Abstract
Candida albicans is an opportunistic human fungal pathogen that normally resides in the gastrointestinal tract and on the skin as a commensal but can cause life-threatening invasive disease. Salmonella enterica serovar Typhimurium is a gram-negative bacterial pathogen that causes a significant amount of gastrointestinal infection in humans. Both of these organisms are also pathogenic to the nematode Caenorhabditis elegans, causing a persistent gut infection leading to worm death. In the present study, we used a previously developed C. elegans polymicrobial infection model to assess the interactions between S. Typhimurium and C. albicans. We observed that when C. elegans is infected with C. albicans and serovar Typhimurium, C. albicans filamentation is inhibited. The inhibition of C. albicans filamentation by S. Typhimurium in C. elegans appeared to be mediated by a secretary molecule, since filter-sterilized bacterial supernatant was able to inhibit C. albicans filamentation. In vitro coculture assays under planktonic conditions showed that S. Typhimurium reduces the viability of C. albicans, with greater effects seen at 37°C than at 30°C. Interestingly, S. Typhimurium reduces the viability of both yeast and filamentous forms of C. albicans, but the killing appeared more rapid for the filamentous cells. The antagonistic interaction was also observed in a C. albicans biofilm environment. This study describes the interaction between two diverse human pathogens that reside within the gastrointestinal tract and shows that the prokaryote, S. Typhimurium, reduces the viability of the eukaryote, C. albicans. Identifying the molecular mechanisms of this interaction may provide important insights into microbial pathogenesis.
Candida albicans, the most common human fungal pathogen, is a prototypical opportunistic organism that lives harmlessly in the human gastrointestinal tract but has the ability to cause life-threatening invasive disease. Bloodstream infection with C. albicans remains the most lethal form (10), with translocation of the gastrointestinal mucosa being an important pathogenic mechanism, especially in hemato-oncology patients and those who have undergone abdominal surgery. A key virulence determinant of C. albicans is its ability to transition from yeast to a filamentous form (16, 17, 19, 22). This morphogenesis appears important for tissue adherence and invasion (22). Furthermore, C. albicans has the ability to form complex biofilms on medical devices (13) and on human mucosal surfaces, such as the gastrointestinal and bronchial mucosa. C. albicans biofilm formation has immense clinical and economic consequences (13).
Recently the interactions between this important fungal pathogen and bacteria were described (11, 12, 18). These studies focus on the interaction between C. albicans and nonfermenting, gram-negative bacteria, such as Pseudomonas aeruginosa and Acinetobacter baumannii, whose interactions are likely found in the clinical environment, especially in the respiratory tracts of critically ill patients and on wounds of patients with burn injuries (7, 20). Of interest, these bacteria show antagonistic properties toward C. albicans, with a predilection toward reducing the viability of C. albicans filaments. In order to study these prokaryote-eukaryote interactions, our laboratory developed a polymicrobial infection model system using Caenorhabditis elegans as a substitute host (18). Previously, we showed that C. albicans causes a persistent lethal infection of the C. elegans intestinal tract (6). This leads to overwhelming C. albicans intestinal proliferation with subsequent filamentation through the worm cuticle (6). Given these characteristics, we decided to use this model to study the interaction of C. albicans with another intestinal pathogen, Salmonella enterica serovar Typhimurium.
S. Typhimurium is a gram-negative organism that belongs to the Enterobacteriaceae family. It is a gastrointestinal tract pathogen of humans, being responsible for approximately 2 million to 4 million cases of enterocolitis each year in the United States (4, 8, 21, 23). During infection, S. Typhimurium competes with normal intestinal flora (23). Its virulence pathways are well described, and it has been shown to cause a persistent and lethal gut infection of the nematode C. elegans, similar to infection seen with C. albicans (1, 14). Given this and the fact that C. albicans is a common inhabitant of the human gastrointestinal tract, we used the C. elegans polymicrobial infection model (18) to study the interactions between S. Typhimurium and C. albicans. Understanding the interactions between these diverse organisms within the complex milieu of an intestinal tract may provide important pathogenic and therapeutic insights.
MATERIALS AND METHODS
Strains and media.
Unless otherwise specified, fungal and bacterial cultures were grown overnight in yeast-peptone-dextrose (YPD) (Difco) broth at 30°C and LB broth at 37°C, respectively, and all experiments with S. Typhimurium were with the wild-type SL1344 strain, which is resistant to streptomycin and rifampin (rifampicin) (1). In the C. elegans assays, we used the green fluorescent protein-linked S. Typhimurium strain SL1344-GFP to help visualize the bacteria with confocal microscopy. This strain was resistant to ampicillin. Bacterial supernatant was filter sterilized using a 0.22-μm filter (Millipore) and was checked for sterility by plating on LB agar. S. Typhimurium supernatant was heat inactivated at 100°C for 30 to 45 min.
C. elegans coinfection assay for Filamentation.
The methodology for this assay was as described previously (18). In brief, C. elegans glp-4;sek-1 nematodes were propagated on E. coli strain OP50 using established procedures. Synchronized, young adult nematodes were preinfected for 4 h on lawns of C. albicans and were then transferred, after washing in M9 medium, into wells of a six-well microtiter dish (Corning) (∼70 to 80 nematodes per well). Each well contained 2 ml of standard liquid medium containing 80% M9 medium and 20% brain heart infusion. Just prior to transferral of the C. albicans-preinfected worms, S. Typhimurium was inoculated into the liquid media at various cell densities. The plates were then incubated at 25°C and examined at 24-h intervals for 5 days for the formation of penetrative filamentation using a Nikon SMZ645 dissecting microscope (TCS NT; Leica Microsystems). Penetrative filamentation was defined as any breach in the worm cuticle by filamentous cells as seen at magnification ×50. All experiments were repeated at least three times. Differences in worm filamentation on day 5 were compared by the Student t test. A P value of less than 0.05 was considered to be statistically significant. Qualitative assessment of C. albicans filamentation in C. elegans was performed using confocal laser microscopy (TCS NT; Leica Microsystems).
In vitro coinfection assay under planktonic conditions.
In vitro coculture assays were performed in 2 ml of LB broth and were incubated in a roller drum at 30°C or 37°C, as appropriate. A starting inoculum of 106 CFU/ml of S. Typhimurium strain SL1344 and 5 × 105 CFU/ml of C. albicans was used for all experiments. YPD agar plates containing kanamycin (45 μg/ml), ampicillin (100 μg/ml), and streptomycin (100 μg/ml) and LB agar plates containing fluconazole (32 μg/ml) were used to select for C. albicans strains and S. Typhimurium, respectively. YPD and LB agar plates were incubated for 48 h at 30°C and 24 h at 37°C, respectively, before colonies were counted. Results were obtained from three independent experiments.
The viability of C. albicans when cocultured with S. Typhimurium was further assessed by using the Live/Dead staining system (Molecular Probes) according to the manufacturer's protocol.
Silicone pad biofilm assay.
The effect of S. Typhimurium on C. albicans biofilm growth was evaluated using a polymicrobial silicone pad assay as described previously (18). Spider medium (10 g of nutrient broth, 10 g of mannitol, and 2 g of K2HPO4 in 1 liter) (15) was used as the medium for C. albicans biofilm development. The quantitative biofilm mass was calculated by subtracting the original weight of the silicone pad from its postincubation (60-h) weight and adjusting for the weight of a control pad exposed to no cells. All experiments were performed at least twice in triplicate. Differences in weight were determined by Student's t test, and a P value of <0.05 was considered to be statistically significant. Confocal laser microscopy was performed on mature biofilms.
RESULTS AND DISCUSSION
S. Typhimurium inhibits C. albicans filamentation in C. elegans.
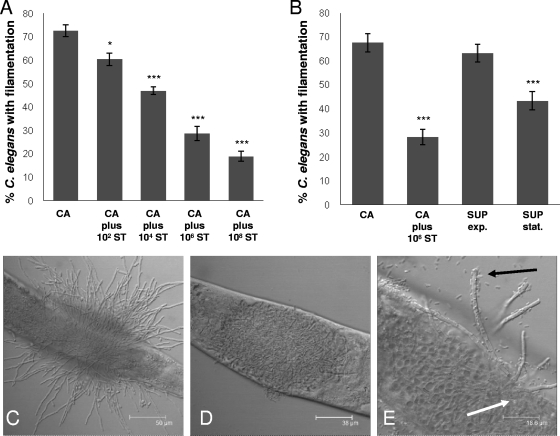
Previously, our laboratory showed that pathogenic gram-negative organisms, such as A. baumannii and P. aeruginosa, are able to inhibit C. albicans filamentation in the C. elegans coinfection model (18). This is in contrast to nonpathogenic gram-negative organisms, such as the auxotrophic Escherichia coli strain OP50, and gram-positive organisms, such as Enterococcus spp., which do not substantially inhibit C. albicans filamentation in C. elegans (18). After infecting young adult nematodes with C. albicans for 4 h and then transferring them into liquid medium containing 106 CFU/ml of S. Typhimurium, we observed a significant reduction in the proportion of worms with C. albicans filaments penetrating through their cuticles (Fig. 1). The effects were inoculum dependent, whereby larger numbers of bacterial cells caused greater inhibition of C. albicans filamentation (Fig. 1A). The antagonistic effect of S. Typhimurium on C. albicans is also shown by confocal microscopy, whereby a marked reduction in C. albicans filamentation was observed with coinfection with S. Typhimurium (Fig. 1D) compared to results without (Fig. 1C). Interestingly, at higher magnification, bacterial cells were seen adhering to the C. albicans filaments (Fig. 1E).
FIG. 1.
S. Typhimurium (ST) inhibits C. albicans (CA) filamentation in the C. elegans coinfection model. (A) The degree of inhibition of C. albicans filamentation was dependent on the initial inoculum of S. Typhimurium in the liquid medium of the assay. (B) S. Typhimurium supernatant taken from stationary-phase growth (16 h of growth) (SUP stat.) caused a significant inhibition of C. albicans filamentation in the C. elegans model, whereas that taken from exponential-phase growth (4 h of growth) (SUP exp.) did not. All images were taken at 24 h. Column bars represent the means, and error bars represent the standard deviations. Confocal microscopy showed the marked phenotypic differences between C. elegans infection with C. albicans alone (C) and infection with C. albicans and S. Typhimurium (D). (E) By using a green fluorescent protein-linked S. Typhimurium strain (SL1344-GFP), we observed not only bacteria and fungal cells within the gut of the worm (white arrow) but also bacteria associating with sparse filaments that had protruded through the worm cuticle (black arrow). Asterisks denote comparison of percentage filamentation with that for C. albicans strain DAY185 alone: ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05 (two-tailed t test). Scale bar: 50 μm (C), 38 μm (D), or 18.6 μm (E).
It has previously been shown that a secretory molecule from P. aeruginosa inhibits filament formation in C. albicans (12). To assess whether a bacterial secretory factor was responsible for the observed inhibition of C. albicans filamentation in C. elegans by S. Typhimurium, we transferred worms that had been infected with C. albicans into S. Typhimurium culture filtrate. As shown in Fig. 1B, when preinfected worms were transferred into bacterial supernatant taken from exponential-phase growth, no inhibition of C. albicans filaments was observed. However, when supernatant was taken from the stationary phase, significant inhibition of penetrative filamentation was seen (Fig. 1B).
These data suggest that S. Typhimurium has an antagonistic interaction with C. albicans and that a bacterial secretory factor plays a significant role in this process. Given the growth-dependent antifungal activity of S. Typhimurium culture filtrate on C. albicans, one may hypothesize that a quorum-sensing molecule may be responsible. However, secretion of a quorum-sensing molecule by S. Typhimurium has not been described thus far (2). Despite this, S. Typhimurium has been shown to have a quorum sensor encoded by sdiA, which responds to quorum-sensing molecules produced by other bacteria and can influence S. Typhimurium virulence (2, 3). We tested the effect of the S. Typhimurium sdiA mutant and found that this mutant caused the same degree of inhibition of C. albicans filamentation in C. elegans as its parent strain, 14028 (data not shown). Thus, the observed antifungal activity of S. Typhimurium supernatant is likely due to a different type of growth-dependent secretory molecule.
S. Typhimurium reduces the viability of C. albicans in vitro.
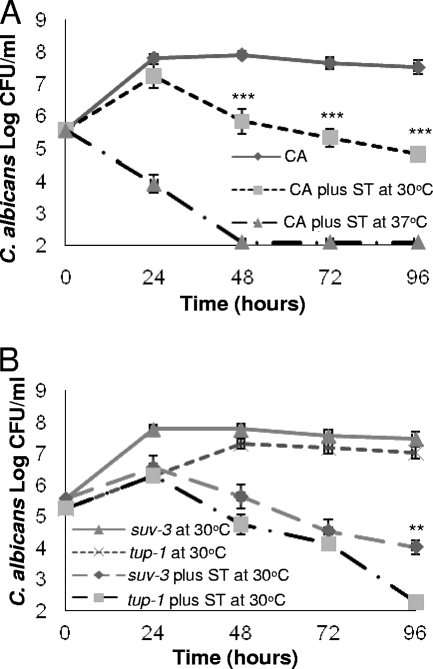
To further define the observed interaction between C. albicans and S. Typhimurium, we performed in vitro coinfection cultures under planktonic conditions. First, we coinfected the C. albicans reference strain DAY185 with the wild-type S. Typhimurium strain SL1344 in Luria-Bertani (LB) medium at temperatures closer to that used in the C. elegans model (30°C). As shown in Fig. 2A, after initial growth of C. albicans, the viability of the fungus declined in the presence of S. Typhimurium. Interestingly, when the coinfection culture was incubated at temperatures more consistent with those of the human body (37°C), the killing of C. albicans by S. Typhimurium was significantly more rapid, with undetectable levels by 48 h (Fig. 2A). Of note, the growth of C. albicans and that of S. Typhimurium when cultured alone were similar when incubated at 30°C and 37°C (data not shown).
FIG. 2.
S. Typhimurium (ST) reduces the viability of C. albicans (CA) in an in vitro environment. (A) The viability of the C. albicans DAY185 strain in the presence of S. Typhimurium is significantly lower at 37°C than at 30°C. The C. albicans suv3 mutant, which is in the yeast form at 30°C, was more resistant to killing by S. Typhimurium than the constitutively filamentous C. albicans tup1 mutant strain at 30°C (B). Error bars represent the standard deviations. In panel A, asterisks denote comparison of log CFU/ml of C. albicans DAY185 at 30°C and 37°C. In panel B, asterisks denote comparison of log CFU/ml of the C. albicans suv3 strain to the C. albicans tup1 strain at 30°C: ***, P ≤ 0.001; **, P ≤ 0.01 (two-tailed t test).
These data suggest that either S. Typhimurium is more pathogenic or C. albicans is more susceptible at 37°C. Temperature is an important environmental stimulus for C. albicans morphogenesis: at 37°C the majority of C. albicans cells form filaments while at 30°C there is only a small ratio of hyphae to yeast cells (9). Given our observations of C. albicans filament inhibition in coinfection with S. Typhimurium in C. elegans and the temperature differences in the viability of C. albicans in the coinfection in vitro cultures, we tested whether S. Typhimurium has morphological specificity in its antagonism toward C. albicans. Furthermore, we have previously shown that A. baumannii has a predilection for killing the filamentous form of C. albicans (18). Using the in vitro assay already described, we coinfected the constitutively filamentous C. albicans tup1 mutant (5, 11) and the C. albicans suv3 mutant, which is impaired in hypha formation and remains in the yeast form at 30°C (18, 19), with S. Typhimurium (SL1344) at 30°C. As shown in Fig. 2B, killing was observed for both C. albicans mutant strains; however, it was more pronounced for the C. albicans tup1 mutant by 96 h of incubation. More-rapid killing was observed with both mutant strains when the experiment was repeated at 37°C (data not shown). Given that the suv3 mutant can form filaments at 37°C, we were unable to detect a statistically significant difference between the killing of these strains. Taken in their totality, these data suggest that the temperature effect may be associated with C. albicans morphology or it could be independent of morphology and associated with increased susceptibility of C. albicans at 37°C. Nevertheless, increased pathogenicity of S. Typhimurium against C. albicans at 37°C cannot be excluded.
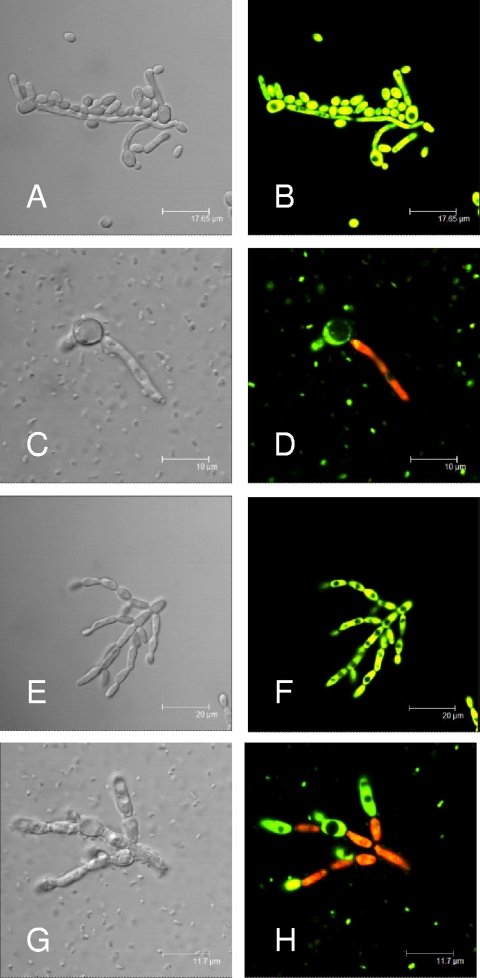
Interestingly, when we performed confocal microscopy of the in vitro coinfection culture with the reference strain of C. albicans (DAY185) (Fig. 3C) and the C. albicans tup1 mutant (Fig. 3G), we did not see significant cell-cell association as has been shown previously with A. baumannii and P. aeruginosa (11, 18). However, after the coculture was stained at 24 h (incubation at 37°C) with the Live/Dead staining system (Molecular Probes), whereby live cells stain green (SYTO9) and dead cells stain red (propidium iodide), the viability of the filaments was reduced from that of the yeast form (Fig. 3D and H). These data suggest that a secretory molecule may be the predominant mechanism by which S. Typhimurium kills C. albicans.
FIG. 3.
Confocal laser microscopy of in vitro cultures after staining with the Live/Dead staining system, whereby dead cells stain red and live cells stain green. C. albicans strain DAY185 cultured alone (A and B) or in the presence of S. Typhimurium (C and D), showing a nonviable filamentous cell when cocultured with S. Typhimurium (D) despite an absence of pronounced cell-cell association. (E and F) The constitutively filamentous C. albicans tup1 mutant cultured alone (E and F) or in the presence of S. Typhimurium (G and H), showing a similar finding of reduced viability (red stain) when cocultured with S. Typhimurium (H). Images were taken after 24 h of incubation in LB broth at 37°C. Scale bar: 17.65 μm (A and B), 10 μm (C and D), 20 μm (E and F), or 11.7 μm (G and H).
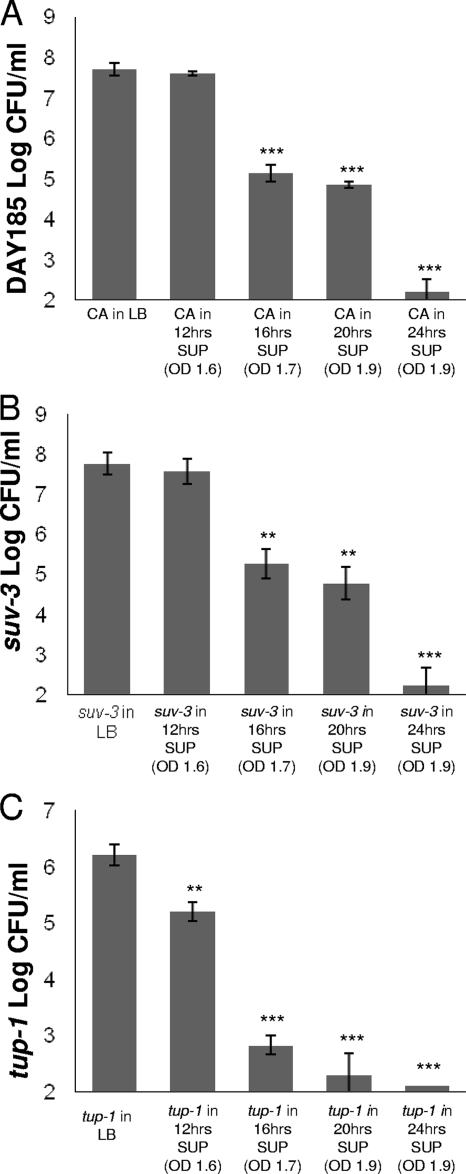
To determine the timing in the bacterial growth phase in which the S. Typhimurium supernatant reduced the viability of C. albicans, we cultured the fungus at 37°C in filter-sterilized supernatant taken from different stages of bacterial growth. As shown in Fig. 4A, the viability of C. albicans strain DAY185 was reduced when it was cultured in supernatant taken from at least 16 h of S. Typhimurium growth (stationary phase). The effects increased as supernatant was taken from bacteria grown for up to 24 h (Fig. 4A). In order to investigate further the effect of the supernatant on C. albicans filamentation, we inoculated the C. albicans tup1 and suv3 mutants in S. Typhimurium supernatant isolated from different time points of S. Typhimurium growth (in LB broth) and incubated them at 37°C. Interestingly, the constitutively filamenting tup1 mutant was more susceptible to the bacterial supernatant isolated at 12 h (early stationary phase) than the C. albicans suv3 mutant (Fig. 4B and C). Both strains were inhibited in supernatants isolated from late stationary phase. Given the fact that an acidic environment decreases C. albicans filamentation (9), we measured the pH of the supernatant medium. Interestingly, the pH of the supernatant isolated from S. Typhimurium grown in LB broth to late stationary phase was alkaline (pH 8.5). Furthermore, there was no difference in the antifungal activity of the S. Typhimurium supernatant after heat inactivation (100°C for up to 45 min).
FIG. 4.
S. Typhimurium supernatant (SUP) isolated at 37°C inhibits C. albicans strain DAY185 (CA) in a growth-dependent fashion (A). Inhibition of C. albicans DAY185 (CA) and suv3 mutant growth was not observed until they were exposed to supernatant taken from S. Typhimurium grown to late stationary phase (16 h, optical density at 600 nm [OD600] of 1.7) (A and B, respectively). tup1 mutant growth was inhibited in S. Typhimurium supernatant (SUP) isolated from early stationary phase (12 h, OD600 of 1) (C). When C. albicans strains DAY185 (CA) and suv3 were grown in S. Typhimurium supernatant taken from a 24-h growth (OD600 of 1.9), the density of C. albicans was more than 7 log CFU/ml lower than that with growth in fresh LB medium (A and B). In the same supernatant, there was almost no growth for the tup1 strain (C). All determinations of C. albicans CFU/ml were performed after 24 h of growth at 37°C. Column bars represent the means and error bars represent the standard deviations. Asterisks denote comparison of log CFU/ml with that of C. albicans in LB medium: ***, P ≤ 0.001; **, P ≤ 0.01 (two-tailed t test).
S. Typhimurium inhibits C. albicans biofilm formation on silicone pads.
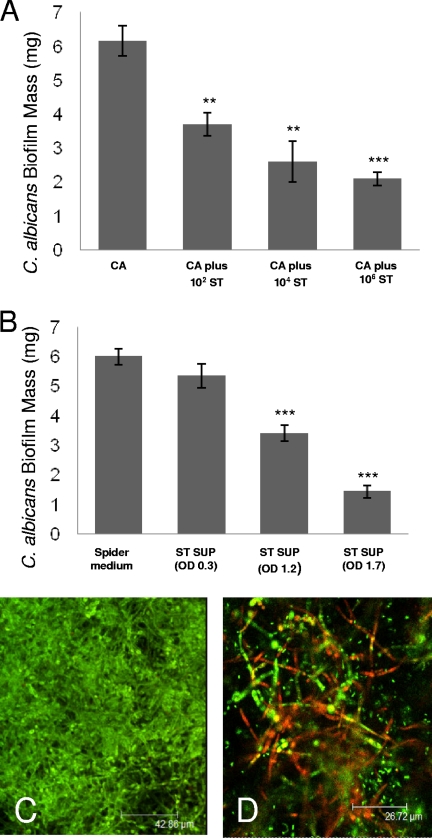
Given our observations of the effects of S. Typhimurium on C. albicans in the C. elegans coinfection model and under planktonic culture conditions in vitro, we assessed its effect on C. albicans biofilm formation on an abiotic surface (silicone pads). As shown in Fig. 5A, S. Typhimurium inhibited the development of C. albicans biofilm in a dose-dependent manner. A significant inhibition of C. albicans biofilm formation was observed after inoculation of as little as 102 CFU/ml of S. Typhimurium into the biofilm medium (spider medium) (Fig. 5A). Consistent with other results presented in this study (Fig. 1E and 4), C. albicans biofilm development was inhibited by filter-sterilized culture filtrate from S. Typhimurium (Fig. 5B). The degree of inhibition was dependent on the bacterial density in which the supernatant was derived (Fig. 5B). Confocal microscopy performed at the completion of the biofilm experiment (60 h) showed the presence of dead filaments with incubation in the presence of S. Typhimurium compared to results in its absence (Fig. 5C and D). These data confirm that the antifungal properties of S. Typhimurium are effective toward C. albicans in a biofilm, a notoriously resistant environment.
FIG. 5.
S. Typhimurium (ST) inhibits the development of C. albicans (CA) biofilm on silicone pads. (A) The degree of inhibition of C. albicans (CA) biofilm formation was dependent on the density of S. Typhimurium (ST) inoculated into the biofilm environment. (B) S. Typhimurium supernatant (ST SUP) isolated at 37°C, when taken from a bacterial growth with an optical density (OD) at 600 nm of at least 1.2, led to a significant reduction in C. albicans biofilm mass compared to its development in fresh spider medium. (C and D) Fluorescent images of mature C. albicans biofilms (48 h of growth) when grown in the absence (C) or presence (D) of S. Typhimurium. with use of the Live/Dead staining system, nonviable (red stain) filaments are observed with incubation in the presence of S. Typhimurium. Column bars represent the means, and error bars represent the standard deviations. Asterisks denote comparison of C. albicans biofilm mass (mg) to C. albicans alone (A) or to C. albicans grown in spider medium (B): ***, P ≤ 0.001; **, P ≤ 0.01 (two-tailed t test). Scale bar: 42.86 μm (C) or 26.72 μm (D).
Concluding remarks.
In the present study, we have utilized the substitute host, C. elegans, to identify an antagonistic interaction between two human pathogens that reside within the gastrointestinal tract. We observed that S. Typhimurium inhibits C. albicans filamentation, a key virulence determinant of this pathogen, in coinfection of C. elegans. Furthermore, after in vitro cocultures are performed in planktonic and biofilm environments, S. Typhimurium is capable of significantly reducing the viability of C. albicans and inhibiting its ability to form biofilm. Interestingly, culture filtrate from stationary-phase S. Typhimurium growth has significant anticandidal activity. The interaction of C. albicans with bacteria might shape microbial virulence and alter filamentation by Candida spp. in the intestinal tract. Identifying the molecular mechanisms of this interaction may provide important insights into microbial pathogenesis.
Acknowledgments
Support was provided by National Institutes of Health R01 award AI075286 (to E.M.) and a University of Queensland postgraduate scholarship award (to A.Y.P.).
We thank Brian Ahmer for the S. Typhimurium sdiA mutant and the parent strain, 14028.
Footnotes
Published ahead of print on 27 March 2009.
REFERENCES
- 1.Aballay, A., P. Yorgey, and F. M. Ausubel. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 101539-1542. [DOI] [PubMed] [Google Scholar]
- 2.Ahmer, B. M. 2004. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 52933-945. [DOI] [PubMed] [Google Scholar]
- 3.Ahmer, B. M., J. van Reeuwijk, C. D. Timmers, P. J. Valentine, and F. Heffron. 1998. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 1801185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle, E. C., J. L. Bishop, G. A. Grassl, and B. B. Finlay. 2007. Salmonella: from pathogenesis to therapeutics. J. Bacteriol. 1891489-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277105-109. [DOI] [PubMed] [Google Scholar]
- 6.Breger, J., B. B. Fuchs, G. Aperis, T. I. Moy, F. M. Ausubel, and E. Mylonakis. 2007. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chim, H., B. H. Tan, and C. Song. 2007. Five-year review of infections in a burn intensive care unit: high incidence of Acinetobacter baumannii in a tropical climate. Burns 331008-1014. [DOI] [PubMed] [Google Scholar]
- 8.Coburn, B., G. A. Grassl, and B. B. Finlay. 2007. Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 85112-118. [DOI] [PubMed] [Google Scholar]
- 9.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 1461763-1774. [DOI] [PubMed] [Google Scholar]
- 10.Gudlaugsson, O., S. Gillespie, K. Lee, J. Vande Berg, J. Hu, S. Messer, L. Herwaldt, M. Pfaller, and D. Diekema. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 371172-1177. [DOI] [PubMed] [Google Scholar]
- 11.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 2962229-2232. [DOI] [PubMed] [Google Scholar]
- 12.Hogan, D. A., A. Vik, and R. Kolter. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 541212-1223. [DOI] [PubMed] [Google Scholar]
- 13.Kojic, E. M., and R. O. Darouiche. 2004. Candida infections of medical devices. Clin. Microbiol. Rev. 17255-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labrousse, A., S. Chauvet, C. Couillault, C. L. Kurz, and J. J. Ewbank. 2000. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 101543-1545. [DOI] [PubMed] [Google Scholar]
- 15.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 2661723-1726. [DOI] [PubMed] [Google Scholar]
- 16.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90939-949. [DOI] [PubMed] [Google Scholar]
- 17.Nobile, C. J., and A. P. Mitchell. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 151150-1155. [DOI] [PubMed] [Google Scholar]
- 18.Peleg, A. Y., E. Tampakakis, B. B. Fuchs, G. M. Eliopoulos, R. C. Moellering, Jr., and E. Mylonakis. 2008. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 10514585-14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richard, M. L., C. J. Nobile, V. M. Bruno, and A. P. Mitchell. 2005. Candida albicans biofilm-defective mutants. Eukaryot. Cell 41493-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 1999. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit. Care Med. 27887-892. [DOI] [PubMed] [Google Scholar]
- 21.Rychlik, I., and P. A. Barrow. 2005. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol. Rev. 291021-1040. [DOI] [PubMed] [Google Scholar]
- 22.Saville, S. P., A. L. Lazzell, C. Monteagudo, and J. L. Lopez-Ribot. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 21053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stecher, B., A. J. Macpherson, S. Hapfelmeier, M. Kremer, T. Stallmach, and W. D. Hardt. 2005. Comparison of Salmonella enterica S. Typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect. Immun. 733228-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]