody signals with the KS sera (Table 1 and data not shown). For example, ORF52 showed weak positive signals with only 3 of the 16 KS sera (Table 1), while ORF74 showed no signal with any of the sera tested (data not shown). However, the latent HHV-8 protein, v-cyclin, displayed positive results with 12 of 16 KS samples (Table 1). The geometric mean titer of the anti-v-cyclin antibody in the 16 KS samples was 14,281 LU (95% confidence interval [CI], 1,318 to 154,759 LU), 1,000-fold higher than the geometric mean titer of 15 LU (95% CI, 0.65 to 56) in the controls (Mann-Whitney U test, P < 0.0068). The anti-v-cyclin antibody-positive KS patients did not simply reflect high anti-HHV8 antibody responding patients, as 2 of 16 anti-v-cyclin-positive samples were from KS sera that were negative for anti-LANA, anti-K8.1, and anti-ORF65 antibodies. These results suggest that anti-v-cyclin antibodies might represent a highly sensitive, independent marker of HHV-8 infection in KS patients.
Selection of an HHV-8 antigen panel.
We focused our efforts on defining a diagnostic antigen panel and utilized the six most informative antigens: LANA-Δ2, LANA-Δ3, v-cyclin, K8.1-Δ1, K8.1-Δ2, and ORF65, with sensitivity values of 63%, 81%, 75%, 75%, 75%, and 63%, respectively. We derived a cutoff value of 9,000 LU from the approximate sum of the values of the means plus 5 SD. Using this parameter and the combined results of each singly tested antigen, LIPS correctly identified 15 of the 16 KS patient sera (94% sensitivity and 100% specificity). The panel missed only one KS patient sample, later identified as an individual with a low CD4 count of 40. In comparison, the HHV-8 ELISA, based on two separate LANA and K8.1 tests (25), detected 13 of the 16 KS samples as positive (81% sensitivity and 100% specificity), with 3 KS samples designated indeterminate. Interestingly, one of the ELISA-indeterminate sera was the same serum sample missed by LIPS.
Initial testing of the training set with an HHV-8 panel.
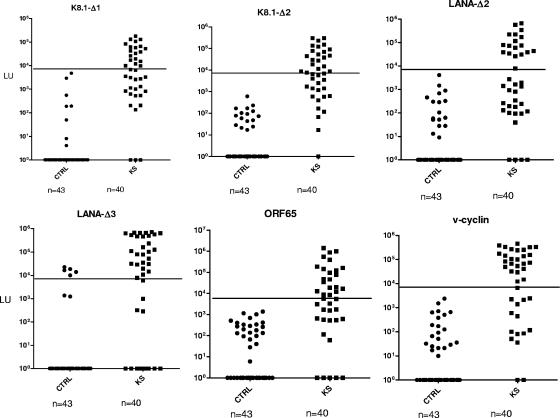
Based on the results of the pilot set, an independent training set of 83 blinded sera was tested with the six-antigen panel identified above. We screened the six antigens separately and also simultaneously as a single antigen mixture. The sum of the six separate tests and the value of the six-antigen mixture were closely correlated (Pearson R = 0.97; P < 0.00001), and using the 9,000-LU cutoff, both formats identified the same 44 potential positives. Following unblinding, LIPS performance showed 95% sensitivity (38/40) and 88.4% specificity (38/43). In addition, all six separate tests showed robust antibody titers in many of the KS samples compared to the volunteers (Fig. 1). Compared to LIPS, the optimized ELISA showed slightly higher performance, with 97.5% (39/40) sensitivity and 88.4% (38/43) specificity.
FIG. 1.
LIPS detection of antibodies to six different HHV-8 protein fragments. Using LIPS, antibodies were evaluated against K8.1-Δ1, K8.1-Δ2, LANA-Δ2, LANA-Δ3, v-cyclin, and ORF65 using 83 sera from a training set. Each symbol represents an individual sample from the training set consisting of 43 controls (CTRL) and 40 KS/HHV-8-infected subjects (KS). Antibody titers are plotted on the y axis using a log10 scale. The 9,000-LU cutoff is shown by the solid lines. A volunteer sample (non-KS) from the training cohort that was found HHV-8 positive by both ELISA and LIPS is included with the KS samples.
Improving the performance of the HHV-8 antigen panel.
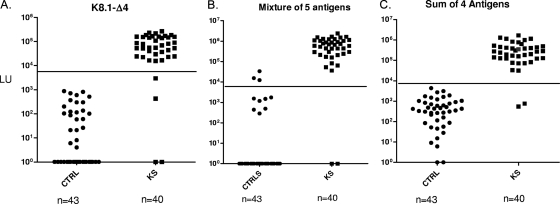
During the course of our studies, a new K8.1 antigen, K8.1-Δ4, was found to have the highest discriminatory potential in comparison to other HHV-8 antigens (Table 1). In an effort to improve the performance of the HHV-8 LIPS panel, we replaced the K8.1-Δ1 and K8.1-Δ2 protein fragments with the full-length K8.1-Δ4. After reblinding the 83 sera from the training set, we reevaluated the sera by LIPS using the K8.1-Δ4 antigen alone and with the new five-antigen mixture (K8.1-Δ4, v-cyclin, LANA-Δ2, LANA-Δ3, and ORF65). Following unblinding, the anti-K8.1-Δ4 antibody test showed promising results, detecting all but 4 of the 40 KS samples (90% sensitivity) with 100% specificity (Fig. 2A). Furthermore, the new five-antigen panel showed improved diagnostic performance, demonstrating 95% (38/40) sensitivity and 93% (40/43) specificity (Fig. 2B). Despite the improved performance of the new five-antigen mixture, three false-positive sera were still found above the cutoff of 9,000 LU. Inspection of the separate results from these five antigen tests revealed that the LANA-Δ3 test was responsible for all the false samples above the 9,000 LU cutoff. Since the anti-LANA-Δ3 antibody-positive signals were also positive for the anti-LANA-Δ2 antibody test, we omitted the LANA-Δ3 result and recalculated the sum of the four antigen tests (LANA-Δ2, K8.1-Δ4, v-cyclin, and ORF65). This new four-antigen panel showed 95% sensitivity and 100% specificity for discriminating KS-positive from KS-negative samples (Fig. 2C). These results suggest that this four-antigen panel assayed separately or potentially as a mixture might be a simple and useful test for diagnosis.
FIG. 2.
Improving the performance of the LIPS test. The 83 sera of the training set, 43 controls (CTRL) and 40 KS/HHV-8-infected subjects (KS), were reblinded and retested with a new five-antigen mixture employing K8.1-Δ4 instead of K8.1-Δ1 and K8.1-Δ2 (A), as well as with K8.1-Δ4 alone (B). Use of the same cutoff of 9,000 LU with the new five-antigen mixture resulted in improved diagnostic performance of 95% (38/40) sensitivity and 93% (40/43) specificity. The sum of antibody titers to only four antigens (LANA-Δ2, K8.1-Δ4, v-cyclin, and ORF65) was recalculated (C), and using the 9,000-LU cutoff (solid lines) with these four antigens markedly improved the test (95% sensitivity and 100% specificity). A volunteer sample (non-KS) from the training cohort that was found HHV-8 positive by both ELISA and LIPS is included with the KS samples.
A four-HHV-8-antigen panel shows 100% sensitivity and 100% specificity.
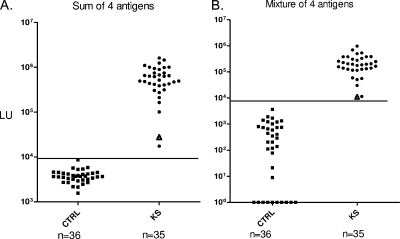
To test the effectiveness of the new four-antigen panel, a new validation cohort of 71 blinded sera were evaluated with the four antigens individually and as a mixture. The four HHV-8 antigens assayed separately or as a mixture perfectly distinguished (100% sensitivity and 100% specificity) the 35 KS-positive samples from the 36 controls (Fig. 3). The sum of the four separate tests and the four-antigen mixture were also closely correlated (Pearson R = 0.95; P < 0.00001). LIPS also detected two more positive samples than the ELISA (94% sensitivity and 100% specificity), but ROC analysis of LIPS and ELISA showed no significant difference in diagnostic performance.
FIG. 3.
A four-HHV-8-antigen panel shows 100% sensitivity and 100% specificity. Antibodies were evaluated by LIPS with a 71-serum validation cohort, 36 controls (CTRL), and 35 KS/HHV-8-infected subjects (KS), using a four-antigen panel consisting of K8.1-Δ4, LANA-Δ2, v-cyclin, and ORF65. These LIPS tests were either evaluated separately and summed (A) or tested as a cocktail (B). As shown, the two tests showed similar results and had 100% sensitivity and specificity. The 9,000-LU cutoff is shown by the solid lines. A volunteer sample (non-KS) from the validation cohort that was found HHV-8 positive by both ELISA and LIPS is denoted by the triangles.
Further analysis of anti-K8.1, anti-v-cyclin, anti-LANA-Δ2, and anti-ORF65 antibody titers measured by LIPS in the assays described above revealed that the patient antibody responses to these different antigens varied markedly. For example, using the Spearman rank test, the correlation, R, between the anti-K8.1 and anti-v-cyclin antibodies was −0.13 (95% CI, −0.4511 to 0.2237), while the correlation of anti-cyclin and anti-LANA antibodies was 0.06 (95% CI, −0.2861 to 0.3964). To easily visualize these differing patient antibody responses to the panel of four HHV-8 antigens, a heat map was employed (Fig. 4). In this graphical format, the marked differences in patient antibody responses to this panel become obvious. For example, several samples, including 513, 327, and 35, showed high level of antibodies to all four antigens, while several samples, including samples 192, 635, and 700, showed only weak antibodies to one or two antigens. Of note, one of the KS patient samples, sample 700, only had significant anti-v-cyclin antibodies and was missed by the ELISA (Fig. 4).
DISCUSSION
This study demonstrated that LIPS can robustly detect antibody titers to a panel of HHV-8 antigens with high diagnostic sensitivity and specificity. Our approach of using a four-antigen panel is consistent with ELISA studies showing the diagnostic usefulness of two or more different HHV-8 antigens (18, 22). Using either single-antigen tests or an HHV-8 antigen mixture, the LIPS assay consistently showed high sensitivity and specificity in identifying individuals with KS. The titer values obtained using both strategies closely matched each other, suggesting that in the LIPS format, diagnosis is highly reproducible regardless of assay conditions. The use of an antigen mixture by LIPS is highly convenient because it simplifies data collection and analysis. Furthermore, increasing the number of antigens within a mixture is generally a favorable condition if each member of the antigen has a comparable low background signal. In contrast, ELISAs perform poorly when more than one antigen is immobilized, and the alternative is to employ complicated multiepitope hybrid molecules. The positive results obtained here by LIPS with the HHV-8 antigen mixture are also consistent with our previous finding that a two-antigen mixture is a simple and sensitive method for diagnosis of Strongyloides stercoralis infection (24). Thus, this general approach of combining antigens in a mixture is likely to be useful for diagnosing many other infections.
Overall, the LIPS assay showed 97% (88/91) sensitivity in detecting the KS samples. The K8.1-Δ4 protein was the most useful antigen in the LIPS format, identifying 94% (86/91) of the KS-positive samples with 100% specificity. While not statistically significant, ROC comparisons of LIPS and ELISA in the detection of anti-K8.1 antibodies in the validation cohort revealed that LIPS performed slightly better than the ELISA (data not shown), which might be due to the detection of more conformational epitopes by LIPS. The anti-ORF65 antibody test did not increase sensitivity, and this antigen could have been omitted without affecting performance. Further modifications of the HHV-8 LIPS tests, such as the addition of new HHV-8 antigens/fragments, leaving out antigens (e.g., ORF65), and adjusting cutoffs, may further simplify testing and improve assay performance. Nevertheless, one limitation of our study is that mainly KS patients were used to assess the diagnostic performance of the LIPS assay. The use of only KS samples may cause performance to be overestimated because of the known higher antibody titers in KS compared to non-KS, HHV-8-infected samples. Future studies directed at testing other cohorts, including asymptomatic HHV-8-infected individuals, should further establish the utility of LIPS screening.
We report here for the first time that many KS patients have high levels of anti-v-cyclin antibodies. The relatively high titer and high prevalence (75%) of anti-v-cyclin antibody responses is consistent with mRNA expression showing that v-cyclin is highly expressed in KS tumors (10). v-Cyclin may be involved in HHV-8 malignancy because it is similar to the human cyclin D oncogene (17). It is worth noting that Katano et al. screened a number of glutathione S-transferase fusions of HHV-8 proteins for their serologic reactivities, including the v-cyclin (ORF72), but did not detect immunoreactivity to v-cyclin by Western blotting (16). The detection of diagnostically useful antibodies to v-cyclin by LIPS, but not with a bacterial recombinant protein, supports the notion that the LIPS antigens may have many more conformational epitopes, because these antigens were produced in mammalian cells. Particularly useful was the finding that some of the positive anti-v-cyclin antibody responses were detected in sera that were negative by ELISA and/or LIPS for anti-LANA, anti-K8.1 and anti-ORF65 antibodies. Nevertheless, 3% (3/91) of the total KS serum samples studied here by LIPS showed undetectable antibodies to v-cyclin, K8.1, LANA, and ORF65, which is likely consistent with the weak or undetectable anti-HHV-8 antibody responses that are associated with low CD4 counts in HIV-coinfected patients (9).
Although a few of the HHV-8 antigens screened in the initial small pilot set did not show immunoreactivity, it is possible that some of these proteins may show informative humoral targets when larger numbers of HHV-8-infected sera are studied. Along these lines, LIPS detection of patient-specific antibody responses to a large panel of HHV-8 antigens might have additional utility for understanding immune responsiveness, the severity and duration of infection, and response to drug therapy. It will also be of interest to determine whether the antibody profiles against v-cyclin and other HHV-8 proteins described here are different in other cohorts, including in asymptomatic HHV-8-infection, KS, and HHV-8 B-cell lymphoproliferative disorders, such as primary effusion lymphoma and multicentric Castleman disease.
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Dental and Craniofacial Research, and the Clinical Center, in part by a Bench to Bedside award from the NIH Clinical Research Center, and in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Burbelo, P. D., K. H. Ching, T. L. Mattson, J. S. Light, L. R. Bishop, and J. A. Kovacs. 2007. Rapid antibody quantification and generation of whole proteome antibody response profiles using LIPS (luciferase immunoprecipitation systems). Biochem. Biophys. Res. Commun. 352889-895. [DOI] [PubMed] [Google Scholar]
- 2.Burbelo, P. D., R. Goldman, and T. L. Mattson. 2005. A simplified immunoprecipitation method for quantitatively measuring antibody responses in clinical sera samples by using mammalian-produced Renilla luciferase-antigen fusion proteins. BMC Biotechnol. 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burbelo, P. D., S. Groot, M. C. Dalakas, and M. J. Iadarola. 2008. High definition profiling of autoantibodies to glutamic acid decarboxylases GAD65/GAD67 in stiff-person syndrome. Biochem. Biophys. Res. Commun. 3661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burbelo, P. D., H. Hirai, H. Leahy, A. Lernmark, S. A. Ivarsson, M. J. Iadarola, and A. L. Notkins. 2008. A new luminescence assay for autoantibodies to mammalian cell-prepared insulinoma-associated protein 2. Diabetes Care 311824-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burbelo, P. D., Y. Hoshino, H. Leahy, T. Krogmann, R. L. Hornung, M. J. Iadarola, and J. I. Cohen. 2009. Serological diagnosis of human herpes simplex virus type 1 and 2 infections by luciferase immunoprecipitation system assay. Clin. Vaccine Immunol. 16366-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burbelo, P. D., E. Meoli, H. P. Leahy, J. Graham, K. Yao, U. Oh, J. E. Janik, R. Mahieux, F. Kasanchi, M. J. Iadarola, and S. Jacobson. 2008. Anti-HTLV antibody profiling reveals an antibody signature for HTLV-I-associated myelopathy/tropical spastic paraparesis (HAM/TSP). Retrovirology 596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burbelo, P. D., R. Ramanathan, A. D. Klion, M. J. Iadarola, and T. B. Nutman. 2008. Rapid, novel, specific, high-throughput assay for diagnosis of loa loa infection. J. Clin. Microbiol. 462298-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coscoy, L. 2007. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat. Rev. Immunol. 7391-401. [DOI] [PubMed] [Google Scholar]
- 9.de Souza, V. A., L. C. Pierrotti, L. M. Sumita, W. S. Freire, A. A. Segurado, and C. S. Pannuti. 2007. Seroreactivity to Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent nuclear antigen in AIDS-associated Kaposi's sarcoma patients depends on CD4+ T-cell count. J. Med. Virol. 791562-1568. [DOI] [PubMed] [Google Scholar]
- 10.Dittmer, D. P. 2003. Transcription profile of Kaposi's sarcoma-associated herpesvirus in primary Kaposi's sarcoma lesions as determined by real-time PCR arrays. Cancer Res. 632010-2015. [PubMed] [Google Scholar]
- 11.Engels, E. A., D. Whitby, P. B. Goebel, A. Stossel, D. Waters, A. Pintus, L. Contu, R. J. Biggar, and J. J. Goedert. 2000. Identifying human herpesvirus 8 infection: performance characteristics of serologic assays. J. Acquir. Immune Defic. Syndr. 23346-354. [DOI] [PubMed] [Google Scholar]
- 12.Ganem, D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1273-296. [DOI] [PubMed] [Google Scholar]
- 13.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat. Med. 2925-928. [DOI] [PubMed] [Google Scholar]
- 14.Hladik, W., S. C. Dollard, J. Mermin, A. L. Fowlkes, R. Downing, M. M. Amin, F. Banage, E. Nzaro, P. Kataaha, T. J. Dondero, P. E. Pellett, and E. M. Lackritz. 2006. Transmission of human herpesvirus 8 by blood transfusion. N. Engl. J. Med. 3551331-1338. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins, F. J., R. B. Hayes, A. Jackson, G. Pizza, G. Mbisa, D. Whitby, and J. J. Goedert. 2007. Human herpesvirus 8 seroprevalence among prostate cancer case patients and control subjects. J. Infect. Dis. 196208-211. [DOI] [PubMed] [Google Scholar]
- 16.Katano, H., T. Iwasaki, N. Baba, M. Terai, S. Mori, A. Iwamoto, T. Kurata, and T. Sata. 2000. Identification of antigenic proteins encoded by human herpesvirus 8 and seroprevalence in the general population and among patients with and without Kaposi's sarcoma. J. Virol. 743478-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koopal, S., J. H. Furuhjelm, A. Jarviluoma, S. Jaamaa, P. Pyakurel, C. Pussinen, M. Wirzenius, P. Biberfeld, K. Alitalo, M. Laiho, and P. M. Ojala. 2007. Viral oncogene-induced DNA damage response is activated in Kaposi sarcoma tumorigenesis. PLoS Pathog. 31348-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laney, A. S., J. S. Peters, S. M. Manzi, L. A. Kingsley, Y. Chang, and P. S. Moore. 2006. Use of a multiantigen detection algorithm for diagnosis of Kaposi's sarcoma-associated herpesvirus infection. J. Clin. Microbiol. 443734-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin, J. N., Z. Amad, C. Cossen, P. K. Lam, D. H. Kedes, K. A. Page-Shafer, D. H. Osmond, and B. Forghani. 2000. Use of epidemiologically well-defined subjects and existing immunofluorescence assays to calibrate a new enzyme immunoassay for human herpesvirus 8 antibodies. J. Clin. Microbiol. 38696-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martro, E., M. J. Cannon, S. C. Dollard, T. J. Spira, A. S. Laney, C. Y. Ou, and P. E. Pellett. 2004. Evidence for both lytic replication and tightly regulated human herpesvirus 8 latency in circulating mononuclear cells, with virus loads frequently below common thresholds of detection. J. Virol. 7811707-11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, P. S., and Y. Chang. 2003. Kaposi's sarcoma-associated herpesvirus immunoevasion and tumorigenesis: two sides of the same coin? Annu. Rev. Microbiol. 57609-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeiffer, R. M., R. J. Carroll, W. Wheeler, D. Whitby, and S. Mbulaiteye. 2008. Combining assays for estimating prevalence of human herpesvirus 8 infection using multivariate mixture models. Biostatistics 9137-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabkin, C. S., T. F. Schulz, D. Whitby, E. T. Lennette, L. I. Magpantay, L. Chatlynne, R. J. Biggar, et al. 1998. Interassay correlation of human herpesvirus 8 serologic tests. J. Infect. Dis. 178304-309. [DOI] [PubMed] [Google Scholar]
- 24.Ramanathan, R., P. D. Burbelo, S. Groot, M. J. Iadarola, F. A. Neva, and T. B. Nutman. 2008. A luciferase immunoprecipitation systems assay enhances the sensitivity and specificity of diagnosis of Strongyloides stercoralis infection. J. Infect. Dis. 198444-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, N. A., C. A. Sabin, R. Gopal, D. Bourboulia, W. Labbet, C. Boshoff, D. Barlow, B. Band, B. S. Peters, A. de Ruiter, D. W. Brown, R. A. Weiss, J. M. Best, and D. Whitby. 1999. Serologic evidence of human herpesvirus 8 transmission by homosexual but not heterosexual sex. J. Infect. Dis. 180600-606. [DOI] [PubMed] [Google Scholar]
- 26.Zhu, L., R. Wang, A. Sweat, E. Goldstein, R. Horvat, and B. Chandran. 1999. Comparison of human sera reactivities in immunoblots with recombinant human herpesvirus (HHV)-8 proteins associated with the latent (ORF73) and lytic (ORFs 65, K8.1A, and K8.1B) replicative cycles and in immunofluorescence assays with HHV-8-infected BCBL-1 cells. Virology 256381-392. [DOI] [PubMed] [Google Scholar]