Abstract
Phylogenetic analyses of sequences generated from portions of three genes coding for the proteins enolase (enoA), β-tubulin (benA), and calmodulin (calM) of a large number of isolates within the section Terrei, genus Aspergillus, revealed the presence of a new cryptic species within this section, Aspergillus alabamensis. Most members of this new cryptic species were recovered as colonizing isolates from immunocompetent patient populations, had decreased in vitro susceptibilities to the antifungal drug amphotericin B, and were morphologically similar to but genetically distinct from Aspergillus terreus isolates.
Invasive infections caused by Aspergillus terreus are often disseminated with increased lethality compared with infections caused by other Aspergillus species and tend to be resistant to treatment with the antifungal drug amphotericin B (6, 14, 17). Despite the clinical significance of this organism, little is known about the epidemiology, genetic diversity, and population structure of A. terreus.
Historically, A. terreus has been identified in the laboratory by conventional methods such as colony morphology and microscopic characteristics. Such morphological studies have placed A. terreus as a single homogenous species within the section Terrei along with two other varieties, A. terreus var. africanus and A. terreus var. aureus (11). Recent studies have shown that morphological characteristics may not be reliable for distinguishing Aspergillus species, as inferred from the demonstration of multiple cryptic species within the section Fumigati by molecular phylogenetic methods (3-5, 13, 18).
In the past, molecular methods largely based on randomly amplified polymorphic DNA-PCR-based assays have shown that A. terreus isolates can have great strain diversity (1, 8, 16). One recent genotyping study of several A. terreus clinical isolates recovered from two different medical centers using this method concluded that nosocomial acquisition of A. terreus infections was highly unlikely given the great genetic diversity observed (7). Another study demonstrated that comparative sequence analyses of the D1 and D2 regions had limited utility to study relationships within the section Terrei, while the internal transcribed spacer regions were useful since there was more nucleotide diversity in this region (16). However, the authors of this study could not resolve species within the section Terrei using these molecular approaches.
In the present study, we have developed a multilocus sequence approach employing three protein-coding regions to study species diversity of the section Terrei using a large panel of isolates from both clinical and environmental origins recovered from various parts of the world. The studies outlined below demonstrate the presence of a new, clinically relevant species, Aspergillus alabamensis, and clarify the taxonomic position of the A. terreus variant A. terreus var. aureus.
MATERIALS AND METHODS
Fungal isolates.
A total of 94 clinical and environmental A. terreus isolates were analyzed in this study, including 30 isolates from University of Alabama at Birmingham (UAB); 23 isolates from the Department of Hygiene, Microbiology and Social Medicine, Medical University of Innsbruck, Innsbruck, Austria; 23 isolates from the CBS Fungal Biodiversity Centre, Utrecht, The Netherlands; 17 isolates from the Center for Microbial Biotechnology, Biocentrum-DTU, Technical University of Denmark, Lyngby; and one isolate from the National Center for Agricultural Utilization Research, U.S. Department of Agriculture, Peoria, IL. Apart from this, two isolates of Aspergillus allahabadii, one isolate each of Aspergillus niveus var. indicus and Fennellia flavipes, and three isolates of A. niveus were also received from the National Center for Agricultural Utilization Research and included in this study. All fungal isolates were well separated in time and location of origin, with the years of recovery ranging from 1926 to 2006, and included isolates from Europe, South America, Asia, and North America (Table 1). Isolates were stored frozen and were subcultured on Sabouraud's dextrose agar plates before DNA isolation.
TABLE 1.
Source and origin of A. terreus isolates used in the study
| Isolate no. | Species | Geographic origin | Source | GenBank identification no. (calM, enoA, benA) | Yr of isolation |
|---|---|---|---|---|---|
| IBT26915 | A. terreus | Panama | Capybara | EU147503, EU147596, EU147689 | Unknown |
| IBT24859 | A. terreus | Slovenia | Saltern | EU147504, EU147597, EU147690 | Unknown |
| IBT13089 | A. terreus | Unknown | Unknown | EU147505, EU147598, EU147691 | Unknown |
| IBT26385 | A. terreus | Unknown | Unknown | EU147506, EU147599, EU147692 | Unknown |
| IBT21125 | A. terreus | Unknown | Unknown | EU147507, EU147600, EU147693 | Unknown |
| IBT12713 | A. terreus | New Mexico | Kangaroo rat | EU147508, EU147601, EU147694 | 1989 |
| IBT6450 | A. terreus | Unknown | Unknown | EU147509, EU147602, EU147695 | Unknown |
| IBT20944 | A. terreus | Unknown | Unknown | EU147510, EU147603, EU147696 | Unknown |
| IBT23544 | A. terreus | Unknown | Hay | EU147511, EU147604, EU147697 | Unknown |
| IBT14590 | A. terreus | Merck strain | Unknown | EU147512, EU147605, EU147698 | Unknown |
| IBT6271 | A. terreus | Unknown | Unknown | EU147513, EU147606, EU147699 | Unknown |
| IBT22563 | A. alabamensisa | Unknown | Unknown | EU147514, EU147607, EU147700 | Unknown |
| IBT26384 (NRRL274) | A. terreus | Brazil | Human ear | EU147515, EU147608, EU147701 | Unknown |
| IBT16744 | A. terreus | Galapagos Islands | Environment | EU147516, EU147609, EU147702 | Unknown |
| IBT16745 | A. terreus | Galapagos Islands | Environment | EU147517, EU147610, EU147703 | Unknown |
| IBT15722 | A. terreus | Unknown | Unknown | EU147518, EU147611, EU147704 | Unknown |
| IBT13121 | A. terreus | Japan | Soil | EU147519, EU147612, EU147705 | Unknown |
| CBS383.75 | A. terreus | India | Soil | EU147520, EU147613, EU147706 | 1975 |
| CBS118.27 | A. terreus | Unknown | Unknown | EU147521, EU147614, EU147707 | 1927 |
| CBS601.65 (NRRL255) | A. terreus (type) | Connecticut | Soil | EU147522, EU147615, EU147708 | 1965 |
| CBS594.65 (NRRL680) | A. terreus | Unknown | Soil | EU147523, EU147616, EU147709 | 1965 |
| CBS106.25 | A. terreus | Unknown | Unknown | EU147524, EU147617, EU147710 | 1920 |
| CBS134.60 | A. terreus | Unknown | Cotton (Gossypium) | EU147525, EU147618, EU147711 | 1926 |
| CBS125.38 | A. terreus | New Zealand | Unknown | EU147526, EU147619, EU147712 | 1938 |
| CBS8G5 | A. terreus | The Netherlands | White soy beans | EU147527, EU147620, EU147713 | 2005 |
| CBS17A1 | A. terreus | The Netherlands | Laboratory medium | EU147528, EU147621, EU147714 | 2006 |
| CBS15F8 | A. alabamensisa | Argentina | Soil | EU147529, EU147622, EU147715 | 2006 |
| CBS15F9 | A. alabamensisa | Argentina | Soil | EU147530, EU147623, EU147716 | 2006 |
| CBS503.65 (NRRL1923) | A. aureoterreusb | Texas | Soil | EU147531, EU147624, EU147717 | Unknown |
| CBS8G3 | A. terreus | The Netherlands | Blended almond pits | EU147532, EU147625, EU147718 | 2005 |
| CBS130.55 (NRRL2399) | A. terreus var. africanus (type) | Tafo, Ghana | Soil | EU147533, EU147626, EU147719 | 1955 |
| 1769-05 | A. terreus | India | Clinical isolate | EU147534, EU147627, EU147720 | 2005 |
| 1686-05 | A. terreus | India | Keratitis | EU147535, EU147628, EU147721 | 2005 |
| 1796-05 | A. terreus | India | Keratitis | EU147536, EU147629, EU147722 | 2005 |
| CBS19F3 | A. terreus | India | Clinical isolate | EU147537, EU147630, EU147723 | 2005 |
| CBS24A3 | A. terreus | The Netherlands | Sucrose tank | EU147538, EU147631, EU147724 | 2006 |
| CBS24A4 | A. terreus | The Netherlands | Sugar silo door | EU147539, EU147632, EU147725 | Unknown |
| CBS2B7 | A. terreus | The Netherlands | Unknown | EU147540, EU147633, EU147726 | 2005 |
| CBS469.81 | A. terreus | Thailand | Cardiac valve | EU147541, EU147634, EU147727 | 1981 |
| CBS6I9 | A. terreus | The Netherlands | Soil | EU147542, EU147635, EU147728 | Unknown |
| P4 | A. terreus | Austria | Tracheal secretions | EU147543, EU147636, EU147729 | 1996 |
| P12 | A. terreus | Austria | Lung | EU147544, EU147637, EU147730 | 1997 |
| P13 | A. terreus | Austria | Brain | EU147545, EU147638, EU147731 | 1997 |
| P14 | A. terreus | Austria | BALc | EU147546, EU147639, EU147732 | 1999 |
| P16 | A. terreus | Austria | BAL | EU147547, EU147640, EU147733 | 1996 |
| P22 | A. terreus | Austria | BAL | EU147548, EU147641, EU147734 | 2001 |
| E5 | A. terreus | Austria | Air, hospital | EU147549, EU147642, EU147735 | 2004 |
| E7 | A. terreus | Austria | Air, hospital | EU147550, EU147643, EU147736 | 2004 |
| E8 | A. terreus | Austria | Air, hospital | EU147551, EU147644, EU147737 | 2005 |
| E9 | A. terreus | Austria | Air, hospital | EU147552, EU147645, EU147738 | 2005 |
| P7 | A. terreus | Austria | BAL | EU147553, EU147646, EU147739 | 2000 |
| P9 | A. terreus | Austria | Tracheal secretions | EU147554, EU147647, EU147740 | 1999 |
| P10 | A. terreus | Austria | Lung | EU147555, EU147648, EU147741 | 2001 |
| P11 | A. terreus | Austria | Sputum | EU147556, EU147649, EU147742 | 2005 |
| P15 | A. terreus | Austria | Lung | EU147557, EU147650, EU147743 | 2002 |
| P20 | A. terreus | Austria | Tracheal secretions | EU147558, EU147651, EU147744 | 2003 |
| P23 | A. terreus | Austria | Sputum | EU147559, EU147652, EU147745 | 2003 |
| P26 | A. terreus | Austria | Lung | EU147560, EU147653, EU147746 | 2001 |
| P29 | A. terreus | Austria | BAL | EU147561, EU147654, EU147747 | 2004 |
| P30 | A. terreus | Austria | Sputum | EU147562, EU147655, EU147748 | 2005 |
| P32 | A. terreus | Austria | Lung | EU147563, EU147656, EU147749 | 2006 |
| P33 | A. terreus | Austria | Brain | EU147564, EU147657, EU147750 | 2005 |
| P38 | A. terreus | Austria | Lung | EU147565, EU147658, EU147751 | 2005 |
| UAB1 | A. alabamensisa | Alabama | Tracheal aspirate | EU147566, EU147659, EU147752 | 1996 |
| UAB2 | A. terreus | Alabama | Sputum | EU147567, EU147660, EU147753 | 1997 |
| UAB3 | A. terreus | Alabama | Bronchial wash | EU147568, EU147661, EU147754 | 1997 |
| UAB4 | A. terreus | Alabama | Bronchial wash | EU147569, EU147662, EU147755 | 1998 |
| UAB5 | A. terreus | Alabama | Bronchial wash | EU147570, EU147663, EU147756 | 1998 |
| UAB6 | A. terreus | Alabama | Sputum | EU147571, EU147664, EU147757 | 1999 |
| UAB7 | A. terreus | Alabama | Bronchial wash | EU147572, EU147665, EU147758 | 1999 |
| UAB8 | A. terreus | Alabama | Thyroid | EU147573, EU147666, EU147759 | 1999 |
| UAB9 | A. terreus | Alabama | Bronchial wash | EU147574, EU147667, EU147760 | 1999 |
| UAB10 | A. terreus | Alabama | BAL | EU147575, EU147668, EU147761 | 1999 |
| UAB11 | A. terreus | Alabama | Sputum | EU147576, EU147669, EU147762 | 2000 |
| UAB12 | A. terreus | Alabama | Sputum | EU147577, EU147670, EU147763 | 2000 |
| UAB13 | A. alabamensisa | Alabama | Sputum | EU147578, EU147671, EU147764 | 2000 |
| UAB15 | A. alabamensisa | Alabama | Sputum | EU147579, EU147672, EU147765 | 2000 |
| UAB17 | A. terreus | Alabama | Catheter tip | EU147580, EU147673, EU147766 | 2000 |
| UAB18 | A. alabamensisa | Alabama | Sputum | EU147581, EU147674, EU147767 | 2000 |
| UAB19 | A. terreus | Alabama | BAL | EU147582, EU147675, EU147768 | 2000 |
| UAB20 | A. alabamensisa | Alabama | Wound | EU147583, EU147676, EU147769 | 2000 |
| UAB21 | A. terreus | Alabama | Sputum | EU147584, EU147677, EU147770 | 2000 |
| UAB22 | A. alabamensisa | Alabama | Tracheal aspirate | EU147585, EU147678, EU147771 | 2000 |
| UAB23 | A. alabamensisa | Alabama | Sputum | EU147586, EU147679, EU147772 | 2000 |
| UAB26 | A. terreus | Alabama | Sputum | EU147587, EU147680, EU147773 | 2000 |
| UAB28 | A. alabamensisa | Alabama | BAL | EU147588, EU147681, EU147774 | 2000 |
| UAB30 | A. alabamensisa | Alabama | Sputum | EU147589, EU147682, EU147775 | 2001 |
| UAB31 | A. terreus | Alabama | BAL | EU147590, EU147683, EU147776 | 2001 |
| UAB32 | A. terreus | Alabama | Fingernail | EU147591, EU147684, EU147777 | 2001 |
| UAB33 | A. alabamensisa | Alabama | Foot tissue | EU147592, EU147685, EU147778 | 2001 |
| UAB34 | A. terreus | Alabama | Sputum | EU147593, EU147686, EU147779 | 2001 |
| UAB37 | A. terreus | Alabama | Tracheal aspirate | EU147594, EU147687, EU147780 | 2001 |
| UAB38 | A. alabamensisa | Alabama | Left ear | EU147595, EU147688, EU147781 | 2001 |
| NRRL4609 | A. terreus var. africanus | Panama | Soil | NAd | Unknown |
| NRRL515 (CBS114.33) | A. niveus | Unknown | Unknown | NA | 1933 |
| NRRL4101 | A. allahabadii | San Salvador | Soil | NA | Unknown |
| NRRL4539 | A. allahabadii | India | Soil | NA | Unknown |
| NRRL4751 | A. niveus | Unknown | Unknown | NA | Unknown |
| NRRL5505 | A. niveus | Unknown | Unknown | NA | Unknown |
| NRRL6134 (CBS444.75) | A. niveus var. indicus | Maharashtra, India | Soil | NA | 1975 |
| NRRL5504 | Fennellia flavipes | Unknown | Unknown | NA | Unknown |
Isolates originally identified by morphology as A. terreus, currently assigned to the new species A. alabamensis based on phylogenetic analyses described in the text.
Originally identified as A. terreus var. aureus.
BAL, bronchoalveolar lavage.
NA, not applicable.
Genomic DNA isolation, PCR amplification, and sequencing.
For genomic DNA isolation, fungi were grown in liquid broth for 48 h, after which the fungal material was disrupted using an Omni-Mixer (Omni International, Warrenton, VA) in the presence of buffer ATL (Qiagen, Valencia, CA) and 55 μl proteinase K; the sonicated material was incubated at 55°C for an hour in a water bath with frequent vortexing. Fungal DNA was isolated using the DNeasy blood and tissue kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations.
For the multilocus analysis, genes encoding enolase (enoA) and β-tubulin (benA) were identified using the publicly available A. terreus genome database (A. terreus Sequencing Project, Broad Institute of Harvard and MIT; http://www.broad.mit.edu). Primers were designed using the Genefisher program to yield a product size of about 300 bp for enoA (enoA F, 5′ CCGTCTACGACTCTCGCGGTA; and enoA R, 5′ TGAGGAACTCGTCAACCTTGGA) and a product size of 400 bp for benA (benA F, 5′ GGGGATAGGATGTTTTGTGACA; and benA R, 5′ GGTCAACGAGGACGGCACGA). For calmodulin (calM), previously described degenerate primers CF1 F (5′ GCCGACTCTTTGACYGARGAR) and CF4 R (5′ TTTYTGCATCATRAGYTGGAC), predicted to yield a 700-bp product, were used (9). PCR amplification was performed with 1 μl of genomic DNA as the template in a final reaction volume of 25 μl consisting of PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl); 0.2 mM each of dATP, dGTP, dCTP, and dTTP; 1.2 mM MgSO4; 0.2 pmol of primers; 1 U of Pfx DNA polymerase (Invitrogen-BRL, Life Technologies, Carlsbad, CA); and 1× PCR enhancer (Invitrogen). Amplification was performed in a GeneAmp PCR system 9700 thermocycler (PE-Applied Biosystems) after initial denaturation at 94°C for 5 min followed by 35 cycles at 94°C for 15 s, 55°C for 30 s, and 68°C for 30 s, and the last cycle was followed by a final extension at 68°C for 2 min.
PCR products were visualized in a 1.2% agarose gel using ethidium bromide and purified using the ExoSAP-IT enzyme system (USB, Cleveland, OH). For sequencing, 1 μl of the purified PCR amplicon was added to the sequencing mixture containing 4 μl Big Dye, 1.6 μl of 1 μM primer (same as the respective PCR primers), and 3.4 μl water. The sequence cycle was 96°C for 5 s, followed by 30 cycles at 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Both strands were sequenced in an ABI 3730 DNA sequencer, and the resultant nucleotide sequences were edited with the Sequencher version 4.7 software (Genecodes, Inc., Ann Arbor, MI).
Phylogenetic analysis.
The number of polymorphic sites, number of genotypes, and parsimony informative sites generated by each of the three loci and all loci combined were determined using PAUP* v4B10 (15). The aligned sequences were used to estimate phylogenetic trees under the maximum likelihood criterion (ML) as employed by PAUP* v4B10. An HKY + I + G model of evolution was used to correct for multiple hits. Specifically, we used a neighbor-joining tree to generate the first tree (neighbor-joining start) and then optimized the start tree with the subtree pruning and regrafting swapping algorithm. Trees were rooted with the midpoint rooting algorithm, as implemented in PAUP* v4B10. Support for nodes was generated by nonparametric bootstrapping of the data with the same ML model of evolution estimated from the complete data set. Sequences of the loci enoA, benA, and calM of A. terreus NIH2624, whose genome has been sequenced, were downloaded from the A. terreus Sequencing Project (http://www.broad.mit.edu/annotation/genome/aspergillus_terreus/Home.html) and included for reference.
Morphological studies.
For macromorphological observations, isolates were grown on Czapek yeast autolysate (CYA), malt extract agar (MEA), and Czapek agar and incubated at 25°C in the dark for 7 days and at 37°C and 42°C on CYA. For micromorphological observations such as size of conidia, phialides, vesicles and conidiophores, and stipe wall morphology, microscopic mounts from MEA colonies were made in lactic acid and a drop of alcohol was added to remove air bubbles and excess conidia.
Nucleotide sequence accession numbers.
Sequences of the enoA, benA, and calM of the section Terrei isolates have been submitted to GenBank and assigned the accession numbers listed in Table 1.
RESULTS
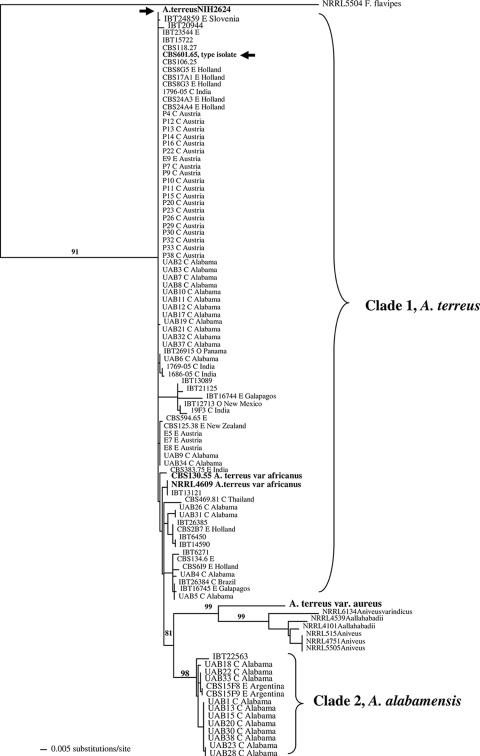
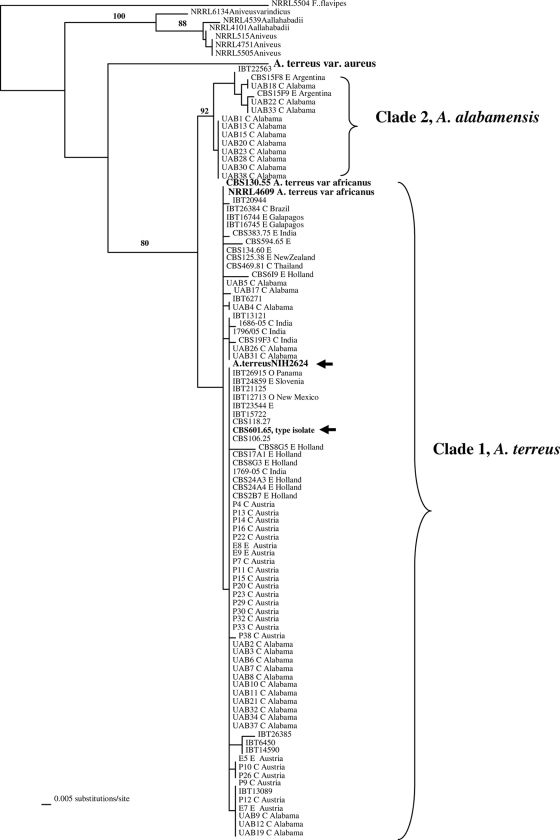
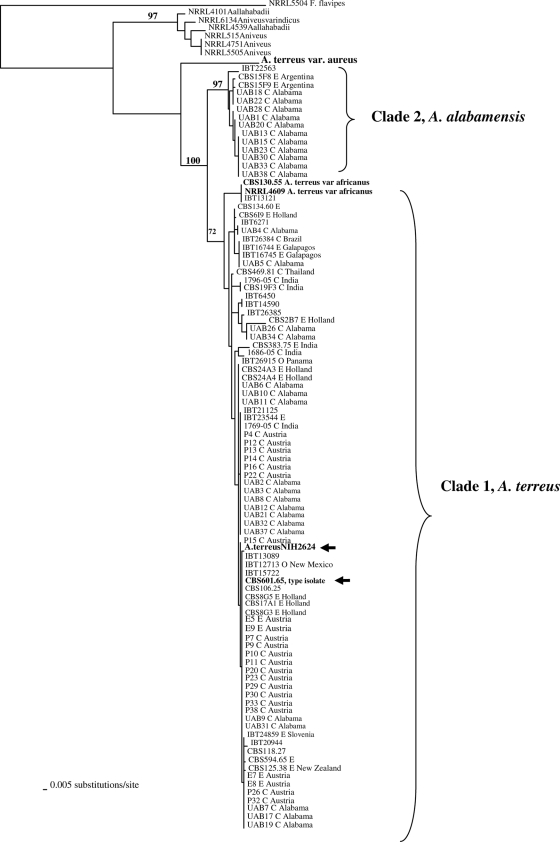
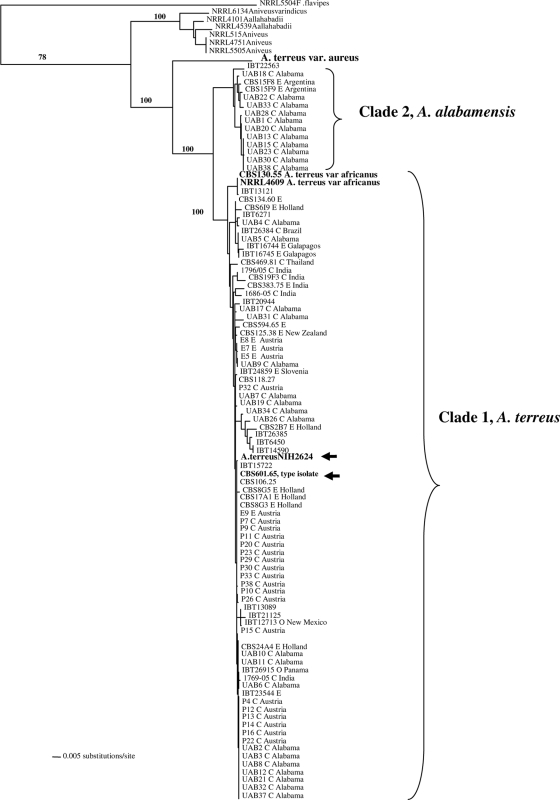
Genomic DNA from all isolates was amenable to PCR amplification and sequencing in the selected gene regions. ML trees generated from the three genes (Fig. 1, 2, and 3) and the combine ML tree generated from the sequences from all the threeloci (Fig. 4) revealed two distinct, well-supported clades within section Terrei. Clade 1 included 85.1% of isolates (80/94) previously identified as A. terreus, including the type isolate (CBS601.65) and the genome sequence isolate (NIH2624), indicating that this clade represents, at least in part, that taxon (Fig. 1 to 3). There was no clustering of genotypes from environmental or clinical origin within clade 1 (Fig. 1 to 3). The two A. terreus var. africanus isolates (CBS130.55 = NRRL2399; NRRL4609) clustered within clade 1 in all three loci. Another isolate, IBT13121, clustered with A. terreus var. africanus in the benA and calM loci.
FIG. 1.
ML tree generated from partial sequences from the calmodulin (calM) gene region. Fennellia flavipes is used as the outgroup. Bootstrap values are shown above the branches. Isolate NIH2624, whose genome has been sequenced, is indicated with an arrow. Environmental isolates are denoted by E, and clinical isolates are denoted by C.
FIG. 2.
ML tree generated from partial sequences of the enolase (enoA) gene region. Arrows indicate A. terreus isolate NIH2624, whose genome has been sequenced. Bootstrap values are shown above the branches. Environmental isolates are denoted by E, and clinical isolates are denoted by C.
FIG. 3.
ML tree generated from partial sequences from the β-tubulin (benA) gene region. Isolate NIH2624, whose genome has been sequenced, is indicated by arrows. Bootstrap values are shown above the branches. Environmental isolates are denoted by E, and clinical isolates are denoted by C.
FIG. 4.
Combined ML tree generated from sequences from the three protein-coding loci calM, enoA, and benA showing A. alabamensis as a separate species with high bootstrap values.
The type strain of A. terreus var. aureus (CBS503.65 = NRRL1923) was distinct from all other isolates in all three loci as well as in the combined-locus tree (Fig. 1 to 4).
Clade 2 was distinct from the A. terreus clade (clade 1) and included 14 isolates, of which 11 were clinical specimens recovered from 11 different patients attending UAB (Fig. 1 to 3). This clade had high bootstrap support in all three loci studied. The combined-gene tree also gave strong support for the reciprocal monophyly of clades 1 and 2 (Fig. 4). Eight of 11 clinical isolates were colonizing isolates as they were recovered from sputum or tracheal aspirates of immunocompetent hosts and not from a sterile body fluid or an invasive site. One isolate each was recovered from a broncheoalveolar lavage sample of a kidney/pancreas transplant patient, a wound from a burn patient, and the ear of a patient with external otitis. This clade also included three soil isolates—one from Florida and two from Argentina. The source and origin of one isolate (IBT22563) are not known.
Members of clade 2 were recognized as a new species within the section Terrei, which is formally described below.
Aspergillus alabamensis Balajee, Baddley, Frisvad & Samson, sp. nov.
Holotype.
Isolate UAB 20, recovered from a clinical specimen received from UAB, is designated as the holotype and has been deposited in the CBS Fungal Biodiversity Centre.
Coloniae in agaris CYA et MEA luteobrunneae vel cinnamomeae, saepe ex strato coacto conidiophororum constantes, etsi auctus floccosus praebentes. Capitula conidialia longa, dense columnaria, in maturitate 30-50 μm diam, 150-500+ μm longa. Conidiophora biseriata, laevia, hyalina, 100-250 × 4.5-6.0 μm; vesiculae subglobosae, 10-16 μm diam; phialidae 5.0-7.0 × 2.0-2.5 μm; metulae arcte contiguae, 5.5-7.5 × 1.5-2.0 μm; conidia globosa vel subelliptica, laevia, 1.8-2.4 μm diam.
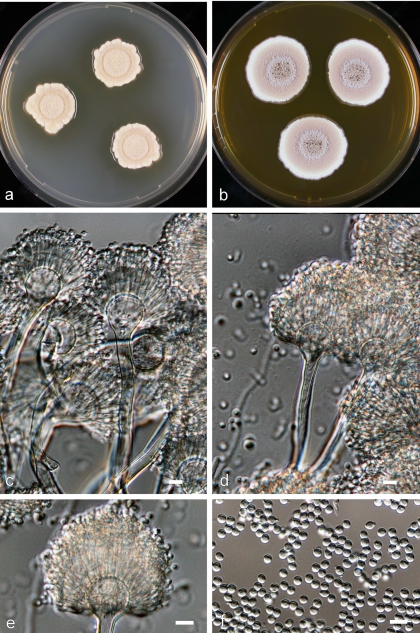
The basic morphological characteristics of A. alabamensis are illustrated in Fig. 5. Colonies on Czapek yeast extract and MEA are yellowish-brown to cinnamon-brown, often consisting of a dense felt of conidiophores but also showing floccose growth. Conidial heads are densely columnar. Conidial heads are long, columnar, 30 to 50 μm in diameter, and 150 to 500 μm or more in length at maturity; conidiophores are biseriate, smooth, colorless, and 100 to 250 μm by 4.5 to 6.0 μm. Vesicles are subglobose and 10 to 16 μm in diameter. Phialides are 5.0 to 7.0 μm by 2.0 to 2.5 μm. Metulae are closely packed and 5.5 to 7.5 μm by 1.5 to 2.0 μm. Conidia are globose to slightly elliptical, smooth, and 1.8 to 2.4 μm in diameter.
FIG. 5.
Aspergillus alabamensis sp. nov. UAB 20T. Shown are colonies on MEA after 7 days at 25°C on CYA (a) and on MEA (b), conidial heads (c and d), a single conidial head (e), and conidia (f). Bar, 10 μm.
Etymology.
The name Aspergillus alabamensis was chosen since most of the members of this new species were recovered as clinical specimens from patients at the UAB.
DISCUSSION
The present study, by means of a multilocus phylogenetic approach, characterized a large number of A. terreus isolates assembled from clinical and environmental origins representing different geographic locations of the world.
Results of the present study demonstrated that 33% of the clinical isolates (11/33) received from the UAB consistently clustered into a separate clade (clade 2) in the single-locus and combined-gene genealogies with strong bootstrap support; this clade was distinct from the A. terreus clade (Fig. 1 to 4), and thus the members of clade 2 are recognized as belonging to a new species, A. alabamensis. Antifungal susceptibility data were available for 7/11 isolates through a previous study (1); all 7 had higher MICs to amphotericin B (1 to 2 μg/ml) and lower MICs to voriconazole (0.25 to 0.5 μg/ml) and itraconazole (0.25 to 1 μg/ml). This susceptibility pattern was similar to the MICs of A. terreus to amphotericin B, voriconazole, and itraconazole (1). Interestingly, most of the clinical isolates in this new species were recovered as colonizing isolates, but given the small sample size of the study population, it is unclear at this time if these isolates have a decreased propensity to cause invasive infection. Apart from clinical isolates, this new species also included isolates recovered from soil in Florida and Argentina. Detailed morphological analyses revealed that A. alabamensis is macroscopically similar to A. terreus, with no striking macroscopical and microscopical differences. A feature common to both A. alabamensis and A. terreus is the colony pattern diversity (variation in color from yellow-brown to cinnamon-buff with or without orange tints) observed in this study as well as documented decades ago (11). Although A. alabamensis and A. terreus share many common secondary metabolites, the characteristic metabolite in A. alabamensis is citrinin, while A. terreus produces mevinolin (none of the A. alabamensis isolates tested produced this lovastatin derivative) and citreoviridin. This metabolite pattern appears to be a distinguishing feature between the species and warrants further detailed examination (J. Frisvad, personal communication).
A. terreus var. aureus has been previously recognized as a variety of A. terreus based on morphological characteristics, and the phenotype of this species is strikingly distinct from that of A. terreus (11). Specifically, this variety presents on MEA as slow-growing, floccose colonies with a bright golden yellow color due to pigmentation of the vegetative mycelium, with slower sporulation than A. terreus. Conidiophores are long, often becoming 500 μm or more in length, and bear rather small but definitely columnar heads that range from white to cream or light buff-colored conidia. Combined phylogenetic and morphological evidence demonstrates that this variant should be considered a new species within the section Terrei.
Aspergillus terreus var. africanus is morphologically distinct from A. terreus in that the colonies grow as bright yellow colonies on both Czapek agar and MEA, with less sporulation and the presence of globose sclerotium-like bodies in light tan shades. Despite this distinctive phenotype, phylogenetic analyses placed these isolates (CBS 130.55 = NRRL 2399; NRRL 4609) in a basal cluster within a broader, well-supported A. terreus clade. Thus, the taxonomic status of A. terreus var. africanus could not be clearly resolved in this study using the three-locus phylogeny. Several A. terreus isolates recovered from diverse geographic origins appeared to share the same multilocus haplotype, with no clear correlation between genotypes of A. terreus isolates, their source (environmental versus clinical), and their geographic origin. These results are similar to the findings that A. fumigatus persists as a single, global phylogenetic population with no evidence of endemism (10, 12). Thus, A. terreus appears to be a cosmopolitan fungus, with several genotypes spread worldwide. Although, in this study, sequences generated from the three loci yielded sufficient diversity to clearly delineate species within the section Terrei, the proposed scheme had limited utility in strain discrimination. It was recently shown that a multilocus sequence typing scheme was not useful for strain discrimination in A. fumigatus because of low genetic diversity (2). Similarly, data from this study indicate that a multilocus sequence typing scheme, albeit using highly conserved genes, may not be suitable to discriminate individuals in a population of A. terreus: other subtyping schemes such as microsatellite marker-based formats may be more useful for A. terreus strain typing.
In conclusion, employing multilocus phylogenetic analyses, the present study describes a new species within the section Terrei: A. alabamensis. Preliminary evidence suggests that members of the newly recognized species A. alabamensis are often colonizing isolates, but a larger screening study and pathogenesis studies with an in vivo model of invasive aspergillosis will be needed to validate this observation.
Acknowledgments
We thank Emory Simmons for help with the Latin description of A. alabamensis.
Current members of the ISHAM Working Group on A. terreus include Robert Samson, János Varga, Jan Dijksterhuis, CBS Fungal Biodiversity Centre, Utrecht, The Netherlands; Cornelia Lass-Flörl, Medical University of Innsbruck, Innsbruck, Austria; Jens Christian Frisvad, Technical University of Denmark, Copenhagen; David Nickle, University of Washington, Seattle; Corné Klaassen, Canisius Wilhelmina Hospital, Nijmegen, The Netherlands; Dimitrios P. Kontoyiannis and Russell Lewis, M. D. Anderson Cancer Center, Houston, TX; Stephen W. Peterson, National Center for Agricultural Utilization Research, U.S. Department of Agriculture, Peoria, IL; and S. Arunmozhi Balajee, Mycotic Diseases Branch, Centers for Disease Control and Prevention, Atlanta, GA.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the CDC.
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Baddley, J. W., P. G. Pappas, A. C. Smith, and S. A. Moser. 2003. Epidemiology of Aspergillus terreus at a university hospital. J. Clin. Microbiol. 415525-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain, J. M., A. Tavanti, A. D. Davidson, M. D. Jacobsen, D. Shaw, N. A. R. Gow, and F. C. Odds. 2007. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. J. Clin. Microbiol. 451469-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balajee, S. A., J. L. Gribskov, E. Hanley, D. Nickle, and K. A. Marr. 2005. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot. Cell 4625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong, S. B., H. S. Cho, H. D. Shin, J. C. Frisvad, and R. A. Samson. 2006. Novel Neosartorya species isolated from soil in Korea. Int. J. Syst. Evol. Microbiol. 56477-486. [DOI] [PubMed] [Google Scholar]
- 5.Hong, S. B., S. J. Go, H. D. Shin, J. C. Frisvad, and R. A. Samson. 2005. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 971316-1329. [DOI] [PubMed] [Google Scholar]
- 6.Iwen, P. C., M. E. Rupp, A. N. Langnas, E. C. Reed, and S. H. Hinrichs. 1998. Invasive pulmonary aspergillosis due to Aspergillus terreus: 12-year experience and review of the literature. Clin. Infect. Dis. 261092-1097. [DOI] [PubMed] [Google Scholar]
- 7.Lass-Flörl, C., K. Grif, and D. P. Kontoyiannis. 2007. Molecular typing of Aspergillus terreus isolates collected in Houston, Texas, and Innsbruck, Austria: evidence of great genetic diversity. J. Clin. Microbiol. 452686-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lass-Florl, C., K. Griff, A. Mayr, A. Petzer, G. Gastl, H. Bonatti, M. Freund, G. Kropshofer, M. P. Dierich, and D. Nachbaur. 2005. Epidemiology and outcome of infections due to Aspergillus terreus: 10-year single centre experience. Br. J. Haematol. 131201-207. [DOI] [PubMed] [Google Scholar]
- 9.Peterson, S. W., F. E. Vega, F. Posada, and C. Nagai. 2005. Penicillium coffeae, a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia 97659-666. [DOI] [PubMed] [Google Scholar]
- 10.Pringle, A., D. M. Baker, J. L. Platt, J. P. Wares, J. P. Latge, and J. W. Taylor. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution Int. J. Org. Evolution 591886-1899. [PubMed] [Google Scholar]
- 11.Raper, B. K., and D. I. Fennell. 1965. The genus Aspergillus. Williams and Wilkins, Baltimore, MD.
- 12.Rydholm, C., G. Szakacs, and F. Lutzoni. 2006. Low genetic variation and no detectable population structure in Aspergillus fumigatus compared to closely related Neosartorya species. Eukaryot. Cell 5650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samson, R. A., S. Hong, S. W. Peterson, J. C. Frisvad, and J. Varga. 2007. Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Stud. Mycol. 59147-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinbach, W. J., D. K. Benjamin, Jr., D. P. Kontoyiannis, J. R. Perfect, I. Lutsar, K. A. Marr, M. S. Lionakis, H. A. Torres, H. Jafri, and T. J. Walsh. 2004. Infections due to Aspergillus terreus: a multicenter retrospective analysis of 83 cases. Clin. Infect. Dis. 39192-198. [DOI] [PubMed] [Google Scholar]
- 15.Swofford, D. L. 1999. PAUP* 4.0: phylogenetic analysis using parsimony (*and other methods). 4.0b2a ed. Sinauer Associates, Inc., Sunderland, MA.
- 16.Varga, J., B. Toth, S. Kocsube, B. Farkas, G. Szakacs, J. Teren, and Z. Kozakiewicz. 2005. Evolutionary relationships among Aspergillus terreus isolates and their relatives. Antonie van Leeuwenhoek 88141-150. [DOI] [PubMed] [Google Scholar]
- 17.Walsh, T. J., V. Petraitis, R. Petraitiene, A. Field-Ridley, D. Sutton, M. Ghannoum, T. Sein, R. Schaufele, J. Peter, J. Bacher, H. Casler, D. Armstrong, A. Espinel-Ingroff, M. G. Rinaldi, and C. A. Lyman. 2003. Experimental pulmonary aspergillosis due to Aspergillus terreus: pathogenesis and treatment of an emerging fungal pathogen resistant to amphotericin B. J. Infect. Dis. 188305-319. [DOI] [PubMed] [Google Scholar]
- 18.Yaguchi, T., Y. Horie, R. Tanaka, T. Matsuzawa, J. Ito, and K. Nishimura. 2007. Molecular phylogenetics of multiple genes on Aspergillus section Fumigati isolated from clinical specimens in Japan. Nippon Ishinkin Gakkai Zasshi 4837-46. [DOI] [PubMed] [Google Scholar]