Abstract
Brucellosis is a bacterial zoonotic disease of major global importance. Natural hosts for Brucella species include animals of economic significance, such as cattle and small ruminants. Controlling brucellosis in natural hosts by high-throughput serological testing followed by the slaughter of seropositive animals helps to prevent disease transmission. This study aimed to convert an existing competitive enzyme-linked immunosorbent assay (cELISA), used for the serodiagnosis of brucellosis in ruminants, to two electrochemiluminescence (ECL) immunoassays on the Meso Scale Discovery (MSD) platform. The first assay employed a conventional plate washing step as part of the protocol. The second was a no-wash assay, made possible by the proximity-based nature of ECL signal generation by the MSD platform. Both ECL wash and no-wash assays closely matched the parent cELISA for diagnostic sensitivity and specificity. The results also demonstrated that both ECL assays met World Organization for Animal Health (OIE) standards, as defined by results for the OIE standard serum (OIEELISASPSS). This report is the first to describe an ECL assay incorporating lipopolysaccharide, an ECL assay for serodiagnosis of a bacterial infectious disease, a separation-free (no-wash) ECL assay for the detection of serum antibodies, and the use of the MSD platform for serodiagnosis. The simple conversion of the cELISA to the MSD platform suggests that many other serodiagnostic tests could readily be converted. Furthermore, the alignment of these results with the multiplex capability of the MSD platform offers the potential of no-wash multiplex assays to screen for several diseases.
Species of the genus Brucella cause serious chronic infections, collectively known as brucellosis. Brucellosis is a mammalian disease infecting many economically important animal species as well as humans. With a global distribution, brucellosis causes considerable animal and human health problems as well as huge economic costs. Brucella species are gram-negative, nonmotile, facultative intracellular coccobaccilli belonging to the α-2 subdivision of proteobacteria. The genus consists of six classical species, namely, Brucella abortus, B. melitensis, B. suis, B. ovis, B. canis, and B. neotomae, plus more recently discovered strains from marine mammals. Of the Brucella species, B. abortus, B. melitensis, and B. suis are of principal human health and economic importance. These species have smooth lipopolysaccharide (sLPS), which is considered a major virulence factor of disease (23), whereas B. ovis and B. canis have rough LPS (1).
The World Organization for Animal Health (OIE) prescribed and alternative serological tests for diagnosis of brucellosis caused by smooth strains rely largely upon the measurement of the host antibody response to the O antigen of the sLPS (8, 22). Classical tests include the Rose Bengal test, the complement fixation test (CFT), and the serum agglutination test (SAT), all of which employ a whole-cell antigen as the key diagnostic reagent. More contemporary techniques, such as indirect enzyme-linked immunosorbent assay (iELISA), competitive ELISA (cELISA), and fluorescent polarization assay (FPA), employ purified LPS or O antigen as the diagnostic reagent. The immunodominance of the LPS O antigen is the basis for the generally excellent sensitivity of these assays (21). However, the use of this antigen can lead to false-positive serological results when animals are infected with bacteria possessing O antigens of similar structure (6), such as Yersinia enterocolitica O:9.
ELISAs are readily amenable to high-throughput testing due to the standardized nature of the technology and reagents. This allows for many efficiency savings compared to the classical assays, including the use of effective automation (16). Despite the advantages of ELISA over the classical tests in this regard, ELISAs still require several steps to complete, including separation steps. Although these steps can be automated, they are a vital part of the assay and are a frequent source of imprecision, error, and mechanical breakdown. Assays which have the advantages of ELISA, such as a 96-well format, objective assessment, and good sensitivity and specificity, but which reduce the burden of work and opportunity for error are clearly desirable.
The Meso Scale Discovery (MSD) electrochemiluminescence (ECL) platform uses electrochemical stimulation of reporter molecules conjugated to biological components to generate a light signal measured by photodetectors (2, 31), such as a charge-coupled device (CCD) camera. Carbon electrodes are integrated into the bottom of 96-well microtiter plates, to which biological components (for example, LPS) from traditional assays such as ELISA can be passively adsorbed. Biological conjugates, for example, monoclonal antibodies (MAbs), can be labeled with the reporter molecule ruthenium(II) tris-bipyridal [Ru(bpy)32+], which upon electrical stimulation emits light at 620 nm if it is within sufficient proximity to the carbon electrode excitation source. The reaction is enhanced by the addition of read buffer, which contains coreactants, including tripropylamine. Nonspecific signals are minimized as the stimulation mechanism (electricity) is decoupled from the signal (light).
MSD ECL assays have the potential for separation-based (“wash”) and non-separation-based (“no-wash”) immunoassays due to the proximity-based nature of signal generation, where only labels near the surfaces of the electrodes are stimulated. The non-separation-based approach allows the addition of read buffer (MSD) directly to the components of the assay; this has an advantage over separation-based assays in that it removes a source of variation and reduces the time and labor required to perform the assay. In addition, MSD ECL assays can be multiplexed by spotting up to 10 different antigens onto discrete areas of the carbon electrode within each well of a 96-well plate; thus, the potential exists for multiplexed no-wash serological assays. The signal from each spot can be detected independently by a CCD camera, providing a quantitative measurement of the amount of light detected. Furthermore, the time taken to read a 96-well plate, approximately 70 s, is comparable to that for ELISAs and compares favorably to those for some other multiplex and ECL systems.
We assessed the feasibility and potential of wash and no-wash MSD ECL assays for veterinary serodiagnosis of brucellosis. Biological components (B. melitensis 16 M sLPS antigen and BM40 [10], an anti-M O-chain epitope MAb) from the brucellosis cELISA produced by the Veterinary Laboratories Agency (26) were applied to the MSD platform. As with the parent cELISA, the wash and no-wash ECL assays allow competition between serum antibodies and the MAb BM40 [labeled with Ru(bpy)32+ for the MSD ECL assay] for sLPS bound to the carbon electrode. This affects the quantity of BM40 able to bind to this target. Thus, a positive result yields a low-intensity signal and a negative result yields a high-intensity signal. The new assays were validated against existing serological methods, using sera from Brucella-infected and noninfected ruminants.
MATERIALS AND METHODS
Serum samples.
To assess the diagnostic specificity (DSp) of the ECL assays, single serum samples from 160 randomly selected cattle from Great Britain (officially brucellosis free since 1985) were collected. In addition, serum samples from 160 randomly selected small ruminants (sheep and goats) from Great Britain were also collected.
To assess the diagnostic sensitivity (DSn) of the assay, single serum samples from 32 cattle and 41 small ruminants from the Veterinary Laboratories Agency (VLA) serum archive were tested. Of the cattle samples, 8 were from naturally infected culture-positive animals, 2 were from culture-positive animals experimentally infected with B. abortus strain 544, 10 were from culture-positive animals, and a further 12 were from serologically positive animals (by CFT and SAT) from a culturally confirmed outbreak of brucellosis. Of the 41 small-ruminant samples, 2 were from naturally infected culture-positive animals, 5 were from culture-positive animals from experimental infection with B. melitensis, 9 were from serologically positive animals (by CFT) from a culturally confirmed outbreak of brucellosis, and the remaining 25 were from a suspected outbreak of brucellosis from an area of endemicity.
Labeling of MAb BM40.
The MAb BM40 was purified from hybridoma culture fluid with a protein G spin trap (GE Healthcare) and subsequently buffer exchanged using Zebra desalting columns (Pierce) into phosphate-buffered saline (PBS), pH 7.9, adjusted by the addition of NaOH in accordance with the manufacturer's instructions.
BM40 was concentrated using an Amicon Ultra-4 centrifugal filter device (Amicon), and the concentration was determined by the Pierce bicinchoninic acid (BCA) assay. The concentrated antibody was diluted with PBS (pH 7.9) to 2 mg/ml for labeling with MSD Sulfo-Tag NHS-ester containing the reporter molecule according to the manufacturer's instructions. Briefly, 100 μl of ice-cold deionized water was added to 300 nmol of Sulfo-Tag NHS-ester to give a 3-nmol/μl solution. Next, 98.7 μl of this solution was added to 1,850 μl of 2-mg/ml BM40. This was shielded from light and incubated for 2 hours at room temperature. The labeled BM40 was then buffer exchanged into PBS-0.05% sodium azide, using the supplied columns (MSD). The final concentration of BM40 was again determined by BCA assay. In addition, the concentration of the MSD Sulfo-Tag was determined by measuring the absorbance at 455 nm of the labeled protein conjugate and dividing the result by the extinction coefficient of the label (15,400 M−1 cm−1). To calculate the MSD Sulfo-Tag label/protein ratio, the concentration of MSD Sulfo-Tag was divided by the protein concentration determined by the BCA assay. This showed each BM40 molecule to be labeled with an average of 9.4 MSD Sulfo-Tag molecules, within the ideal range given by the manufacturer.
MSD ECL method.
sLPS antigen (100 μl of sLPS diluted in PBS per well), derived from B. melitensis 16 M by hot phenol extraction (28), was passively absorbed onto MSD standard-bind 96-well plates by overnight incubation at 4°C. Plates were washed four times with 200 μl of PBS plus Tween 20 (PBST), air dried, and stored in the dark at 4°C until use. The labeled MAb, BM40, was used at a final concentration of 4 nM.
To perform the assay, 5 μl of undiluted serum was added to each well and incubated with 45 μl of labeled BM40, diluted in PBS plus 1% bovine serum albumin (Sigma), for 2 hours. Plates with samples to be tested by the wash protocol were washed four times with 200 μl PBST before the addition of 75 μl of read buffer (MSD) to each well. Plates with samples to be tested by the no-wash protocol had 75 μl of read buffer added directly to wells containing serum and BM40. All samples for both assays were tested in duplicate. Plates were mixed briefly using a rotary plate shaker and read using a Sector Imager 6000 reader (MSD) containing a CCD camera.
All MSD plates had a positive serum control, a weak-positive (cutoff) serum control, a negative serum control, and a conjugate control (serum replaced with 5 μl of buffer). The weak-positive (cutoff) serum control was prepared to match the titer of a 1/16 predilution of the OIE standard serum (OIEELISASPSS) in negative serum measured by the MSD wash and no-wash assays.
The MSD ECL wash and no-wash assays were also performed on sLPS-coated plates without serum to assess the homogeneity of the sLPS coating procedure. Each well of a single plate was processed as a conjugate control, and plates were then incubated and processed as described previously to complete the assays.
To measure non-antibody-mediated serum effects, two MSD plates were directly coated with 100 μl per well of conjugated BM40 MAb at a concentration of 2.5 mg/ml in PBS. The plates were incubated overnight at 4°C and washed four times with 200 μl PBST. One plate was used for the wash assay, and the other was used for the no-wash assay. In each plate, 5 μl of serum (from noninfected cattle) and 45 μl of PBS were added to each well in duplicate for 32 different samples. For the remaining 32 wells, 50 μl of PBS only was added. These plates were then incubated and processed as described previously to complete the assays.
The analytical sensitivities of the ECL methods were compared to that of the parent cELISA by measuring the inhibitory effect of unlabeled BM40. The performance of the ECL assays was also assessed using OIEELISASPSS diluted in negative cattle serum.
cELISA.
Blank Nunc Polysorb plates were coated with B. melitensis 16 M sLPS (28) antigen overnight at 4°C and subsequently washed five times with distilled H2O (dH2O). Samples were tested individually by adding 20 μl of serum to each well. Optimal dilutions of serum and antigen were identified by checkerboard titration. Each plate had high-titer and low-titer positive serum controls, a negative serum control, and conjugate-only controls. Horseradish peroxidase (HRP)-labeled BM40 was added (100 μl per well), and plates were incubated on an orbital shaker at 160 rpm for 30 min at room temperature before being washed five times with dH2O. Plates were developed with H2O2 substrate and OPD chromogen. Plates were analyzed using a Thermo Multiskan Ascent reader at 450 nm. A result was considered positive if the percent inhibition of the conjugate control was >40%.
iELISA.
To test the cattle samples, blank Nunc Polysorb plates were coated with B. abortus S99 sLPS (28) antigen overnight at 4°C and subsequently washed five times with dH2O. Samples and controls were added to the test plate at a 1/200 dilution in PBST. Optimal dilutions of serum and antigen were identified by checkerboard titration. Each plate had high-titer and low-titer positive serum controls and a negative serum control. Plates were incubated for 30 min at room temperature on an orbital shaker (160 rpm) before being washed five times with dH2O. An HRP-labeled rabbit anti-bovine polyclonal antibody (Dako) was diluted to working strength in PBST, and 100 μl was added to each well. Plates were incubated for 30 min at room temperature on an orbital shaker (160 rpm) before being washed five times with dH2O. Plates were developed with H2O2 substrate and 2,2′-azinobis-3-ethylbenzthiazoline-6-sulfonic acid (ABTS) chromogen. Plates were analyzed using a Thermo Multiskan Ascent reader at 405 nm. A result was considered positive if it was >10% of the high-titer positive control value.
The small-ruminant iELISA was performed as for the cattle iELISA, but with B. melitensis 16 M sLPS, not B. abortus S99 sLPS, and the assay used an HRP-labeled rabbit anti-sheep/goat polyclonal antibody conjugate. All assays conformed to OIE ELISA requirements, including the use of the OIE ELISA standard serum (8, 22).
Data analysis.
Serological results for both MSD ECL assays were expressed as percentages of the weak-positive (cutoff) control value. These results were used to plot two-way receiver operator curves (TW-ROC) (9) for the cattle and small-ruminant samples tested by both the wash and no-wash assays. These curves were used to determine the positive/negative cutoffs for both assays and both validation sample sets in order to maximize DSn and DSp. Confidence intervals for DSn and DSp were calculated using binomial distribution models (3).
Correlation coefficients were calculated using the Pearson product-moment equation and were tested for significance using Student's t test. The significance of the differences between serum and nonserum wells in the conjugated BM40-coated plates was determined using paired t tests, as was also the case for the analysis of differences between the intensities of the wells in the sLPS-coated wash and no-wash plates processed entirely as conjugate controls. The significance of the differences between sera in the conjugated BM40-coated plates was determined by analysis of variance. The significance of the difference in the coefficients of variance (CVs) of the wash and no-wash plates processed entirely as conjugate controls was determined using the method of Miller (19). All data analysis was performed in Microsoft Excel.
RESULTS
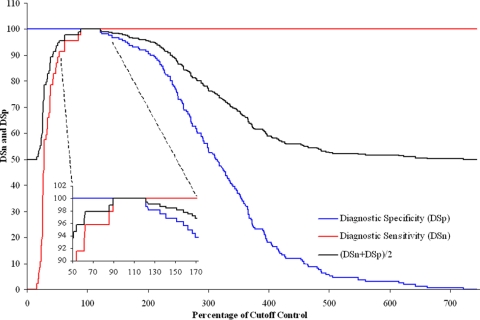
Individual TW-ROCs of the results were calculated for the cattle and small-ruminant samples tested by the wash and no-wash methods. The positive/negative cutoffs were determined from these by selecting the test value that maximized the sum of the DSn and DSp, giving a performance index (22). Optimal cutoffs were determined individually for cattle and small-ruminant samples tested by the wash and no-wash protocols. It was found that the same cutoff values provided maximum DSn and DSp for the two sample types tested by the same assay. A cutoff of 110% of the weak-positive (cutoff) control value for the no-wash protocol was established. This provided the maximum performance index of 200% for both cattle and small-ruminant samples. The TW-ROC for the cattle samples tested by the no-wash protocol is shown in Fig. 1 as an example. For the wash protocol, a cutoff of 127% was established, and this provided a maximum performance index of 196.3% for cattle samples and 200% for small-ruminant samples.
FIG. 1.
TW-ROC for cattle samples tested by the no-wash assay. The enlarged area shows 100% DSn and DSp to occur from 89.3 to 121.1% of the weak-positive (cutoff) control value. From these data and those for the small-ruminant samples tested by the no-wash assay, a cutoff of 110% was determined.
A summary of the comparative serological results for both MSD assays plus iELISA and cELISA is shown in Table 1. DSp and DSn for the small-ruminant samples tested by the wash and no-wash assays were all 100%, which matched the results of the cELISA and iELISA. DSn for cattle samples tested by the wash and no-wash protocols was 100% and exceeded the results for cELISA for both assays. DSp for cattle samples tested by the wash assay was 96.3%, which was lower than those for the other three assays. The iELISA results for cattle samples had the best results, which were 100% for both DSn and DSp.
TABLE 1.
Summary of DSn and DSp results for the wash and no-wash ECL methods, plus cELISA and iELISA results
| Group and assay | Assay performance (% [95% confidence limits])
|
Performance index | |
|---|---|---|---|
| DSn | DSp | ||
| Small ruminants (n = 41 for DSn and 160 for DSp) | |||
| Wash assay | 100.0 (91.4-100.0) | 100.0 (97.7-100.0) | 200 |
| No-wash assay | 100.0 (91.4-100.0) | 100.0 (97.7-100.0) | 200 |
| cELISA | 100.0 (91.4-100.0) | 100.0 (97.7-100.0) | 200 |
| iELISA | 100.0 (91.4-100.0) | 100.0 (97.7-100.0) | 200 |
| Cattle (n = 32 for DSn and 160 for DSp) | |||
| Wash assay | 100.0 (89.1-100.0) | 96.3 (92.0-98.6) | 196.3 |
| No-wash assay | 100.0 (89.1-100.0) | 100.0 (97.7-100.0) | 200 |
| cELISA | 96.9 (83.73-99.9) | 100.0 (97.7-100.0) | 196.9 |
| iELISA | 100.0 (89.1-100.0) | 100.0 (97.7-100.0) | 200 |
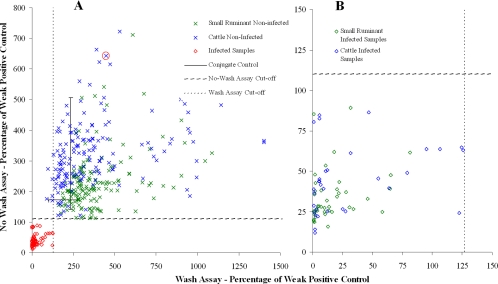
The ECL wash and no-wash results for the validation panel are shown as scatter plots (Fig. 2A and B). These demonstrate a clear differentiation between nearly all positive and negative samples. However, variation within the negative samples is clearly evident, and there was a significant positive correlation between the negative results for the two assays (r = 0.276; P < 0.001). The positive correlation between the positive results was higher than this and was also significant (r = 0.317; P < 0.001).
FIG. 2.
Scatter plots of samples tested by the wash and no-wash assays. (A) The mean results for two replicates are shown. (B) Results for the positive samples in increased detail. The sample that is ringed is the one that was subsequently spiked with OIEELISASPSS to help determine the analytical sensitivities of the wash and no-wash assays.
The scatter plots in Fig. 2 show positive samples tested by the wash protocol to have lower results than those by the no-wash assay. The mean positive result for samples tested by the wash protocol was 31.1%, and the most positive result was 0.7%. The mean positive result for samples tested by the no-wash protocol was 47.2%, and the most positive result was 11.8%.
Conjugate control values are shown in Fig. 2 as a solid black cross. The center of the cross represents the average value for the conjugate control (232.4% for the wash assay and 172.6% for the no-wash assay), and the ends of the lines represent the maximum and minimum values. These data show that there was some variation within the conjugate control data, especially for the no-wash assay. It also demonstrates that many negative results exceeded the conjugate control values, especially for the wash assay.
The results for the sLPS-coated plates developed with BM40 conjugate and buffer alone showed differences in CV and intensity between the wash and no-wash assays. The CV for the no-wash assay was 4.71%, and that for the wash assay was 16.39%. This difference was statistically significant (P < 0.001). The mean intensity for the no-wash assay was 6,865, and that for the wash assay was 4,719. This difference was also statistically significant (P < 0.001).
The data from the plates coated with conjugated BM40 show that there was a significant difference between the wells incubated with serum and those without. This difference was also dependent on the method. With the no-wash method, the serum wells had, on average, a 7.1% lower intensity (P = 0.06), whereas the serum wells in the wash method had an average intensity that was 29.0% higher than that of wells incubated with buffer alone (P < 0.001). These results also showed that there was a significant difference in results between serum samples (P < 0.001). There was also a significant positive correlation between the wash and no-wash results for the serum samples (r = 0.408; P < 0.001).
Analytical sensitivity results, expressed as percentages of the conjugate control value (PCC), are shown in Fig. 3. As the concentration of unlabeled BM40 decreased, the PCC increased. The results show the wash assay to be able to discriminate between 0.5 and 0.25 nM BM40, the no-wash assay to discriminate between 0.25 and 0.125 nM BM40, and the cELISA to discriminate between 0.125 and 0.0625 nm BM40. Figure 3 also shows that with concentrations of 128 nM and 0.0625 nM, the PCCs for the cELISA are 9.7% and 87.0%, respectively. The no-wash assay had equivalent results, of 30.9% and 90.4%, respectively, and the wash assay also had equivalent values, of 3.3% and 98.1%, respectively.
FIG. 3.
Analytical sensitivities of ECL wash and no-wash assays compared to that of the parent cELISA. The data points represent the PCC at different concentrations of free BM40 MAb.
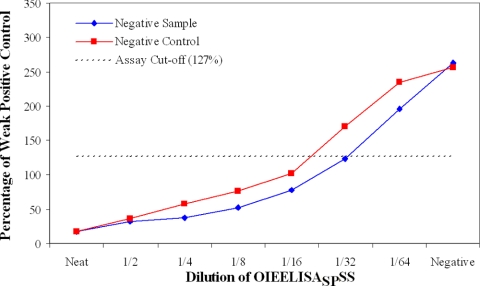
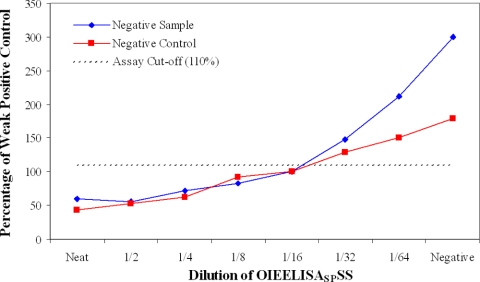
The results for two negative serum samples spiked with OIEELISASPSS at doubling dilutions are shown in Fig. 4 and 5. The two samples, a cattle negative control and a noninfected cattle field sample with high results in the wash and no-wash assays (this sample has been ringed in Fig. 2A), were tested by both the wash (Fig. 4) and no-wash (Fig. 5) assays. Both samples were positive at a 1/16 dilution of OIEELISASPSS and negative at a 1/64 dilution for both methods. The significance of this is described below.
FIG. 4.
Analytical sensitivity of ECL wash assay, determined with negative samples spiked with OIEELISASPSS.
FIG. 5.
Analytical sensitivity of ECL no-wash assay, determined with negative samples spiked with OIEELISASPSS.
DISCUSSION
The proximity-based detection system of the MSD ECL platform enables the development of non-separation-based (no-wash) assays along with more conventional plate-washing approaches. The appeal of a no-wash assay is the reduction in labor and equipment required and the potential for improving assay precision. The aim of this study was to develop and evaluate wash and no-wash ECL serodiagnostic protocols for brucellosis with a view to their interpretation as model systems to indicate the general feasibility of performing serological assays on the MSD ECL platform. This work was intended as a pilot study rather than a full-scale validation.
The no-wash protocol required adaptation from a parent cELISA rather than an iELISA. Indirect techniques would require the labeling of large and variable quantities of non-antigen-specific complexes which would remain in the test well for the stimulation and read phases. This could result in suboptimal specific binding of the label within proximity of the excitation source and in nonspecific diffusion-enhanced excitation of unbound label. The competitive approach allowed for greater control of the labeled element, in this case the conjugated BM40 MAb. The use of a competitive format had the added advantage of generating a non-species-specific assay, which is important for brucellosis diagnosis because the disease occurs in a range of livestock species.
The conversion of the VLA Brucella cELISA to both MSD ECL protocols was simple and straightforward. As expected, optimization of parameters such as reagent concentration and incubation and wash times was required. These optimization studies were conducted using control sera.
The TW-ROC analysis demonstrated that the two ECL assays, the wash and no-wash assays, required different cutoffs to maximize DSn and DSp, with a higher cutoff for the wash assay reflecting the higher average negative result for this assay. The cutoff values calculated by TW-ROC analysis for the wash and no-wash assays were applicable for the two sample groups, i.e., cattle and small ruminants.
The results from the validation sera demonstrated that both ECL assays developed had a performance index of 200% for small-ruminant samples, while for cattle samples the no-wash assay also had a performance index of 200% but the wash assay had a performance index of 196.3%. Owing to the relatively small nature of the validation panel, it is more appropriate to compare the diagnostic performances relative to those of the cELISA and iELISA rather than to consider the presented data in isolation. The performance characteristics of the ELISAs themselves are strong, with performance indexes in excess of 194% (17), and the developed ECL assays are comparable to them. The results also show that the wash and no-wash assays have very similar diagnostic attributes.
The scatter plots in Fig. 2A and B show detailed serological results for the wash and no-wash ECL assays. The strong diagnostic performance is indicated by the segregation of the data points from the infected and noninfected sources. However, there are many other interesting features. Most of the positive samples had very-low-intensity values and were well under the positive/negative threshold (cutoff). This reflects the high anti-sLPS antibody titers found in the sera of most infected animals and the optimization of both assays to favor differentiation of weak-positive (cutoff) from negative samples rather than strong-positive from negative samples. There is still evidence for a dose-response effect, which in most cases can be observed for each sample in both assays. There was a high frequency of positive samples for the wash protocol where the results were very close to zero, but this was not the case for the no-wash protocol. This is probably due to the low-level signal in the no-wash assay due to nonbound MAb randomly diffusing close to the excitation source. The high-titer samples for the no-wash protocol tended to cluster closer to 25%, compared to 10% or less for the wash protocol. Despite this, there is still a highly significant positive correlation between the results of the two protocols for the positive samples.
The well-to-well variation was assessed for both methods by incubating antigen-coated plates with no serum, just labeled BM40 MAb. The results from this showed that there was significantly less well-to-well variation with the no-wash method than with the wash method. This may be expected, as the wash step introduces an additional source of error into the protocol. The CV for the no-wash plate was 4.71, which can be considered acceptable. The CV for the wash plate was 16.39, which would be high for routine use compared to ELISA (25). The highest result on this plate was 2.47 times the magnitude of the lowest.
The most obvious feature in Fig. 2A is the large amount of variation in the negative results. This was unexpected and unusual given that there should be no dose-response effect in such samples and that they should therefore be clustered much more closely together. Many of these negative results were also greater than the results for the conjugate controls, which should present the theoretical maximum signal.
To investigate the possibility of a serum-specific non-antibody-based effect, plates were coated directly with labeled MAb and incubated with serum or buffer. The results showed that there was a significant difference between the results from wells incubated with buffer and those incubated with serum. The investigation also showed that there were significant differences between sera. Although serum caused an average decrease in the no-wash protocol and an average increase in the wash protocol, there was also a significant positive correlation in the sample results for the two protocols. These results suggest that there may be two antibody-independent serum effects taking place. The first may be that serum constituents, to a greater or lesser extent, can permanently enhance the signal generated by the Ru(bpy)32+ label. The second may be that when serum is still present and when read buffer is added (i.e., as for the no-wash protocol), there is a general suppression of the signal. This could explain why data from the serum wells in the no-wash plate were generally lower but still positively correlated with the results from the wash plate.
Despite the variation and non-antibody-mediated serum effects, both ECL assays still demonstrated good diagnostic performance. In positive samples, sLPS-specific antibodies prevented the labeled BM40 from binding close to the excitation source, and therefore any effect that the serum had on the Ru(bpy)32+ label was proportionally reduced. This may therefore be the reason that the discrimination of positive and negative samples was able to transcend these issues to some extent and why they are only apparent in the negative population. For indirect methods, the label would be in proximity to the excitation source in positive samples, but serum is typically added in higher dilutions than is the case in cELISAs. This is also likely to be the case with indirect ECL assays, and therefore any non-serum-antibody-mediated effects would be diluted proportionally.
The analytical sensitivities of both ECL assays were investigated by two approaches. The first used two negative samples, a negative control and a sample with high results for both ECL assays, to dilute the OIEELISASPSS. The results demonstrated that both assays met the OIE requirements for the detection of this standard (22) and that specific antibodies can be detected with the necessary sensitivity, even in samples that would otherwise have high negative results. These results show once again that high-titer positive samples have lower results in the wash protocol. The analytical sensitivity study conducted using unlabeled BM40 MAb as a competing agent showed that there were differences in the maximum sensitivities and dynamic ranges of the assays. However, all assays, including the cELISA, are capable of detecting changes in the 32 to 0.25 nM range, and this appears to be the critical indicator of effective serodiagnostic performance in this instance.
Two other no-wash technologies that are suited to high-throughput testing have been adapted to the serodiagnosis of brucellosis, the FPA (21) and the AlphaLISA (16). Both of these assays are homogeneous, as the binding reactions take place within the wells rather than on the well surfaces (although the AlphaLISA reaction takes place on a bead surface). Neither of these protocols requires a plate coating or wash step, although some upstream manipulation of the reagents is still required, such as reagent labeling and conjugation. Both would represent more cost-effective and efficient means for conducting high-throughput serodiagnosis using single antigens than the two new assays developed here. However, neither the FPA nor the AlphaLISA currently has any effective multiplexing potential.
A number of ECL assays have been described previously, as reviewed below, but this is the first description of the MSD ECL platform being utilized for serum antibody detection. The no-wash assay is the first report of a separation-free ECL assay for the detection of serum antibodies.
Bead-based ECL assays to detect a variety of targets have previously been described. These include ECL assays that measure the quantity of a specific PCR product produced (4, 5, 20). Previous reports of multiplex systems of this type (14) refer to the amplification step, not the ECL assay. Bead-based ECL tests have also been described for the detection of bacteria (32), bacterial toxins (7, 18, 24), and protozoans (15). Antibody detection by bead-based ECL has been reported previously for humans (30) and cats (12). A competitive one-step ECL assay for the detection of human anti-TNF-α antibody in serum has also been developed (13).
These bead-based ECL assays rely upon magnetic separation of the beads from the surrounding unbound components, and this takes place inside the analyzer. This wash and the subsequent read step are applied sequentially for each sample and add a considerable amount of time to the analysis. The MSD ECL assays described here are read directly from signals emitted in the test plate. This is a faster, simpler, and more versatile system (the bead-based assay does not offer a multiplex capability). Some examples of the use of the MSD platform have already been published, such as for the detection of ricin B chain (11) in a variety of substrates (but not serum) and for the multiplexed detection of cytokines in human serum (27). Other systems, such as the Pierce SearchLight and Quansys, offer multiplexing capability (27, 29), but neither appears suitable for conversion to a no-wash format, as they are not proximity based.
The wash and no-wash assays developed as part of this study are the first to use LPS for serodiagnosis on an ECL platform and the first to detect antibodies raised against infectious bacterial disease. The capability of the MSD ECL system to perform multiplex analysis coupled to the data presented here offers the exciting prospect of performing no-wash multiplex tests using antigen and labeled MAb partners. Such a system could prove itself to be a highly efficient alternative to current serological testing regimens.
Acknowledgments
This work was funded by the VLA Test Development Programme and the Department for Environment, Food, and Rural Affairs, Great Britain.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Alton, G., G. L. M. Jones, R. D. Angus, and J. M. Verger. 1988. Techniques for the brucellosis laboratory, p. 37-42. INRA, Paris, France.
- 2.Best, J., D. M. T. Jay, F. Out, J. Ma, A. Nadin, S. Ellis, H. D. Lewis, C. Pattison, M. Reilly, T. Harrison, M. S. Shearman, T. L. Williamson, and J. R. Atack. 2005. Quantitative measurement of changes in amyloid-beta(40) in the rat brain and cerebrospinal fluid following treatment with the gamma-secretase inhibitor LY-411575. J. Pharmacol. Exp. Ther. 313902-908. [DOI] [PubMed] [Google Scholar]
- 3.Brownlee, K. A. 1965. Statistical theory and methodology in science and engineering, 2nd ed., p. 148-149. John Wiley, New York, NY.
- 4.Collins, R. A., L. S. Ko, K. Y. Fung, L. T. Lau, J. Xing, and A. C. Yu. 2002. A method to detect major serotypes of foot-and-mouth disease virus. Biochem. Biophys. Res. Commun. 297267-274. [DOI] [PubMed] [Google Scholar]
- 5.Collins, R. A., L. S. Ko, K. L. So, T. Ellis, L. T. Lau, and A. C. Yu. 2002. Detection of highly pathogenic and low pathogenic avian influenza subtype H5 (Eurasian lineage) using NASBA. J. Virol. Methods 103213-225. [DOI] [PubMed] [Google Scholar]
- 6.Corbel, M. J. 1985. Recent advances in the study of Brucella antigens and their serological cross-reactions. Vet. Bull. 55927. [Google Scholar]
- 7.Cote, C., K. C. A. Rossi, A. S. Kang, P. R. Morrow, J. S. Lee, and S. L. Welkos. 2005. The detection of protective antigen (PA) associated with spores of Bacillus anthracis and the effects of anti-PA antibodies on spore germination and macrophage interactions. Microb. Pathog. 38209-225. [DOI] [PubMed] [Google Scholar]
- 8.Garin-Bastuji, B., and J. M. Blasco. 2008. Caprine and ovine brucellosis, p. 974-982. In OIE manual of diagnostic tests and vaccines for terrestrial animals, 6th ed. Office International des Epizooties, Paris, France.
- 9.Greiner, M., D. Sohr, and P. Gobel. 1995. A modified ROC analysis for the selection of cut-off values and the definition of intermediate results of serodiagnostic tests. J. Immunol. Methods 185123-132. [DOI] [PubMed] [Google Scholar]
- 10.Greiser-Wilke, I., V. Moennig, D. Thon, and K. Rauter. 1985. Characterization of monoclonal antibodies against Brucella melitensis. Zentralbl. Veterinarmed. B 32616-627. [DOI] [PubMed] [Google Scholar]
- 11.Guglielmo-Viret, V., and P. Thullier. 2007. Comparison of an electrochemiluminescence assay in plate format over a colorimetric ELISA, for the detection of ricin B chain (RCA-B). J. Immunol. Methods 32870-78. [DOI] [PubMed] [Google Scholar]
- 12.Horii, Y., N. P. Garcia, D. Noviana, F. Kone, T. Sawada, T. Naraki, and K. Yamaguchi. 2001. Detection of anti-borna disease virus antibodies from cats in Asian countries, Japan, Philippines and Indonesia using electrochemiluminescence immunoassay. J. Vet. Med. Sci. 63921-923. [DOI] [PubMed] [Google Scholar]
- 13.Horninger, D., E. Eirikis, C. Pendley, J Giles-Komar, H. M. Davis, and B. E. Miller. 2005. A one-step, competitive electrochemiluminescence-based immunoassay method for the quantification of a fully human anti-TNF-α antibody in human serum. J. Pharm. Biomed. Anal. 38703-708. [DOI] [PubMed] [Google Scholar]
- 14.Jean, J., D. H. D'Souza, and L. A. Jaykus. 2004. Multiplex nucleic acid sequence-based amplification for simultaneous detection of several enteric viruses in model ready-to-eat foods. Appl. Environ. Microbiol. 706603-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuczynska, E., D. G. Boyer, and D. R. Shelton. 2003. Comparison of immunofluorescence assay and immunomagnetic electrochemiluminescence in detection of Cryptosporidium parvum oocysts in karst water samples. J. Microbiol. Methods 5317-26. [DOI] [PubMed] [Google Scholar]
- 16.McGiven, J. A., J. Sawyer, L. L. Perrett, S. D. Brew, N. J. Commander, A. Fisher, S. McLarnon, K. Harper, and J. A. Stack. 2008. A new homogeneous assay for high throughput serological diagnosis of brucellosis in ruminants. J. Immunol. Methods 3377-15. [DOI] [PubMed] [Google Scholar]
- 17.McGiven, J. A., J. D. Tucker, L. L. Perrett, J. A. Stack, S. D. Brew, and A. P. MacMillan. 2003. Validation of FPA and cELISA for the detection of antibodies to Brucella abortus in cattle sera and comparison to SAT, CFT, and iELISA. J. Immunol. Methods 278171-178. [DOI] [PubMed] [Google Scholar]
- 18.Merrill, G., A. V. R. Rivera, D. D. Neal, C. Young, and M. A. Poli. 2006. A quantitative electrochemiluminescence assay for Clostridium perfringens alpha toxin. Anal. Biochem. 357181-187. [DOI] [PubMed] [Google Scholar]
- 19.Miller, G. E. 1991. Asymptotic test statistics for coefficients of variation. Commun. Stat. Theor. Methods 203351-3363. [Google Scholar]
- 20.Munshi, V., M. Lu, P. Felock, R. Barnard, D. Hazuda, M. Miller, and M. Lai. 2008. Monitoring the development of non-nucleoside reverse transcriptase inhibitor-associated resistant HIV-1 using an electrochemiluminescence-based reverse transcriptase polymerase assay. Anal. Biochem. 374121-132. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, K., and D. Gall. 2001. Fluorescence polarization assay for the diagnosis of brucellosis: a review. J. Immunoassay Immunochem. 22183-201. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen, K., and D. Ewalt. 2008. Bovine brucellosis, p. 634-659. In OIE manual of diagnostic tests and vaccines for terrestrial animals, 6th ed. Office International des Epizooties, Paris, France.
- 23.Porte, F., A. Naroeni, S. Ouahrani-Bettache, and J. P. Liautard. 2003. Role of the Brucella suis lipopolysaccharide O antigen in phagosomal genesis and in inhibition of phagosome-lysosome fusion in murine macrophages. Infect. Immun. 711481-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivera, V. R., F. J. Gamez, W. K. Keener, J. A. White, and M. A. Poli. 2006. Rapid detection of Clostridium botulinum toxins A, B, E, and F in clinical samples, selected food matrices, and buffer using paramagnetic bead-based electrochemiluminescence detection. Anal. Biochem. 353248-256. [DOI] [PubMed] [Google Scholar]
- 25.Shekarchi, I. C., J. L. Sever, Y. L. Lee, G. Castellano, and D. L. Madden. 1984. Evaluation of various plastic microtiter plates with measles, toxoplasma, and gamma globulin antigens in enzyme-linked immunosorbent assays. J. Clin. Microbiol. 1989-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stack, J. A., L. L. Perrett, S. D. Brew, and A. P. MacMillan. 1999. Competitive ELISA for bovine brucellosis suitable for testing poor quality samples. Vet. Rec. 145735-736. [PubMed] [Google Scholar]
- 27.Toedter, G., K. Hayden, C. Wagner, and C. Brodmerkel. 2008. Simultaneous detection of eight analytes in human serum by two commercially available platforms for multiplex cytokine analysis. Clin. Vaccine Immunol. 1542-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westphal, O., O. Luderitz, and F. Bister. 1952. Uber die extraction von bakterien mit phenol/wasser. Z. Naturforsch. 7148. [Google Scholar]
- 29.Whelan, C., E. Shuralev, G. O'Keeffe, P. Hyland, H. F. Kwok, P. Snoddy, A. O'Brien, M. Connolly, P. Quinn, M. Groll, T. Watterson, S. Call, K. Kenny, A. Duignan, M. J. Hamilton, B. M. Buddle, J. A. Johnston, W. C. Davis, S. A. Olwill, and J. Clarke. 2008. Multiplex immunoassay for the serological diagnosis of Mycobacterium bovis-infected cattle. Clin. Vaccine Immunol. 151834-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan, G., D. Xing, S, Tan, and Q. Chen. 2004. Rapid and sensitive immunomagnetic-electrochemiluminescent detection of p53 antibodies in human serum. J. Immunol. Methods 28847-54. [DOI] [PubMed] [Google Scholar]
- 31.Yang, H., J. K. Leland, D. Yost, and R. J. Massey. 1994. Electrochemiluminescence: a new diagnostic and research tool. ECL detection technology promises scientists new “yardsticks” for quantification. Biotechnology (New York) 12193-194. [DOI] [PubMed] [Google Scholar]
- 32.Yu, H., and J. G. Bruno. 1996. Immunomagnetic-electrochemiluminescent detection of Escherichia coli O157 and Salmonella typhimurium in foods and environmental water samples. Appl. Environ. Microbiol. 62587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]