Abstract
Recently, molecular screening for pathogenic agents has identified a partial genome of a novel parvovirus, called human bocavirus (HBoV). The presence of this newly described parvovirus correlated with upper and lower respiratory tract infections in children. Lower respiratory tract infections are a leading cause of hospital admission in children, and the etiological agent has not been identified in up to 39% of these cases. Using baculovirus expression vectors (BEVs) and an insect cell system, we produced virus-like particles (VLPs) of HBoV. The engineered BEVs express the HBoV capsid proteins stoichiometrically from a single open reading frame. Three capsid proteins assemble into the VLP rather than two proteins predicted from the HBoV genome sequence. The denatured capsid proteins VP1, VP2, and VP3 resolve on silver-stained sodium dodecyl sulfate-polyacrylamide gels as three bands with apparent molecular masses of 72 kDa, 68 kDa, and 62 kDa, respectively. VP2 apparently initiates at a GCT codon (alanine) 273 nucleotides downstream from the VP1 start site and 114 nucleotides upstream from the VP3 initiation site. We characterized the stable capsids using physical, biochemical, and serological techniques. We found that the density of the VLP is 1.32 g/cm3 and is consistent with an icosahedral symmetry with approximately a 25-nm diameter. Rabbit antiserum against the capsid of HBoV, which did not cross-react with adeno-associated virus type 2, was used to develop enzyme-linked immunosorbent assays (ELISAs) for anti-HBoV antibodies in human serum. Using ELISA, we tested 404 human serum samples and established a range of antibody titers in a large U.S. adult population sample.
Among the family Parvoviridae, the genus Parvovirinae has many pathogenic species such as feline panleukopenia virus (38, 46), canine parvovirus (39), and Aleutian disease virus of mink (7). However, the only human-pathogenic parvovirus is the sole member of the Erythrovirinae, strain B-19 (3, 26, 52). Recently, a second potentially pathogenic human parvovirus was isolated and assigned the species named human bocavirus (HBoV) (2). Although HBoV DNA was detected in clinical isolates of children with lower respiratory tract infections (4, 5, 12, 30), it is unclear whether HBoV was the etiological agent or contributed to the pathogenicity of the respiratory infection (34, 45). Until recently, the presence of HBoV in respiratory secretions relied on PCR (13, 16, 32, 35, 37, 40). Using PCR, HBoV DNA has been detected worldwide with 5 to 10% prevalence among children with upper or lower respiratory tract infections (6, 22, 24, 27, 33, 36, 41, 51). However, 80% of the HBoV DNA-positive patients were coinfected with common human respiratory viruses (8, 10, 17). Thus, whatever role HBoV plays in lower respiratory tract infection remains unclear.
Using the baculovirus expression vector (BEV) system, we produced virus-like particles (VLPs) of HBoV. Two recent reports developed enzyme-linked immunosorbent assays (ELISAs) using HBoV antigen that associated into a homomeric particle with apparent parvovirus-like size and symmetry. As reported previously by Kahn et al. (23), 270 serum specimens, mostly from infants, were tested using a VP2 VLP-based ELISA. Using a cutoff of 0.150 absorption units at a dilution of 1:80, 90% of infants ≤2 months old were considered to be HBoV seropositive. The percentage dropped to 25% in the 4-month-old age group but increased to 85% for individuals in the >48-month- to 20-year-old age group. In a previously reported study (29) also using VP2 VLP-HBoV, 394 sera of children under 15 years old were tested. Using a cutoff of 0.3 absorption units and a 1:200 serum dilution, those authors reported an age-related increase in seroprevalence in healthy children and an even higher seroprevalence in children (birth to 3 years old) with lower respiratory tract infection. The incomplete VLP described in recent reports (23, 29, 31) might have resulted in the relatively low titers perhaps due to a lack of native epitopes derived from tertiary protein interactions. Based on precedence with adeno-associated virus (AAV), proper capsid formation requires the major coat protein and VP2 for AAV VLP assembly using baculovirus and Sf9 cells (44).
Based on the reported HBoV sequence, we designed a BEV to express the HBoV capsid proteins from a single open reading frame. Using a non-AUG initiation codon for VP1, the ratio of the smaller, major capsid protein VP2 to the minor, larger capsid protein VP1 was approximately 5:1. However, in both the BEV-infected insect cell lysates and purified VLPs, three capsid proteins were observed, not two as predicted from the HBoV genomic sequence. Rabbit antiserum produced in response to immunizations with purified VLP developed high-titer immunoglobulins (Igs) specific for HBoV. Thus, we developed an ELISA and tested 404 serum samples from adults. The data obtained from these sera produced a broad range of titers, suggesting that the prevalence of prior exposure to HBoV in the United States adult population is 59 to 67%.
MATERIALS AND METHODS
Bocavirus sequence and modification.
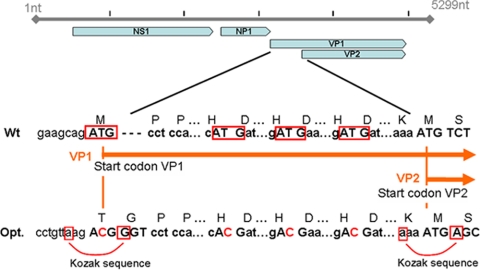
The open reading frame of the bocavirus capsid proteins (or virus proteins), encoded by the cap gene, is based on the previously published HBoV-st2 sequence (GenBank accession number DQ000496) (2). By introducing several sequence modifications into the capsid protein genes, a bicistronic mRNA was utilized to produce VP1 and VP2 in the baculovirus system (Fig. 1). A single open reading frame encodes both the large and small capsid proteins, with the larger capsid protein (VP1) and internal translational initiation producing the smaller, major capsid protein. To prevent translation initiation at a noninitiating AUG, the three out-of-frame ATG triplets in the VP1 unique sequence were altered without changing the amino acid. Thus, the first methionine codon that scanning ribosomes encounter is the initiation codon for the major coat protein. Achieving the typical parvovirus-like stoichiometry of the major and minor capsid proteins in the heterologous insect cell system was accomplished by changing the predicted VP1 initiation codon from AUG to ACG. Members of the Dependovirinae (AAV) utilize non-AUG initiation codons to regulate the level of VP2 produced from a VP2/VP3 bicistronic mRNA, and this strategy has proven successful for recombinant AAV produced in BEV-insect cell cultures (50). The threonine (ACG) codon requires the proper context for translational initiation; therefore, the nonanucleotide immediately upstream of the wild-type AAV2 VP2 ACG initiation codon (CCTGTTAAG, corresponding to nucleotides 2606 to 2613 of AAV2) was inserted upstream of the VP1 initiation site in the bocavirus VP expression cassette (Fig. 1). In addition, a glycine codon (GGT) was inserted in the second codon, creating a Kozak-compatible motif. A silent transversion in the first position of the second codon was introduced to improve the translational initiation of the major coat protein (Fig. 1). To facilitate the cloning in vector pVDF, two restriction enzyme sites (EcoRI and NotI) were included at either end of the VP gene. The new HBoV capsid gene was synthesized (BioBasic, Inc., Markham, Ontario, Canada) and cloned into plasmid pUC 59.
FIG. 1.
Optimization of the HBoV genome for production of virus proteins in a baculovirus system. The VP1 unique region and a VP1 and VP2 amino terminus common region were encoded in a single open reading frame. To prevent translation initiation at a noninitiating AUG, three out-of-frame ATG triplets in the VP1 unique sequence were altered (red boxes). Consensus Kozak elements were introduced to improve translation initiation at the VP1 and VP2 translation initiation sites, as indicated. nt, nucleotides; Opt., optimization; Wt, wild type.
Cloning and baculovirus production.
The synthetic capsid gene was amplified by PCR using 5′-CGCACCACAAAACACCTCAGG and 5′-GGTGACCATTCTGAATTGTG as the upstream and downstream primers, respectively, yielding a 2,200-bp fragment. An aliquot of the PCR mixture was digested with EcoRI and NotI and then purified (PCR purification kit; Qiagen, Inc., Valencia, CA). The digested PCR product was ligated (Fast Ligation; New England BioLabs, Ipswich, MA) with EcoRI- and NotI-digested pFastBac (Invitrogen Corp., Carlsbad, CA), yielding pFB-Boca. Recombinant baculovirus was generated using the Bac-to-Bac system, which produces infectious, recombinant baculovirus DNA in Escherichia coli strain DH10Bac (Invitrogen). The “bacmids” from several DH10Bac colonies were isolated and used to transfect Sf9 cells according to the manufacturer's protocol (Invitrogen). Briefly, 30 μg of bacmid DNA was mixed with 600 μl of Grace's medium (Invitrogen) and combined with 36 μl of Cellfectin (Invitrogen) premixed in 600 μl of Grace's medium. The final mixture (1.2 ml) was added to each well of six-well plates containing approximately 1 × 106 Sf9 cells per well. After 3 days, the supernatant of each well was collected, presumably containing BEV generation P1. The P1 stock was amplified (1:100, by volume) in Sf9 cell suspension cultures (2 × 106 cells/ml) for 3 days, generating P2 BEV. Infectious BEV titers were determined by plaque assays. The P2 stock, titers of >1 × 108 PFU/ml, were filtered (0.22 μm) and stored at 4°C in opaque, dark-sided, 50-ml conical tubes (Greiner Bio-One, Monroe, NC).
VLP production and purification.
The capsid or virus proteins of HBoV were produced by infecting 100 ml of Sf9 cells (2 × 106 cells/ml) with clonally isolated baculovirus at a multiplicity of infection of 3 PFU/cell. At 72 h postinfection, cells and supernatant were separated by centrifugation (900 × g for 15 min). The VLP in the supernatant was recovered by precipitation in 2.5% (final concentration) polyethylene glycol (PEG) (5 ml of 50% PEG 8000 [Sigma-Aldrich, St. Louis, MO] added to 95 ml of supernatant) and incubated for 3 h at 4°C with gentle agitation. Cell pellets were resuspended in 7 ml of phosphate-buffered saline with 2 mM MgCl2 and disrupted using a 7-ml Dounce homogenizer. The cell lysates were clarified by centrifugation (2,000 × g for 15 min) and concentrated by precipitation in 2.5% PEG as described above. The PEG-precipitated material was recovered by centrifugation (45 min at 2,600 × g), and the pellets were resuspended in 11 ml of CsCl solution (refractive index [RI] = 1.372 or ρ = 1.41 g/cm3). The CsCl solutions were centrifuged to equilibrium (72 h at 222,000 × g) in a swinging-bucket rotor (SW41 rotor; Beckman Coulter, Inc., Fullerton, CA). Each centrifuge tube was fractionated dropwise via bottom puncture using a 26-gauge butterfly needle set, 0.5-ml fractions were collected, and the RI of each fraction was measured with a digital refractometer (AR 200; Leica Microsystems, Inc., Buffalo, NY). Typical parvovirus empty-particle densities ranged from 1.30 to 1.32 g/cm3, corresponding to an RI of 1.362 to 1.364 (21). Size exclusion column chromatography (Sephadex 200, 10/300; GE Healthcare Bioscience Division, Piscataway, NJ) provided the final purification step. The homogeneity of the HBoV VLP was assessed by silver-staining sodium dodecyl sulfate (SDS)-polyacrylamide gels.
Larger amounts of VLP were produced using 5-liter bioreactors (Wave mixer; GE Healthcare Bioscience Division, Piscataway, NJ). The purification protocol described above was modified for scale but otherwise remained the same.
The theoretical, 280-nm molar extinction coefficient (117,855 M−1 cm−1) was determined using ProtParam software (18) and used to calculate the protein concentration by UV absorption.
Antibody production and purification.
Rabbit anti-bocavirus VLP immune serum was produced using a standard 70-day prime-boost regimen. In brief, 200 μg of purified HBoV VLP was administrated intramuscularly to the rabbit followed by three intramuscular boosts at 21, 35, and 49 days. Initially, a preimmunization sample was obtained, and subsequently, serum samples were collected at 44, 59, and 63 days postimmunization. The mean specific antibody concentration was estimated to be 0.15 to 0.5 mg/ml.
Purification of HBoV-specific antibody from human serum and rabbit serum was performed with either an affinity column prepared by covalently attaching HBoV-VLP to a 1-ml HiTrap N-hydroxysuccinimide-activated HP Sepharose column (GE Healthcare Biosciences) or using a 1-ml protein G-Sepharose column (GE Healthcare Biosciences) as indicated.
PAGE and Western blotting.
Polyacrylamide gel electrophoresis (PAGE) was used for determining the apparent molecular mass and estimating VLP homogeneity. Ten microliters of loading buffer and 4 μl of reducing agent (NuPage system; Invitrogen Corp.) were added to 26 μl of sample and heated at 70°C for 10 min prior to electrophoresis. Samples were applied (10 μl per lane) to a precast 4 to 20% polyacrylamide gel and electrophoretically fractionated in a morpholineethanesulfonic acid-SDS buffer system (NuPage; Invitrogen) at 150 V (constant voltage) for 60 min. Protein bands were visualized with either Coomassie brilliant blue (SimplyBlue SafeStain; Invitrogen) or silver staining (SilverQuest; Invitrogen) according to the recommendations of the manufacturer. For Western blot analysis, proteins were electroblotted from the gel onto a nitrocellulose membrane according to the manufacturer's protocols (iBlot system; Invitrogen).
Electron microscopy.
The Electron Microscopy Core Facility of the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) (Mathew P. Daniels, Director) supported the ultrastructure analysis of the HBoV VLP specimens using a transmission electron microscope (JEM1200EX; Jeol, Ltd., Tokyo, Japan) equipped with an AMT XR-60 digital camera (Advanced Microscopy Techniques, Danvers, MA). The VLP samples were diluted, and a small aliquot, e.g., 10 to 20 μl, was pipetted onto paraffin film. An aliquot of each sample (5 μl) was placed onto a carbon-coated 200-mesh copper grid for 1 min and then washed with 10 drops of distilled water. Staining was achieved by adding 5 drops of 2% (wt/vol) uranyl acetate. Excess staining solution was immediately wicked away with blotting paper, and the grids were then air dried. The grids were examined with a transmission electron microscope, and micrographs of randomly selected fields were taken at various magnifications.
VLP-based ELISA.
The optimum conditions for ELISA, including coating concentration of the VLP protein, serum dilution, conjugate dilution, incubation times, temperature, and blocking reagent, were determined in preliminary checkerboard titration experiments. To optimize ELISA, bocavirus-VLP proteins were diluted to 2 mg/ml in 0.2 M carbonate-bicarbonate buffer, pH 9.6 (Sigma-Aldrich Corp., St. Louis, MO), and 100 μl was added to each well of 96-well polystyrene flat-bottom plates (MaxiSorp; Nunc) and incubated overnight at 4°C. After the antigen solution was removed, 200 μl of blocking solution (2% bovine serum albumin in carbonate-bicarbonate buffer [pH 9.6]) was added and incubated overnight at 4°C. Sufficient plates were prepared for human serum tests in advance and stored at 4°C. Human serum and preimmune rabbit serum samples were serially fourfold diluted from 1/100 to 1/409,600, whereas bocavirus rabbit antiserum was serially fourfold diluted from 1/1,600 to 1/6,553,600 in PBS-T (phosphate-buffered saline, 1% bovine serum albumin, 0.05% Tween 20). The plates were incubated at 37°C for 1 h on a platform shaker with the diluted sera and then washed four times with PBS-T. The secondary goat anti-human IgG antibody for the human serum samples (Chemicon Division, Millipore, Corp., Billerica, MA) and goat anti-rabbit IgG (Sigma-Aldrich, St. Louis, MO) were obtained as horseradish peroxidase (HRP) conjugates and used after 1:30,000 dilution in PBS-T. One hundred microliters of the diluted appropriate secondary IgG-HRP was added to each well. The nonadsorbed IgG-HRP was removed by washing with PBS-T. After five washes, 100 μl of 3,3′,5,5′-tetramethylbenzidine liquid substrate (Sigma-Aldrich) was added to each well and incubated at room temperature in the dark for 15 min. Peroxidase cleaves 3,3′,5,5′-tetramethylbenzidine, and by adding 1 N sulfuric acid the mixture (100 μl to each well), the peroxidase reaction was terminated, producing a chromogenic end product that absorbs at 450 nm. An automated plate reader (Spectramax M2 and Softmax Pro software; Molecular Devices) measured the absorption of each well at 450 nm with a reference wavelength set at 570 nm.
Human sera.
Human sera were obtained from healthy blood bank donors at the Warren G. Magnuson Clinical Center of the NIH under a protocol approved by the Internal Review Board of the National Cancer Institute. Patient identifiers were removed, and only age and sex data were accessible.
AAV VLP.
The AAV VLPs used in this study were produced in insect cells using the baculovirus expression system. The AAV cap gene expression constructs were described previously (50). The production and purification of AAV-VLPs using a 5-liter-bag Wave bioreactor were performed for HBoV-VLP production.
Statistical testing.
Chi-squared or Fisher's exact test was performed to determine differences between females and males and as a function of age of the donors (1, 15).
RESULTS
HBoV VLP purification.
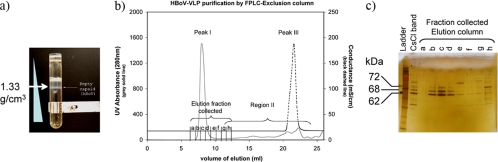
Modifying the capsid protein sequence (Fig. 1) allowed us to produce HBoV capsid proteins using a single BEV in Sf9 cells. In BEV-infected Sf9 cells, the capsid proteins assemble into particles that are physically separable from other cellular components. Using CsCl-isopycnic gradients, an obvious opalescent band appeared approximately midway in the gradient at a density of 1.33 g/cm3 (Fig. 2a). Assuming that the VLP consists of approximately 60 capsid proteins, the molecular mass is therefore approximately 4 MDa, making the VLP much larger than other soluble cellular components, allowing separation by size exclusion chromatography. The VLP fraction elutes from a Sephadex 200 sizing column in the void volume well separated from the smaller cell and baculovirus proteins and CsCl salt, as indicated by the UV (VLP) and conductivity (salt) traces (Fig. 2b). The protein composition of the column fractions was assessed by SDS-PAGE and silver staining (Fig. 2c). Three distinct bands appeared in the VLP fractions, with masses of 72 kDa, 68 kDa, and 62 kDa, corresponding to VP1, VP2, and VP3, respectively. Only two capsid proteins were predicted from the HBoV genomic sequence (2); therefore, the VP2 gel band was excised, and the first eight N-terminal residues were determined by N-terminal sequencing analysis using Edman degradation and mass spectroscopy. Based on protein sequence analysis, the first residue of VP2 is the alanine corresponding to VP1 residue 92.
FIG. 2.
Purification step and partial characterization of the HBoV capsid. (a) Capsid proteins are purified using a CsCl isopycnic gradient. The opalescent band approximately midway in the gradient at a density of 1.33 g/cm3 was collected and purified using an exclusion Superdex 200 sizing-column chromatography system. (b) A UV detector (gray solid line) and conductivity detector (black dashed line) were used to monitor the purification of the proteins. The VLP-containing fraction elutes in the void volume (peak I [gray solid line]), separated from smaller proteins (region II [gray solid line]) and CsCl salt (peak III [black dashed line]). FPLC, fast protein liquid chromatography. (c) PAGE and protein silver staining of fractions a to h from b. Protein composition of the HBoV-VLP was assessed by silver staining of the gel. Three distinct bands appeared in the VLP fractions at 72 kDa, 68 kDa, and 62 kDa, corresponding to VP1, VP2, and VP3, respectively.
VLP physical characterization.
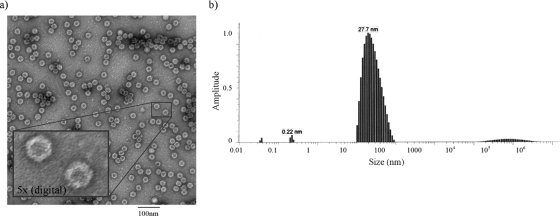
Electron microscopy performed on purified samples confirmed the formation of VLPs (Fig. 3a). Consistent with the Parvoviridae, the coexpressed VP polypeptides assembled into uniformly shaped particles with apparent icosahedral symmetry. According to the electron microscopy analysis, the estimated particle diameter is between 18 and 25 nm. Dynamic light-scattering (DLS) analysis provides a means for measuring nanoparticle sizes in solution and characterizing the state of the VLP in solution (model number 802; Viscotek, Houston, TX). The DLS data indicate that the solution contains a range of particles that average 27.7 nm in diameter. Although the peak is not perfectly symmetrical, the steep leading edge suggests that monodisperse particles are a prominent component in the solution. The more gradual trailing edge suggests that particle dimerization and possibly higher-order interactions exist in solution, at least under these conditions (Fig. 3b).
FIG. 3.
Physical characterization of VLPs. (a) Electron microscopy was performed on purified samples and photographed at a magnification of ×60,000. The VLPs appear as uniformly shaped particles and are consistent with icosahedral symmetry. Estimates of particle size range from 18 to 25 nm by direct measurement. In the insert, the digitally magnified image (about fivefold) reveals fine structure and hexagonal symmetry of the VLP. (b) DLS analysis provided a means for estimating nanoparticle sizes in solution. DLS indicates that the solution contains particles ranging from monodisperse to small aggregates with an average diameter of 27.7 nm.
Polyclonal antibody cross-reactivity.
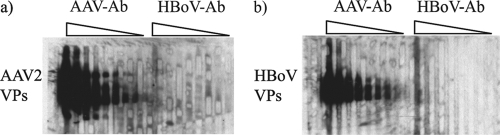
Rabbit immune serum was tested for cross-reactivity to AAV, an unrelated parvovirus. Using single-lane PAGE, AAV or HBoV capsid proteins were transferred onto a membrane and incubated with serially diluted rabbit immune sera in a multilane blotting apparatus. The HBoV and AAV antisera were tested for cross-reactivity to either AAV (Fig. 4a) or HBoV (Fig. 4b) capsid antigens using serum dilutions of 1/1,000 to 1/250,000. The Western blots indicate that the antisera did not react with the heterologous virus capsid antigens. A sequence alignment of HBoV capsid protein and AAV capsid proteins indicates that there are three regions of similarity greater than 66% consisting of 49, 33, and 11 amino acids. The largest region of similarity is located in a unique region of VP1 that overlaps with (or contains) the phospholipase A2 motif (49 to 53 residues). The phospholipase A2 motif is required for infectivity by facilitating endosomal escape (19) and is highly conserved among the Parvoviridae (53). Structural analysis predicts that these residues are located internally (25) and are unlikely to interact with circulating neutralizing Igs. The two other regions are located in VP3, 91 and 471 residues after the VP3 starting codon.
FIG. 4.
Test for polyclonal antibody (Ab) cross-reactivity. A single-well gel loaded with either 5 mg of AAV-VLP (a) or 5 mg of HBoV-VLP (b) was electroblotted onto a nitrocellulose membrane. Using a multilane blotting manifold, 700 μl of twofold-serially-diluted rabbit immune sera (from 1/1,000 to 1/256,000 [lanes 1 to 9 and 10 to 18, respectively]) was added and incubated for 1 h at room temperature. After washing each lane two times with a washing buffer, the membranes were removed from the manifold, washed again two times with larger volumes of washing buffer, and incubated with a secondary HRP anti-rabbit conjugate (1/10,000 dilution) for 1 h. Very low background was detected using this technique.
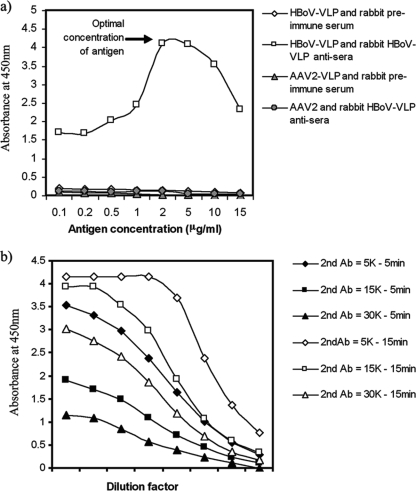
VLP-based ELISA optimization.
To determine the optimal concentration of antigen for the ELISA, serially diluted HBoV-VLP (0.1 to 15 μg/ml) was used to coat the plate. The optimum concentration, defined as the antigen concentration producing the maximum signal in the colorimetric assay using the least amount of antigen, was 2 μg/ml (Fig. 5a). Conditions for detecting anti-HBoV antibodies in human serum samples were established using rabbit immune serum as a surrogate. Microtiter plates were coated with HBoV VLP (2 μg/ml), and using a twofold dilution series of rabbit immune serum (1:100 to 1:12,800), a single concentration of rabbit serum was added to each row of wells. Each dilution of the secondary antibody goat anti-rabbit IgG-HRP (1:5,000, 1:15,000, or 1:30,000) was then added to a column of wells, incubated for either 5 or 15 min, and processed as described in Materials and Methods. The best response over the greatest range of immune serum dilutions (Fig. 5b) without saturation of the color and in a reasonable incubation time was produced using a 1:30,000 dilution of secondary antibody with a 15-min incubation time.
FIG. 5.
Optimization of VLP-based ELISA. (a) We tested a serially diluted HBoV-VLP-coated plate from 0.1 to 15 μg/ml with a 1/1,000-diluted rabbit HBoV-VLP antiserum. The optimum concentration allowing a higher result with rabbit HBoV-VLP antisera was found to be 2 μg/ml. (b) Optimization of the secondary antibody (Ab) concentration. Several conditions were evaluated to determine the optimum concentration of the secondary HRP-conjugated antibody and incubation time for anti-HoBV in rabbit antiserum samples. Microtiter plates coated with HBoV VLP (2 μg/ml) were tested with serially diluted rabbit HBoV-VLP antisera (1/100 to 1/12,800). The data indicate that the best response over the greatest range of immune serum dilutions occurred with 1:30,000-diluted secondary antibody and a 15-min incubation time (open triangles) (see text).
VLP-based ELISA of human samples.
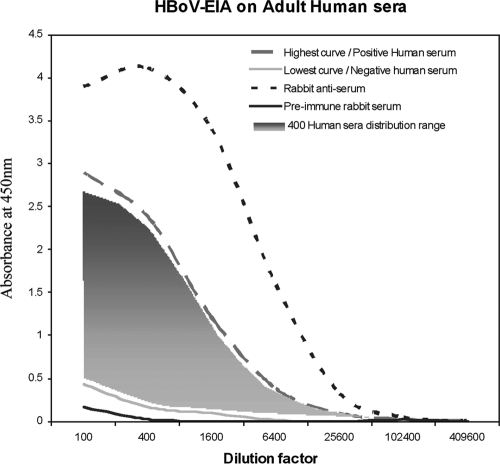
Serum samples from 404 adults were tested for the presence of HBoV-Ig using an ELISA method. The response ranges were uniformly distributed between the highest serum response and the lowest serum response and are presented graphically in Fig. 6. Every clinical sample value fell between values for the rabbit preimmune serum (negative control) and the rabbit immune serum (positive control). From these results, we categorized the higher values obtained with human sera as positive samples and, conversely, the lower values obtained with human sera as negative samples. The test produced typical sigmoid ELISA curves with an end point (optical density value below 0.3) at a 1:1600 dilution for negative sera and an end point at 1:25,600 for positive sera. With these parameters and under these conditions, in a cohort of 404 adults, we found that 63% were serologically positive for HBoV. In Table 1, we categorized the response results by age and sex. The prevalence tended to increase with age and was slightly higher in women. Age groups of men between 18 and 65 years of age had 59% to 65% seropositivity, whereas women between 18 and 65 years of age had 57 to 73% seropositivity.
FIG. 6.
Adult human sera were tested for the presence of HBoV-Ig using an ELISA established as described in the text. The values of 404 serum samples diluted from 1/100 to 1/409,600 are presented. Each sample was tested twice, and the average point is plotted on the graph: the dashed gray line represents the highest human serum response, and the solid gray line represents the lowest human serum response; all other human sera are presented graphically as the gray shaded area. The solid black line represents values of rabbit preimmune serum (negative control), and the dashed black line represents rabbit immune serum (positive control). EIA, enzyme immunoassay.
TABLE 1.
Categorization of ELISA results by age and sexa
| Age range | Men
|
Women
|
||||||
|---|---|---|---|---|---|---|---|---|
| Avg age (yr) | Total no. of patients | No. of positive patients | % Positive patients | Avg age (yr) | Total no. of patients | No. of positive patients | % Positive patients | |
| Total | 31 | 246 | 147 | 59.76 | 30.79 | 158 | 106 | 67.09 |
| 18-25 | 22 | 92 | 55 | 59.78 | 22.25 | 71 | 40 | 56.34 |
| 26-35 | 30.21 | 82 | 47 | 57.32 | 29.94 | 48 | 36 | 75 |
| 36-45 | 39.26 | 55 | 34 | 61.82 | 40.09 | 27 | 22 | 81.48 |
| 46-65 | 53.91 | 17 | 11 | 64.71 | 51.75 | 11 | 8 | 72.73 |
Curves with an end point (optical density value below 0.3) at a dilution 1:1,600 are scored as negative sera, whereas curves with an end point at a dilution of 1:25,600 are scored as positive sera. Between 59 and 81% of ELISAs were found to be positive, increasing with age and being slightly higher in women.
ELISA using purified polyclonal antibody.
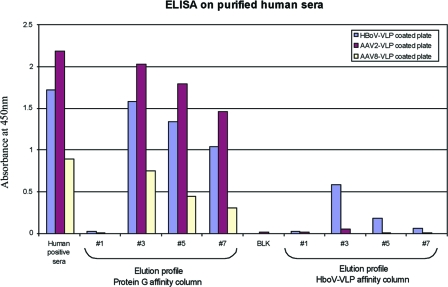
To confirm that specific HBoV antibodies were present in human sera, we fractionated total Ig antibodies or specific HBoV-Ig antibodies using a protein G column or an HBoV-VLP affinity column, respectively. The eluted Ig fractions were tested for the presence of HBoV-Ig, AAV2-Ig, and AAV8-Ig using plates coated with HBoV-VLP, AAV2-VLP, or AAV8-VLP. In Fig. 7, we plotted the ELISA results for the crude human serum, the elution profiles of protein G Ig purification, and the elution profiles of the HBoV-VLP affinity column for the three different viral-protein-coated plates. Crude human serum was positive for HBoV, AAV2, and AAV8. After protein G purification, the samples remained positive for the three different viruses. However, after purification with the HBoV-VLP affinity column, only the HBoV-VLP-coated plate gave a strong signal compared to the signals of the AAV2- and AAV8-coated plates. These results confirm that the presence of specific antibody against HBoV in human sera did not result from artifactual reactivity or cross-reactivity with other nonspecific antibodies.
FIG. 7.
ELISA of affinity column-purified polyclonal antibody. The ELISA values of 1/100-diluted serum samples processed as described in the text are shown. Unpurified human serum reacts positively by ELISA with HBoV-VLP-, AAV2 VLP-, or AAV8 VLP-coated plates. The same human serum purified with a protein G affinity column produces results similar to those of the total serum. However, serum fractions eluted from an HBoV VLP affinity column showed reduced reactivity in the AAV2 and AAV8 ELISA but retained reactivity in the HBoV ELISA.
DISCUSSION
VLP of numerous virus types have been produced using the BEV-insect cell system for structural studies (9, 48) or vaccines (20, 28, 42). In addition, recombinant AAV produced in insect cells has been used for preclinical and clinical studies (43, 47). Regulation of the amounts of AAV structural and nonstructural proteins transcriptionally or posttranscriptionally was a critically important development for producing capsids with a defined stoichiometry of structural proteins (11, 50). Therefore, the regulation of posttranscriptional protein levels was adapted for HBoV VLP production. Members of the Dependovirinae naturally regulate the stoichiometry of the capsid proteins by alternative splice acceptors and translational initiation at non-AUG codons (49). The relatively inefficient translational initiation of VP2 at a non-AUG codon caused by rRNA readthrough produces high levels of VP3 translation from the initiating AUG codon. We adapted this strategy for HBoV VLP production by substituting an ACG for the predicted VP1 AUG starting codon. Although there are no consensus Kozak elements bordering this presumptive initiation codon, it is the upstream AUG codon proximal to the Parvoviridae invariant phospholipase-like domain. Unexpectedly, we found that in addition to VP1 and the major coat protein, a third intermediate-size capsid protein was present in the VLP. Protein sequence determination of the first 8 amino acids allowed us to characterize the amino-terminal residue of this protein. This capsid protein, VP2, starts at codon 91 of VP1, which codes for alanine. The preceding codon is a GTG, which in a Kozak context has the potential to initiate protein translation as follows, where “seq” is sequence and “Prot” is protein:

However, because no mammalian tissue culture system for HBoV propagation is available and there are no sources of wild-type HBoV, the possibility that VP2 expression results from idiosyncratic baculovirus expression remains. Thus, the strategy for regulating the expression levels of the capsid proteins remains unresolved. Alternative splicing is a common method used by cells to produce two or more proteins from the same primary transcript. However, we were not able to identify a splicing region between VP1 and VP2, nor were sequence-specific protease cleavage sites identified at the site of the new capsid protein, limiting the possibility of posttranslational modification. Typically, AAVs use a noncanonical starting codon to produce several proteins in the same reading frame. In the case of HBoV, the same strategy is possibly used to produce two or three proteins derived from a common open reading frame. A weak AUG VP1 initiation codon lacking both the upstream and downstream Kozak flanking motifs followed by the putative VP2 GUG initiation codon in a Kozak context and, finally, the VP3 AUG initiation codon might be used. If there is one mature transcript derived from the capsid protein gene, this strategy may result in the production of three capsid proteins at a ratio of 1:(1 to 5):10.
The BEV constructs allowed us to produce and purify a stable HBoV VLP composed of VP1, VP2, and VP3 for use as an antigen and ELISA substrate. Using this VLP, we conducted a seroepidemiology study of a sample of 404 adults living in the United States. We observed a broad distribution of randomly distributed serology results. As observed in others studies using VP2 or VP1 HBoV ELISA (14, 23, 29), we found a high level of seropositivity: 63% of the human samples that we tested were positive for HBoV antibody, suggesting prior exposure, e.g., infection, with HBoV. The small sample size for the female cohort, ages 46 to 65 years, is not statistically representative. However, the seropositivity prevalence tended to increase with age and was statistically slightly higher in women. Because we have no information about the medical history of the serum donors, it is impossible to correlate the serology results with disease. However, the development of an ELISA using an HBoV VLP should enable a prospective study to develop correlations between HBoV infection and human disease or associations with other viral infections.
Acknowledgments
We acknowledge the professional skills and advice of Mathew P. Daniels (Electron Microscopy Core Facility, National Heart, Lung, and Blood Institute, National Institutes of Health) regarding electron microscopy performed in this study.
This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute and the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Abramowitz, M. 1965. Handbook of mathematical functions with formulas, graphs, and mathematical tables. Dover, New York, NY.
- 2.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 10212891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, L. J. 1987. Role of parvovirus B19 in human disease. Pediatr. Infect. Dis. J. 6711-718. [DOI] [PubMed] [Google Scholar]
- 4.Arden, K. E., P. McErlean, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2006. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J. Med. Virol. 781232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold, J. C., K. K. Singh, S. A. Spector, and M. H. Sawyer. 2006. Human bocavirus: prevalence and clinical spectrum at a children's hospital. Clin. Infect. Dis. 43283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastien, N., N. Chui, J. L. Robinson, B. E. Lee, K. Dust, L. Hart, and Y. Li. 2007. Detection of human bocavirus in Canadian children in a 1-year study. J. Clin. Microbiol. 45610-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloom, M. E., H. Kanno, S. Mori, and J. B. Wolfinbarger. 1994. Aleutian mink disease: puzzles and paradigms. Infect. Agents Dis. 3279-301. [PubMed] [Google Scholar]
- 8.Calvo, C., M. L. Garcia-Garcia, F. Pozo, O. Carvajal, P. Perez-Brena, and I. Casas. 2008. Clinical characteristics of human bocavirus infections compared with other respiratory viruses in Spanish children. Pediatr. Infect. Dis. J. 27677-680. [DOI] [PubMed] [Google Scholar]
- 9.Canady, M. A., H. Tsuruta, and J. E. Johnson. 2001. Analysis of rapid, large-scale protein quaternary structural changes: time-resolved X-ray solution scattering of Nudaurelia capensis omega virus (NomegaV) maturation. J. Mol. Biol. 311803-814. [DOI] [PubMed] [Google Scholar]
- 10.Canducci, F., M. Debiaggi, M. Sampaolo, M. C. Marinozzi, S. Berre, C. Terulla, G. Gargantini, P. Cambieri, E. Romero, and M. Clementi. 2008. Two-year prospective study of single infections and co-infections by respiratory syncytial virus and viruses identified recently in infants with acute respiratory disease. J. Med. Virol. 80716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, H. 2008. Intron splicing-mediated expression of AAV Rep. and Cap genes and production of AAV vectors in insect cells. Mol. Ther. 16924-930. [DOI] [PubMed] [Google Scholar]
- 12.Choi, E. H., H. J. Lee, S. J. Kim, B. W. Eun, N. H. Kim, J. A. Lee, J. H. Lee, E. K. Song, S. H. Kim, J. Y. Park, and J. Y. Sung. 2006. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000-2005. Clin. Infect. Dis. 43585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi, J. H., Y. S. Chung, K. S. Kim, W. J. Lee, I. Y. Chung, H. B. Oh, and C. Kang. 2008. Development of real-time PCR assays for detection and quantification of human bocavirus. J. Clin. Virol. 42249-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endo, R., N. Ishiguro, H. Kikuta, S. Teramoto, R. Shirkoohi, X. Ma, T. Ebihara, H. Ishiko, and T. Ariga. 2007. Seroepidemiology of human bocavirus in Hokkaido Prefecture, Japan. J. Clin. Microbiol. 453218-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher, R. A. 1922. On the interpretation of χ2 from contingency tables, and the calculation of P. J. R. Stat. Soc. 8587-94. [Google Scholar]
- 16.Freymuth, F., A. Vabret, J. Dina, J. Petitjean, and S. Gouarin. 2007. Techniques used for the diagnostic of upper and lower respiratory tract viral infections. Rev. Prat. 571876-1882. (In French.) [PubMed] [Google Scholar]
- 17.Fry, A. M., X. Lu, M. Chittaganpitch, T. Peret, J. Fischer, S. F. Dowell, L. J. Anderson, D. Erdman, and S. J. Olsen. 2007. Human bocavirus: a novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. J. Infect. Dis. 1951038-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 19.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83973-978. [DOI] [PubMed] [Google Scholar]
- 20.Halsey, R. J., F. L. Tanzer, A. Meyers, S. Pillay, A. Lynch, E. Shephard, A. L. Williamson, and E. P. Rybicki. 2008. Chimaeric HIV-1 subtype C Gag molecules with large in-frame C-terminal polypeptide fusions form virus-like particles. Virus Res. 133259-268. [DOI] [PubMed] [Google Scholar]
- 21.Hoggan, M. D., N. R. Blacklow, and W. P. Rowe. 1966. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proc. Natl. Acad. Sci. USA 551467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacques, J., H. Moret, F. Renois, N. Leveque, J. Motte, and L. Andreoletti. 2008. Human bocavirus quantitative DNA detection in French children hospitalized for acute bronchiolitis. J. Clin. Virol. 43142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahn, J. S., D. Kesebir, S. F. Cotmore, A. D'Abramo, Jr., C. Cosby, C. Weibel, and P. Tattersall. 2008. Seroepidemiology of human bocavirus defined using recombinant virus-like particles. J. Infect. Dis. 19841-50. [DOI] [PubMed] [Google Scholar]
- 24.Kesebir, D., M. Vazquez, C. Weibel, E. D. Shapiro, D. Ferguson, M. L. Landry, and J. S. Kahn. 2006. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J. Infect. Dis. 1941276-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kronenberg, S., J. A. Kleinschmidt, and B. Bottcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lacaze, T., Y. de Prost, J. J. Lefrere, and A. M. Courouce. 1987. Parvovirus B19 and the fifth disease. Arch. Fr. Pediatr. 44619-620. (In French.) [PubMed] [Google Scholar]
- 27.Lau, S. K., C. C. Yip, T. L. Que, R. A. Lee, R. K. Au-Yeung, B. Zhou, L. Y. So, Y. L. Lau, K. H. Chan, P. C. Woo, and K. Y. Yuen. 2007. Clinical and molecular epidemiology of human bocavirus in respiratory and fecal samples from children in Hong Kong. J. Infect. Dis. 196986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao, S., S. Wang, L. Xu, D. Deng, Q. Xu, W. Wang, T. Zhu, X. Bai, J. Zhou, G. Xu, Y. Lu, L. Meng, and D. Ma. 2008. Production and verification of human papillomavirus type 18 vaccine in vitro. Oncol. Rep. 20211-217. [PubMed] [Google Scholar]
- 29.Lin, F., W. Guan, F. Cheng, N. Yang, D. Pintel, and J. Qiu. 2008. ELISAs using human bocavirus VP2 virus-like particles for detection of antibodies against HBoV. J. Virol. Methods 149110-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, F., A. Zeng, N. Yang, H. Lin, E. Yang, S. Wang, D. Pintel, and J. Qiu. 2007. Quantification of human bocavirus in lower respiratory tract infections in China. Infect. Agents Cancer 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindner, J., L. Karalar, S. Zehentmeier, A. Plentz, H. Pfister, W. Struff, M. Kertai, H. Segerer, and S. Modrow. 2008. Humoral immune response against human bocavirus VP2 virus-like particles. Viral Immunol. 21443-449. [DOI] [PubMed] [Google Scholar]
- 32.Lu, X., M. Chittaganpitch, S. J. Olsen, I. M. Mackay, T. P. Sloots, A. M. Fry, and D. D. Erdman. 2006. Real-time PCR assays for detection of bocavirus in human specimens. J. Clin. Microbiol. 443231-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma, X., R. Endo, N. Ishiguro, T. Ebihara, H. Ishiko, T. Ariga, and H. Kikuta. 2006. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J. Clin. Microbiol. 441132-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackay, I. M. 2007. Human bocavirus: multisystem detection raises questions about infection. J. Infect. Dis. 196968-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning, A., V. Russell, K. Eastick, G. H. Leadbetter, N. Hallam, K. Templeton, and P. Simmonds. 2006. Epidemiological profile and clinical associations of human bocavirus and other human parvoviruses. J. Infect. Dis. 1941283-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naghipour, M., L. E. Cuevas, T. Bakhshinejad, W. Dove, and C. A. Hart. 2007. Human bocavirus in Iranian children with acute respiratory infections. J. Med. Virol. 79539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neske, F., K. Blessing, F. Tollmann, J. Schubert, A. Rethwilm, H. W. Kreth, and B. Weissbrich. 2007. Real-time PCR for diagnosis of human bocavirus infections and phylogenetic analysis. J. Clin. Microbiol. 452116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parrish, C. R. 1995. Pathogenesis of feline panleukopenia virus and canine parvovirus. Baillières Clin. Haematol. 857-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollock, R. V., and M. J. Coyne. 1993. Canine parvovirus. Vet. Clin. N. Am. Small Anim. Pract. 23555-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi, Z. Y., X. W. Qu, W. P. Liu, Z. P. Xie, H. C. Gao, L. S. Zheng, Z. Z. Kuang, J. P. Yu, and Z. J. Duan. 2007. Genome cloning and phylogenetic analysis of human bocavirus capsid gene. Bing Du Xue Bao 23447-453. (In Chinese.) [PubMed] [Google Scholar]
- 41.Qu, X. W., Z. J. Duan, Z. Y. Qi, Z. P. Xie, H. C. Gao, W. P. Liu, C. P. Huang, F. W. Peng, L. S. Zheng, and Y. D. Hou. 2007. Human bocavirus infection, People's Republic of China. Emerg. Infect. Dis. 13165-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose, R. C., R. C. Reichman, and W. Bonnez. 1994. Human papillomavirus (HPV) type 11 recombinant virus-like particles induce the formation of neutralizing antibodies and detect HPV-specific antibodies in human sera. J. Gen. Virol. 752075-2079. [DOI] [PubMed] [Google Scholar]
- 43.Ross, C. J., J. Twisk, A. C. Bakker, F. Miao, D. Verbart, J. Rip, T. Godbey, P. Dijkhuizen, W. T. Hermens, J. J. Kastelein, J. A. Kuivenhoven, J. M. Meulenberg, and M. R. Hayden. 2006. Correction of feline lipoprotein lipase deficiency with adeno-associated virus serotype 1-mediated gene transfer of the lipoprotein lipase S447X beneficial mutation. Hum. Gene Ther. 17487-499. [DOI] [PubMed] [Google Scholar]
- 44.Ruffing, M., H. Zentgraf, and J. A. Kleinschmidt. 1992. Assembly of virus-like particles by recombinant structural proteins of adeno-associated virus type 2 in insect cells. J. Virol. 666922-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schildgen, O., A. Muller, T. Allander, I. M. Mackay, S. Volz, B. Kupfer, and A. Simon. 2008. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin. Microbiol. Rev. 21291-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott, F. W., C. K. Csiza, and J. H. Gillespie. 1970. Feline viruses. IV. Isolation and characterization of feline panleukopenia virus in tissue culture and comparison of cytopathogenicity with feline picornavirus, herpesvirus, and reovirus. Cornell Vet. 60165-182. [PubMed] [Google Scholar]
- 47.Stroes, E. S., M. C. Nierman, J. J. Meulenberg, R. Franssen, J. Twisk, C. P. Henny, M. M. Maas, A. H. Zwinderman, C. Ross, E. Aronica, K. A. High, M. M. Levi, M. R. Hayden, J. J. Kastelein, and J. A. Kuivenhoven. 2008. Intramuscular administration of AAV1-lipoprotein lipase S447X lowers triglycerides in lipoprotein lipase-deficient patients. Arterioscler. Thromb. Vasc. Biol. 282303-2304. [DOI] [PubMed] [Google Scholar]
- 48.Taylor, D. J., J. A. Speir, V. Reddy, G. Cingolani, F. M. Pringle, L. A. Ball, and J. E. Johnson. 2006. Preliminary X-ray characterization of authentic providence virus and attempts to express its coat protein gene in recombinant baculovirus. Arch. Virol. 151155-165. [DOI] [PubMed] [Google Scholar]
- 49.Trempe, J. P., and B. J. Carter. 1988. Alternate mRNA splicing is required for synthesis of adeno-associated virus VP1 capsid protein. J. Virol. 623356-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urabe, M., C. Ding, and R. M. Kotin. 2002. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 131935-1943. [DOI] [PubMed] [Google Scholar]
- 51.Weissbrich, B., F. Neske, J. Schubert, F. Tollmann, K. Blath, K. Blessing, and H. W. Kreth. 2006. Frequent detection of bocavirus DNA in German children with respiratory tract infections. BMC Infect. Dis. 6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young, N. 1988. Hematologic and hematopoietic consequences of B19 parvovirus infection. Semin. Hematol. 25159-172. [PubMed] [Google Scholar]
- 53.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1291-302. [DOI] [PubMed] [Google Scholar]