Abstract
Candida albicans is an important opportunistic human fungal pathogen that can cause both mucosal and systemic infections in immunocompromised patients. Critical for the virulence of C. albicans is its ability to undergo a morphological transition from yeast to hyphal growth mode. Proper induction of filamentation is dependent on the ubiquitination pathway, which targets proteins for proteasome-mediated protein degradation or activates them for signaling events. In the present study, we evaluated the role of ubiquitination in C. albicans by impairing the function of the major ubiquitin-ligase complex SCF. This was done by depleting its backbone, the cullin Cdc53p (orf19.1674), using a tetracycline downregulatable promoter system. Cdc53p-depleted cells displayed an invasive phenotype and constitutive filamentation under conditions favoring yeast growth mode, both on solid and in liquid media. In addition, these cells exhibited an early onset of cell death, as judged from propidium iodide staining, suggesting that CDC53 is an essential gene in C. albicans. To identify Cdc53p-dependent pathways in C. albicans, a genome-wide expression analysis was carried out that revealed a total of 425 differentially expressed genes (fold change, ≥2; P ≤ 0.05) with 192 up- and 233 downregulated genes in the CDC53-repressed mutant compared to the control strain. GO term analysis identified biological processes significantly affected by Cdc53p depletion, including amino acid starvation response, with 14 genes being targets of the transcriptional regulator Gcn4p, and reductive iron transport. These results indicate that Cdc53p enables C. albicans to adequately respond to environmental signals.
Candida albicans is an important human fungal pathogen that can cause both superficial and systemic infections in immunocompromised patients (57). The ability of this fungus to grow in and change between different morphological forms such as budding yeast (round or oval cells), true hyphae (filamentous cells without constrictions at the sites of septation), and pseudohyphae (chains of elongated cells with constrictions at the sites of septation) (66) is considered critical for its virulence (12, 41). Several environmental conditions are known to trigger a switch from yeast to filamentous growth mode, such as neutral pH, nutrient starvation, contact to solid surfaces, and growth in the presence of serum at 37°C, the latter mimicking the bloodstream and other environments in the host (reviewed in reference 21).
Various signal transduction pathways contribute to the regulation of the yeast-filament growth switch, forming a complex network that converges on a common set of hypha-specific genes (35). As in Saccharomyces cerevisiae, a mitogen-activated protein kinase (MAPK) pathway is involved in filamentation in C. albicans, consisting of the kinases Cst20p, probably Ste11p, Hst7p, Cek1p, and the executing transcription factor Cph1p (reviewed in references 11 and 47). Further filamentation-inducing pathways include the cyclic AMP-dependent protein kinase A pathway with its downstream effector Efg1p, the Cph2p/Tec1p pathway, the Czf1p-dependent matrix induced pathway, and the pH-responsive pathway which acts through the transcriptional regulator Rim101p (reviewed in references 11, 40, and 74). This network, although complex, is still being refined, as can be seen by the recent discovery of the transcriptional regulator, Ume6p, recently shown to regulate hypha extension and virulence (6). Additional complexity is added by negative transcriptional regulation, which is carried out by heterodimeric protein complexes, consisting of Tup1p and one of the DNA-binding proteins Rfg1p or Nrg1p (reviewed in references 11, 40, and 74).
The cytoskeleton reorganization that is necessary for serum-induced filamentation in C. albicans is mediated by the Rho-like GTPase Cdc42p and its exchange factor Cdc24p (8, 46, 70, 71). Recently, an additional Rho G-protein, Rac1p, and its activator Dck1p have been identified in C. albicans, which are required for filamentous growth in an agar matrix (7, 29).
Interestingly, mutations in the ubiquitination pathway have been shown to interfere with the proper regulation of filamentation in C. albicans (3, 38, 39, 60). Ubiquitination, the covalent attachment of a ubiquitin moiety (Ub) onto a target protein, is carried out by an enzymatic cascade consisting of the Ub-activating enzyme E1, the Ub-conjugating enzyme E2, and Ub-ligase E3, either assigning the target proteins for proteasome-mediated degradation or activating them for transcriptional activation or signaling events (16, 26). In S. cerevisiae, the ubiquitination system consists of a single E1, 11 E2s, and 42 E3s, which can be divided into RING or HECT domain-containing proteins (37). Some of the E3 ligases are embedded in SCF complexes, which are highly conserved in eukaryotes and consist of the linker protein Skp1; a cullin, which acts as a scaffold protein enabling spatial proximity of substrate and ligase; and a substrate-recognizing F-box protein (65, 75). A well-characterized example is the S. cerevisiae SCFCdc4-E2-E3 complex, which consists of the scaffold protein Cdc53p, the substrate-recognition F-box protein Cdc4p, the linker protein Skp1p, the E2-conjugating enzyme Cdc34p, and the RING-E3 enzyme Hrt1p (75). This complex is responsible for multiple cellular processes, including the degradation of the cell cycle inhibitor Sic1p, as well as the proper induction of amino acid starvation by degradation of the transcription factor Gcn4p in rich media (75).
In contrast to S. cerevisiae, not much is known about the ubiquitination system of C. albicans. Two C. albicans ubiquitin genes have been identified: UBI4 codes for a polyubiquitin precursor which is processed to three ubiquitin units and is the major source of ubiquitin upon stress conditions, whereas UBI3 encodes a ribosomal protein fused to ubiquitin (59, 60). Depletion of UBI4 and RAD6 (E2), as well as CDC4 and GRR1 (F-box proteins), has been shown to increase filamentation, indicating that ubiquitin-dependent pathways negatively regulate the yeast-to-hypha transition in C. albicans (3, 38, 39, 60). We therefore assumed that impairment of the SCF ubiquitin-ligase function, by depleting the cullin subunit Cdc53p, would help characterize this complex filamentation regulon, as well as other SCF-dependent pathways in C. albicans. Obtained data indicate that impairment of SCF-dependent ubiquitination leads to filamentation and early cell death in C. albicans and suggest that proper transduction of environmental signals via degradation of transcriptional factors is necessary for accurate induction of the amino acid starvation response, as well as the reductive iron transport across the membrane.
MATERIALS AND METHODS
Strains and culture conditions.
All of the strains used in the present study are listed in Table 1. Strains were cultured in YPD medium (2% peptone, 1% yeast extract, 2% glucose) at 30°C, including 2% agar for solid media. To repress the Tet promoter of the pTet-CDC53/cdc53Δ strain KTY31, a YPD overnight culture was diluted into fresh YPD medium containing 20 μg of doxycycline (Sigma)/ml (50) at an initial optical density of 0.005 or streaked for single colonies onto YPD plates supplemented with 20 μg of doxycycline (stock concentration of 50 mg/ml in ethanol)/ml.
TABLE 1.
C. albicans strains used in this study
| Strain | Genotype | Parent | Source or reference |
|---|---|---|---|
| CaSS1 | his3::hisG/his3::hisG leu2::tetR-GAL4AD-URA3/LEU2 | CAI4 | 58 |
| KTY3 | his3::hisG::HIS3/his3::hisG leu2::tetR-GAL4AD-URA3/LEU2 | CaSS1 | This study |
| KTY25 | his3::hisG/his3::hisG leu2::tetR-GAL4AD-URA3/LEU2 cdc53Δ::HIS3/CDC53 | CaSS1 | This study |
| KTY27 | his3::hisG/his3::hisG leu2::tetR-GAL4AD-URA3/LEU2 CDC53/cdc53Δ::HIS3 | CaSS1 | This study |
| KTY30 | his3::hisG/his3::hisG leu2::tetR-GAL4AD-URA3/LEU2 cdc53Δ::HIS3/pTet-CDC53 | KTY25 | This study |
| KTY31 | his3::hisG/his3::hisG leu2::tetR-GAL4AD-URA3/LEU2 pTet-CDC53/cdc53Δ::HIS3 | KTY27 | This study |
Construction of a conditional C. albicans CDC53 mutant.
Mutant construction was carried out using the GRACE method (58). For the creation of heterozygote mutant strains, wild-type C. albicans strain CaSS1, which expresses a tetracycline-dependent transactivation fusion (TetR-ScGal4AD) protein, was transformed using a sequential PCR-generated disruption cassette that contains a HIS3 selectable marker flanked with CDC53 upstream and downstream homologous sequences to replace one CDC53 allele (orf19.1674, Contig19-10123 or orf19.9243, Contig19-20123; chromosome 3) by homologous recombination. To increase the frequency of homologous recombination, two successive rounds of PCR were used to extend the homologous flanking sequences from 79 bp to ∼160 bp each. The primer pair used for the first round of PCR was MR1034 (5′-CGTATCATAACTTTCAATTTACATTATTCAATTTCATCCTCCTTTTCCTCCTCCTCCTCCTACTCACATCAAATAATCAAGATCTTTCTGTGACTCAATTC) and MR1035 (5′-GGAAAGTAAAAGCAAGAAAAAAGATAGTTTTGTAAAACTGTTTAGTCGGAGGAGGAGGAGCAGCAGCAGGAGG TATAATATGGATTTTAGTCAGTAAC), containing both a region complementary to the template plasmid pHIS3 (58) (plasmid-derived sequences are underlined) and to the CDC53 locus. The primer pair used for the second round of PCR was MR1046 (5′-GAAACTTTTCTTTTTTACCTTCTCCCTTTTAACAAGTTATCTTTTTTTATTTTTGTCTTGTTTTTGTTTTGTTTGATTACGTATCATAAC TTTCAATTTAC) and MR1047 (5′-CTATTTATTTGTAAGATACACCAACATACATCAACATATTTAGATTTATACTTATACTTATATATATATATATTAGGACATGGAAAGTAAAAGCAAGAAAAAAG), containing both a region complementary to the first set of primers (underlined nucleotides) and an extended region of the CDC53 locus. To replace the endogenous CDC53 promoter of the remaining wild-type allele with a repressible tetracycline promoter, the heterozygous strains were transformed using a PCR-generated Tet promoter replacement cassette containing the SAT-1 dominant selectable marker, flanked by sequences homologous to the CDC53 locus, using the primer pair MR1063 (5′-GTTTTGAAGAGTGAGAGAGAGAGAGAGAAAGACTTTAAAATTTCGCCCTTCAAATTTTTGCTCTCTCTTTTTACTTTCAACAATTGTGCAC TCCAGCGTCAAAACTAGAG) and MR1064 (5′-CACTGTGGCGGTAACACCTTGTTCTCATGAGCACCCAAAATATATTCAAGTCCTGGTTGGATGAATGTCCAAGTGGCATTTAAATCACTGAATGGTGGAAGAGTAGATGACATTGATTAATTTAGTGTGTGTATTTG). Since flanking sequences already consisted of at least 94 bp due to enhanced primer synthesis, only one PCR was used for creating the promoter replacement cassette. Due to allelic variances in the CDC53 promoter region, primer MR1063 is homologous to nucleotides −385 to −292 of orf19.1674 and to nucleotides −447 to −354 of orf19.9243, whereas primer MR1064 is homologous to nucleotides −4 to +104 of both orf19.1674 and orf19.9243. Plasmid pSAT1-Tet (58) was used as a template, and the plasmid-derived sequences are underlined. Strain KTY3 was created as a wild-type control for growth and transcriptional expression comparisons by reintroduction of the HIS3 cassette back into one his3-deleted allele of strain CaSS1, using plasmid pHIS3. All strains were analyzed for correct integration by Southern blot analysis.
Transformation of C. albicans.
Transformation of C. albicans strains was carried out by using the improved lithium acetate method (73) with minor modifications. Briefly, strains were cultured in 100 ml of YPD to an optical density of 0.5, washed with 1× lithium acetate (LiAc) solution (100 mM lithium acetate, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA [pH 8.0]), and resuspended in 500 μl of 1× LiAc. Portions (100 μl) of this cell suspension were mixed with 1 to 5 μg of DNA, 100 μg of denatured salmon sperm DNA used as carrier, and 600 μl of 40% (wt/vol) polyethylene glycol 4000-1× LiAc and then incubated overnight at 30°C on an overhead rotor. Cell suspensions were then heat shocked for 15 min at 44°C, and selection for histidine prototrophy was performed on synthetic solid medium lacking histidine and uridine (0.67% nitrogen base, 2% glucose, 2% agar, and 0.2% amino acid mix without histidine and uridine). For the replacement of the CDC53 promoter using the SAT-1 cassette, post-heat shock cells were regenerated for 4 h in 2 ml of YPD at 30°C prior to selecting them on YPD plates containing 200 μg of nourseothricin (clonNAT; Werner BioAgents, Jena, Germany)/ml as described by Reuss et al. (56). Compared to the conditions used by Roemer et al. (58), these conditions (reduction in the regeneration time, from overnight to 4 h, and in the amount of nourseothricin used, from 400 to 200 μg/ml) were found to be sufficient for efficient mutant selection (56).
Southern and Northern blot analyses.
C. albicans genomic DNA was purified by using the glass-bead method as described for S. cerevisiae (61). Total RNA was extracted by using the hot phenol method (76). Southern and Northern blot analyses were performed as described previously (63). Probes used for both Southern and Northern analyses were generated by PCR, gel purified, radiolabeled with [α-32P]dATP using random primers (22), and purified by using Sephadex G-50 QuickSpin columns (Roche) according to the supplier's protocol. Primer sequences are listed in Table S1 in the supplemental material. The signals were visualized and quantified using a Fuji Film imaging plate screen and the MultiGauge v2.3 software (Fuji Film).
Microscopy.
Cdc53p-depleted cells were visualized using a Zeiss Axio-Imager Z1 microscope equipped with a ×63 objective lens. For visualization of nuclei and chitin, cells were fixed in 70% ethanol for 30 min and stained with 1 μg of DAPI (4′,6′-diamidino-2-phenylindole; Sigma) or calcofluor white (Fluorescent Brightener 28; Sigma)/ml for 20 min, respectively. Washings were carried out using 1× phosphate-buffered saline. Stained cells were then visualized by both differential interference contrast and epifluorescence microscopy using a DAPI filter, and pictures were taken with an Axiocam. Wild-type and Cdc53p-depleted colonies were visualized with a Leica MZ FLIII fluorescence stereomicroscope in bright and dark fields with a 0.63 objective lens using magnifications of 0.8 and 6.3, and pictures were taken with an Axiocam.
Assessment of dead cells by propidium iodide staining.
For propidium iodide staining, 100-μl aliquots of cell suspensions were pelleted and resuspended in 200 μl of 1× phosphate-buffered saline, and 2 μl of a 1-mg/ml propidium iodide solution (Sigma) was added. After incubation for 15 min at 30°C, the cells were visualized by using a Zeiss Axio-Imager Z1 microscope equipped with a ×20 objective by both differential interference contrast and epifluorescence microscopy using a rhodamine filter, and pictures were taken with an Axiocam. The percentage of stained cells was assessed by determination of the total cell count, as well as the amount of stained cells per picture. In the case of yeast cells, a total of 240 to 480 cells were analyzed, whereas for filaments, due to a lesser (i.e., 7 to 62) filament count per picture, a total of 11 to 15 pictures were analyzed per time point, totaling 306 to 509 filaments per time point. For a positive control, yeast cells, as well as filaments were heated to 65°C for 40 min prior to treatment with propidium iodide.
Microarray analysis.
Transcription profiling and analysis were performed using long oligonucleotide microarrays as described elsewhere (52). Cy3- and Cy5-labeled cDNA probes were prepared from 25 μg of total RNA each and hybridized on microarrays spotted with 70mer oligonucleotide probes for each of the 6,354 genes of Assembly 19 (http://www.candidagenome.org/) that were confirmed and characterized by the Candida Annotation Working Group (13). In all, four hybridizations of independently produced RNA preparations with reciprocal labeling were carried out. Microarrays were scanned by using a ScanArray Lite microarray scanner (Packard Bioscience), and fluorescence intensities were quantified by using QuantArray. Lowess normalization and statistical analysis were performed by using Genespring v7 (Agilent Technologies). Genes were considered to be differentially expressed if their average fold change in expression was ≥2 and if the average fold change was statistically significant as determined by Student t test (P ≤ 0.05). The complete microarray data produced in the present study are available in Table S2 in the supplemental material. It has also been deposited in the NCBI Gene Expression Omnibus (20) and is accessible through GEO series accession number GSE13976 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13976). Gene standard names and GO terms used for the analysis were taken from Assembly 21 of the Candida Genome Database (http://www.candidagenome.org/), as well as S. cerevisiae orthologs/best hits for differentially expressed C. albicans genes in Table S2 in the supplemental material.
RESULTS
Identification of C. albicans CDC53.
To identify the CDC53 gene in C. albicans, a BLAST search of the S. cerevisiae cullin Cdc53p (815 amino acids) against the C. albicans genome was carried out, using BLASTP (protein/protein) at the CandidaDB web server (http://genodb.pasteur.fr/cgi-bin/WebObjects/CandidaDB). This search revealed as closest homologue of the ScCdc53p protein the gene product of orf19.1674, with an overall amino acid identity of 38% and similarity of 57%. A protein domain search in the Protein Knowledgebase UniProtKB (http://beta.uniprot.org/) confirmed a cullin domain represented by the Pfam accession number PF00888. Detailed analysis revealed that C. albicans orf19.1674 encodes a 752-amino-acid protein that, like S. cerevisiae Cdc53p, contains an N-terminal region, consisting of three cullin repeats necessary for interaction with the F-box linking protein Skp1p, and a C-terminal globular α/β domain containing the E3 ligase binding motif (79).
Construction of a conditional CDC53 C. albicans mutant strain.
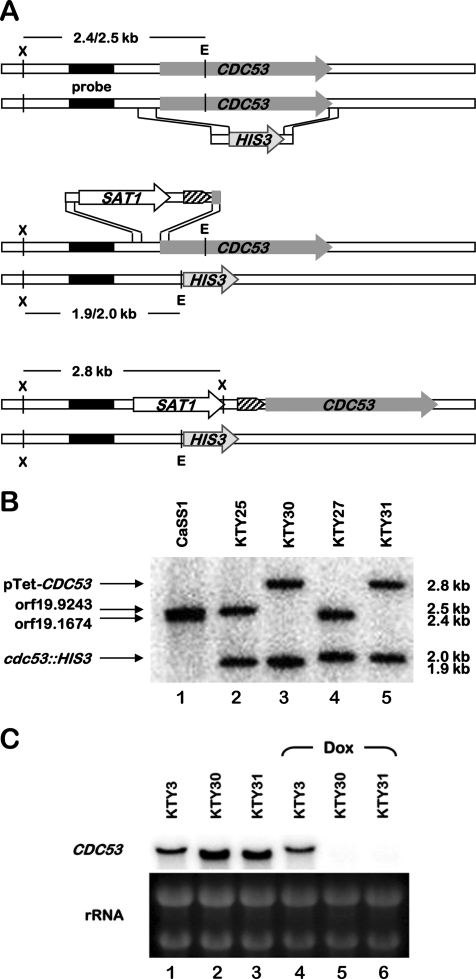
To investigate the role of the cullin Cdc53p in C. albicans and given the essentiality of S. cerevisiae Cdc53p (43), we constructed a conditional CDC53 mutant using the GRACE technology (58) (Fig. 1). This consisted of deleting one allele of CDC53 and exchanging the endogenous CDC53 promoter of the remaining wild-type allele with a repressible tetracycline promoter, using a PCR-based methodology (58). For the deletion of one CDC53 allele, a deletion cassette composed of the HIS3 marker flanked by CDC53 upstream and downstream regions was amplified and was used to transform strain CaSS1 which expresses a chimeric tetracycline transactivator (Fig. 1A, upper panel, and see Materials and Methods). The resulting His+ transformants were analyzed by Southern blotting, along with the CaSS1 strain, using EcoRV- and XbaI-digested genomic DNA and a CDC53 upstream probe (Fig. 1B). In strain CaSS1, the probe detected a doublet band of 2.5 kb (Fig. 1B, lane 1). Genome sequence analysis of strain SC5314, from which CaSS1 is derived (58), showed that these two XbaI-EcoRV fragments (2,428 bp for orf19.1674 and 2,488 bp for orf19.9243) result from allelic variations (base deletion and/or addition) in the promoter region of CDC53. The Southern blot confirmed that one allele of CDC53 was properly deleted in each of the two heterozygous mutants analyzed (KTY25 and KTY27), as judged by the appearance of either a 1.9- or a 2.0-kb fragment (Fig. 1B, lanes 2 and 4). orf19.1674 was deleted in strain KTY25 (cdc53Δ::HIS3/CDC53), and orf19.9243 was deleted in strain KTY27 (CDC53/cdc53Δ::HIS3), based upon the disappearance of the shorter and longer fragments from the doublet, respectively (Fig. 1B, lanes 2 and 4). A second round of transformation was carried out in KTY25 and KTY27, using a PCR amplicon that consisted of the tetracycline-repressible promoter linked to the SAT1 dominant marker and flanking regions of the CDC53 promoter region (Fig. 1A, middle panel, and see Materials and Methods). Two independent Nour transformants, KTY30 (cdc53Δ::HIS3/pTet-CDC53, derived from KTY25) and KTY31 (pTet-CDC53/cdc53Δ::HIS3, derived from KTY27), were analyzed by Southern blot using the same restriction enzymes and probe as for the first round of strain construction. The disappearance of the remaining wild-type fragment of the 2.5-kb doublet and the appearance of a 2.8-kb fragment confirmed a correct promoter replacement (Fig. 1B, lanes 3 and 5). To evaluate CDC53 gene function independent of auxotrophy differences, the control strain KTY3 was created through reconstitution of the HIS3 locus in CaSS1 (see Materials and Methods), and correct integration was confirmed by Southern blot analysis (data not shown).
FIG. 1.
Creation of a conditional CDC53 mutant strain. (A) For the creation of heterozygote mutant strains, one allele of CDC53 of wild-type C. albicans strain CaSS1 was replaced by the HIS3 marker (top panel). The SAT1 dominant selectable marker was used to replace the endogenous CDC53 promoter of the remaining wild-type allele by a repressible tetracycline promoter (block arrow with diagonal pattern) (middle panel). The two alleles of the resulting strain are depicted in the bottom panel. The probe used for Southern blot analysis is indicated (filled rectangle) as well as the expected restriction fragment lengths for the different alleles. E, EcoRV; X, XbaI. (B) Southern blot analysis of the parental strain CaSS1 and the mutants KTY25 (cdc53Δ::HIS3/CDC53), KTY30 (cdc53Δ::HIS3/pTet-CDC53), KTY27 (CDC53/cdc53Δ::HIS3), and KTY31 (pTet-CDC53/cdc53Δ::HIS3) using the restriction enzymes and probe shown in panel A. The different alleles and the sizes of the corresponding restriction fragments are indicated at the left and the right sides of the figure, respectively. (C) Northern blot analysis of the control strain KTY3 and the mutants KTY30 and KTY31 after 12 h of growth at 37°C in YPD in the absence or presence of 20 μg of doxycycline/ml (Dox). rRNAs were used as loading controls.
To confirm conditional CDC53 expression in strains KTY30 and KTY31, both strains, as well as the control strain KTY3, were analyzed by Northern blotting after growth in the absence or the presence of doxycycline, a tetracycline derivative (Fig. 1C). For strains grown in the absence of doxycycline, an increase in signal intensity of CDC53 transcripts was detected for strains KTY30 and KTY31 compared to the control strain KTY3, suggesting that the constitutively expressed tetracycline-repressible promoter is stronger than the endogenous CDC53 promoter. In addition, the length of the CDC53 transcripts derived from the Tet-repressible promoter appeared to be slightly shorter compared to those derived from the endogenous promoter (Fig. 1C, compare lanes 2 and 3 to lane 1). This is potentially due to different transcription start sites giving rise to shorter 5′-untranslated regions in the mRNA species encoded by the pTet-CDC53 allele. After 12 h of growth in YPD in the presence of 20 μg of doxycycline/ml, no CDC53 transcript could be detected in strains KTY30 and KTY31 (even after prolonged exposure [data not shown]), while the intensity of the CDC53 transcript remained unchanged in the control strain KTY3, confirming that these strains and growth conditions were suitable for the characterization of CDC53 in C. albicans.
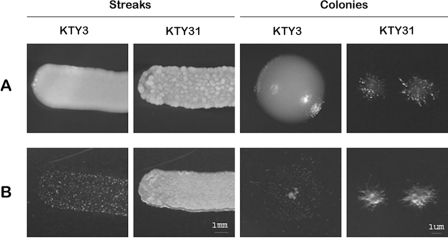
Suppression of CDC53 of C. albicans promotes an invasive phenotype.
On rich medium (YPD) at 30°C, the control strain KTY3, as well as the conditional CDC53 strain KTY31, typically exhibited round, smooth colonies under nonrepressing conditions (data not shown). Under identical growth conditions but in the presence of doxycycline, strain KTY3 still grew as round soft colonies, whereas strain KTY31 displayed a deeply wrinkled surface if plated as a single streak and grew as very small single colonies, which were of a solid consistency and formed protrusions into the agar (Fig. 2A). Upon removal of both streaks and single colonies by washing, a nearly clean agar surface with rare single spots of cells was obtained for strain KTY3, whereas for strain KTY31, only the upper layer of the streaks could be washed off, leaving a complete layer of cells imprinted in the agar (Fig. 2B). This phenotype was even stronger with single colonies, since no cells whatsoever could be washed off, confirming an invasive phenotype of the Cdc53p-depleted strain. Strain KTY30 behaved as strain KTY31 (data not shown); therefore, only one strain (KTY31) was used in the subsequent experiments.
FIG. 2.
Depletion of Cdc53p promotes invasive growth in C. albicans. (A) Macroscopic morphologies of streaks and single colonies of strains KTY3 and KTY31 grown on YPD for 24 h in the presence of doxycycline. (B) To confirm invasiveness for strain KTY31, both streaks and single colonies of strains KTY3 and KTY31 were washed with water and photographed in a dark field.
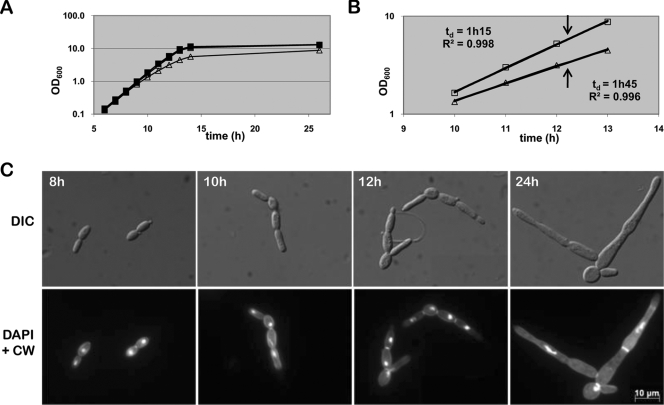
Cdc53p depletion leads to filament formation under yeast growth conditions.
A wrinkled colony surface and agar invasiveness of colonies is often displayed by cells growing as filaments. To evaluate whether these features exhibited by Cdc53p-depleted cells under conditions favoring yeast growth mode were caused by filament formation, we determined the morphology of both control and CDC53-repressing cells during exponential growth mode in rich medium at 30°C by microscopic analysis. Overnight cultures of strains KTY3 and KTY31 were diluted to an initial optical density of 0.005 into fresh YPD medium in the absence or the presence of doxycycline and were cultured at 30°C. After 8 h of induction, the growth curves of strain KTY31 under repressing conditions, and the controls (KTY3 with or without doxycycline and KTY31 without doxycycline) started to separate, reflected in the doubling time of 1 h and 45 min for the mutant strain compared to 1 h and 15 min for the controls. Furthermore, the optical density of the Cdc53p-depleted strain in stationary phase reached only about two-thirds that of the controls (optical density at 600 nm = 8 compared to 12, Fig. 3A and B). To determine whether the reduced growth rate of strain KTY31 in the presence of doxycycline was due to a change in morphology, we investigated the growth of Cdc53p-depleted cells by microscopic analysis (Fig. 3C). After 7 to 8 h of cultivation, when growth curves started to separate, we detected the occurrence of initial cell elongation, with subsequent filament extension taking place well into the stationary phase. Calcofluor white and DAPI staining of exponential growing cells displayed one nucleus per filament, as well as septation at the mother bud neck and at further constrictions, two characteristics of pseudohyphae (66). Cells in stationary phase revealed filament compartments partly missing nuclei, whereas septa were located both at constriction sites and in nonconstricted filaments, with the latter being characteristic for real hyphae (66), thus indicating a mixed population of pseudohyphae and true hyphae in one filament cluster (Fig. 3C).
FIG. 3.
Cdc53p functions as a repressor of filament development. (A) Growth curve of strains KTY3 (squares) and KTY31 (triangles) in liquid YPD. Closed shapes indicate growth in the absence of doxycycline, whereas open shapes represent growth in the presence of doxycycline. Due to superimposition of the three growth curves of KTY3 with or without doxycycline and KTY31 without doxycycline, only the closed squares of KTY3 without doxycycline can be seen. (B) Determination of exponential growth phase and growth rate of the control strain KTY3 (squares) and the cdc53 mutant strain KTY31 (triangles), both in the presence of doxycycline. The coefficients of determination (R2) were calculated using the R-square function of Excel. The arrows indicate the time point of cell harvest of both strains for microarray analysis. (C) Time course microscopic analysis of cells of strain KTY31 under Cdc53p-depleting conditions. Calcofluor white and DAPI staining of exponential grown cells display septa at mother bud necks and at further constrictions, indicating pseudohyphal growth mode. Stationary-phase cells (24 h) are more elongated, with location of septa indicating a mix of pseudohyphae and hyphae.
Depletion of the cullin Cdc53p leads to an early onset of cell death.
The maldistribution of nuclei per filament as detected by DAPI and calcofluor white staining as well as the single colony growth phenotype of Cdc53p-depleted cells suggested early cell death. The growth arrest of single CDC53- repressed colonies after 24 h was only observed if they were not in contact with other colonies. If plated as a streak, cells kept growing as filaments, forming a deeply wrinkled surface, possibly due to some Doxycycline depletion effect by neighboring colonies (Fig. 2). Therefore, we assessed cell viability of the conditional CDC53 strain under repressing conditions (KTY31 plus doxycycline) in comparison to the controls (KTY3 with or without doxycycline and KTY31 without doxycycline) by propidium iodide staining, a fluorescent compound that cannot penetrate viable cells (19). The percentage of stained and therefore dead cells/filaments was determined every 24 h over a time-period of 5 days (Table 2). Heat-killed C. albicans yeast and hyphal cells were used as positive control, for which 100% staining was obtained. The percentage of dead filaments in Cdc53p-depleted cells rose continually from 3% after 24 h to 28% after 96 h, during which time no significant cell death could be observed in the controls (Table 2). The assessment of dead cells was abandoned after 96 h since excessive filament clustering in the Cdc53p-depleted cells hindered accurate filament counting after that time point. These data clearly confirm an early onset of cell death for cells depleted in Cdc53p.
TABLE 2.
Propidium iodide staining of Cdc53p-depleted cells (KTY31 doxycycline) and controls
| Strain | % Cells dead or alivea at:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 24 h
|
48 h
|
72 h
|
96 h
|
|||||
| Dead | Alive | Dead | Alive | Dead | Alive | Dead | Alive | |
| KTY3 | 0 | 100 | 0 | 100 | 0 | 100 | 0 | 100 |
| KTY3 Dox | 0 | 100 | 0 | 100 | 0 | 100 | 0 | 100 |
| KTY31 | 0 | 100 | 0 | 100 | 0 | 100 | 0 | 100 |
| KTY31 Dox | 3 | 97 | 8 | 92 | 15 | 85 | 28 | 72 |
The numbers of dead (propidium iodide-positive) and alive (propidium iodide-negative) cells are given as a percentage. Dox, doxycycline.
Genome-wide transcriptional analysis upon Cdc53p depletion.
In order to characterize the consequences of CDC53 downregulation at the molecular level and to identify potential Cdc53p-dependent pathways in C. albicans, we analyzed the genome-wide expression profile of the mutant strain KTY31 compared to the control strain KTY3, using DNA microarrays. Four independent cell cultures were grown in YPD in the presence of doxycycline and harvested after 12 h of growth, a condition under which CDC53-repressed cells displayed a continuous pseudohyphal growth phenotype, distinct from both the initial cell elongation phase occurring after 8 h and the mix of pseudohyphae and real hyphae of later time points (Fig. 3C). To ensure that the harvested cells were in exponential growth mode, the growth of both strains KTY31 and KTY3 was monitored between 10 and 13 h. A linear regression was performed on the log of the optical density at 600 nm values, returning coefficients of determination (R2) of greater than 0.996, thus confirming exponential growth (Fig. 3B). Microarray analysis was carried out with a total of four hybridizations of independently produced RNA preparations, including reciprocal labeling. Genes were considered differentially expressed if the calculated modulation was at least twofold and statistically significant by t test (P < 0.5). Using these criteria, a total of 423 genes were found to be differentially expressed with 190 upregulated and 233 downregulated genes when comparing the conditional CDC53 mutant strain KTY31 to the control strain KTY3.
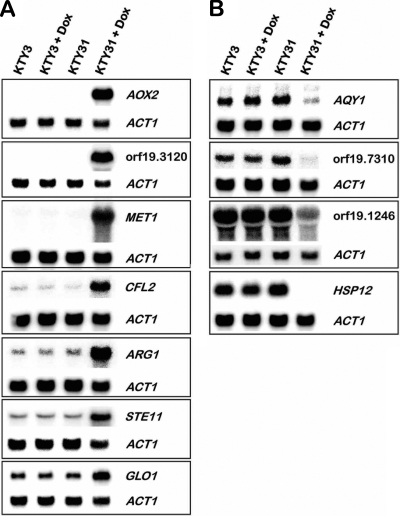
Differential regulation of selected genes was tested by Northern blot analysis (Fig. 4). In order to obtain an independent validation, a distinct set of RNA preparations than the one used for the microarray hybridizations was prepared. The resulting signals revealed a strong up- or downregulation of the selected genes, confirming the expression data obtained by the DNA microarray experiment. Included in the Northern blot analysis were strains KTY3 and KTY31 grown in the absence of doxycycline, which displayed the same gene expression levels as the control strain KTY3 in the presence of doxycycline, indicating that this drug did not affect the expression of genes monitored by Northern blotting under the conditions used.
FIG. 4.
Northern blot validation of microarray results. (A) Upregulation of indicated genes comparing strains KTY3 and KTY31 in the absence or the presence of doxycycline. ACT1 expression was used as loading control. (B) Downregulation of indicated genes comparing strains KTY3 and KTY31 in the absence or presence of doxycycline. ACT1 expression was used as a loading control.
The differentially regulated genes were further analyzed for significantly overrepresented GO terms of the ontology biological process. By applying a P value of <0.01, eight GO term clusters could be distinguished (I to VIII) (Table 3). The highest significance was obtained for the cluster containing the parental terms sulfur metabolic process, nitrogen compound metabolic process, amino acid and derivative metabolic process, and organic acid metabolic process (Table 3, cluster I). Following in significance were several daughter terms of the GO terms transport (ion, lipid, oligopeptide, vitamin, and basic amino acid transport) and response to stimulus (response to temperature stimulus and response to oxidative stress), although both parental terms were not significantly overrepresented (P values of 0.3 and 0.31, respectively).
TABLE 3.
GO terms of the ontology biological process that are significantly overrepresented by Cdc53p depletion
| Cluster | GO term description | Pa | No. of regulated genesb | Background frequencyc |
|---|---|---|---|---|
| I | Sulfur metabolic process | 1.32E−09 | 18 | 48 |
| Nitrogen compound metabolic process | 7.41E−09 | 42 | 226 | |
| Amino acid and derivative metabolic process | 7.94E−09 | 36 | 178 | |
| Organic acid metabolic process | 3.54E−08 | 47 | 278 | |
| II | Transport | 0.3 | 54 | 687 |
| Ion transport | 1.09E−05 | 19 | 90 | |
| Lipid transport | 4.56E−04 | 6 | 16 | |
| Oligopeptide transport | 7.94E−04 | 5 | 12 | |
| Vitamin transport | 1.21E−03 | 3 | 4 | |
| Basic amino acid transport | 9.08E−03 | 3 | 7 | |
| III | Response to stimulus | 0.31 | 33 | 409 |
| Response to temp stimulus | 3.52E−04 | 7 | 21 | |
| Response to oxidative stress | 5.90E−04 | 11 | 51 | |
| IV | Respiratory chain complex IV assembly | 1.23E−03 | 4 | 8 |
| V | Ethanol metabolic process | 2.09E−03 | 4 | 9 |
| VI | Cell separation during cytokinesis | 3.47E−03 | 5 | 16 |
| VII | Fermentation | 3.47E−03 | 5 | 16 |
| VIII | Chromatin assembly or disassembly | 3.91E−03 | 10 | 55 |
That is, the significance of overrepresentation of listed GO terms comparing the transcriptional profiles of strains KTY31 to KTY3 in the presence of doxycycline.
That is, the number of differentially regulated genes belonging to the listed GO terms.
That is, the number of genes belonging to listed GO terms on a genome-wide level.
Analysis of the complex cluster I revealed that all four parental terms include as most significant daughter term the GO term amino acid metabolic process or its daughter term sulfur amino acid metabolic process. Considering that Cdc53p is the backbone of several SCF ligase complexes, downregulation of the CDC53 gene would most likely result in a defect in ubiquitination, therefore either stabilizing specific proteins (including transcriptional regulators) since they would be no longer marked for proteasomal degradation or failing to activate them by monoubiquitination. To analyze the transcriptional response of the amino acid metabolism genes to Cdc53p depletion, we searched the Candida Genome Database for known transcriptional regulators of those genes (http://www.candidagenome.org). Interestingly, we found that 14 of the 36 genes in that category are regulated by the transcriptional regulator Gcn4p under amino acid starvation conditions (68), with all 14 genes being exclusively upregulated in Cdc53p-depleted cells (Table 4), suggesting that Cdc53p negatively regulates Gcn4p in C. albicans as in S. cerevisiae (33) and that failed degradation of Gcn4p could be responsible for the amino acid starvation-like transcriptional response caused by C. albicans Cdc53p depletion (see Discussion).
TABLE 4.
Regulation by known transcription factors of genes differentially expressed upon Cdc53p depletion of the GO term amino acid metabolic process
| Gene group | No. of regulated genes (n = 36)a | Regulation of KTY31 vs KTY3 | Gene name (open reading frame no.) | Regulation typeb |
|---|---|---|---|---|
| 1 | 14 | Up | ARG1 (orf19.7469), ARG3 (orf19.5610), ARO8 (orf19.2098), ARG8 (orf19.3770), ARG5,6 (orf19.4788), ECM42 (orf19.6500), MET15 (orf19.5645), LYS9 (orf19.7448), LYS22 (orf19.4506), orf19.2092, MET6 (orf19.2551), BAT21 (orf19.797), BAT22 (orf19.6994), HIS7 (orf19.5505) | Gcn4p |
| 2 | 4 | Up | MET1 (orf19.5811), MET3 (orf19.5025), CYS3 (orf19.6402), MAE1 (orf19.3419) | Other |
| 3 | 3 | Down | PUT1 (orf19.4274), CAR1 (orf19.3934), CHA1 (orf19.1996) | |
| 4 | 9 | Up | CPA1 (orf19.4630), orf19.5842, orf19.3221, ZCF33 (orf19.5992), ARG4 (orf19.6689), MET14 (orf19.946), orf19.1159, LEU2 (orf19.7080), IDP1 (orf19.5211) | ? |
| 5 | 6 | Down | CBF1 (orf19.2876), MET28 (orf19.7046), CAR2 (orf19.5641), GAD1 (orf19.1153), PUT2 (orf19.3974), ARO10 (orf19.1847) |
That is, the number of differentially regulated genes belonging to the listed GO terms.
That is, the regulating factor for this group of genes. “Other” includes Hog1p, Rim101p, Ssn6p, Sfu1p, Nrg1p, Mig1p, and Cap1p.
The second most significantly overrepresented GO term cluster is represented by the parental term transport with five significantly overrepresented subcategories totaling 35 genes (Table 5), suggesting a major role for Cdc53p in regulating the homeostasis of the cell in response to its environment. Interestingly, 13 of the 19 ion transport genes belonged to the subcategory metal ion transport, of which 11 were upregulated. Further analysis of these genes identified nine genes potentially involved in the copper-dependent reductive iron transport across the membrane: the ferric reductases CFL2 and CFL5, the plasma membrane multicopper oxidases FET3 and FET34, the high-affinity iron permeases FTR1 and FTR2, the cytosolic metallochaperone ATX1, and the copper transporters CTR1 and P-type ATPase CRP1 (Table 5). Two more ion transport genes, orf19.2006.1 and orf19.4967, are homologues of the S. cerevisiae genes COX17 and COX19, respectively, coding for two putative metallochaperones involved in the transfer of copper to the cytochrome c oxidase. The upregulation of these genes under CDC53-repressed conditions indicates an important role for Cdc53p in regulating intracellular copper transport and iron acquisition (see Discussion).
TABLE 5.
Subclassification of the GO term transport of the transcriptional profile of Cdc53p-depleted cells
| GO term | Pa | No. of regulated genesb | Background frequencyc | Regulation of KTY31 vs KTY3 | Gene name (open reading frame no.) |
|---|---|---|---|---|---|
| Transport | 0.3 | 54 | 687 | ||
| Ion transport | 1.09E-05 | 19 | 90 | ||
| Metal ion transport | 1.08E-04 | 11 | 57 | Up | CFL2 (orf19.1264), CTR1 (orf19.3646), FTR1 (orf19.7219), CRP1 (orf19.4784), orf19.2006.1, FET3 (orf19.4211), FET34 (orf19.1206), COX19 (orf19.4967), CFL5 (orf19.1930), FTR2 (orf19.7231), ATX1 (orf19.2369.1) |
| 2 | Down | orf19.4940, ENA21 (orf19.5170) | |||
| Other | 4 | Up | PHO87 (orf19.2454), SSU1 (orf19.7313), SUL2 (orf19.2738), MEP1 (orf19.1614) | ||
| 2 | Down | orf19.2959.1, PHO84 (orf19.1172) | |||
| Lipid transport | 4.56E-04 | 1 | 16 | Up | orf19.4342 |
| 5 | Down | RTA2 (orf19.24), GIT2 (orf19.1978), RTA3 (orf19.23), GIT1 (orf19.1979), orf19.34 | |||
| Oligopeptide transport | 7.94E-04 | 5 | 12 | Down | OPT4 (orf19.176), PTR2 (orf19.6937), IFC3 (orf19.3749), OPT9 (orf19.2584), OPT1 (orf19.2602) |
| Vitamin transport | 1.21E-03 | 1 | 4 | Up | TNA1 (orf19.4335) |
| 2 | Down | PHO113 (orf19.2619), PHO112 (orf19.3727) | |||
| Basic amino acid transport | 9.08E-03 | 1 | 7 | Up | CAN2 (orf19.111) |
| 2 | Down | ALP1 (orf19.2337), orf19.4940 |
That is, the significance of overrepresentation of the listed GO terms comparing the transcriptional profiles of strains KTY31 to KTY3 in the presence of doxycycline.
That is, the number of differentially regulated genes belonging to the listed GO terms.
That is, the number of genes belonging to the listed GO terms on a genome-wide level.
Even though Cdc53p-depleted cells grew clearly as pseudohyphae at the time point of cell harvest and RNA preparation, neither the GO term filamentous growth nor any of its daughter terms were significantly overrepresented. However, we identified several differentially regulated genes linked to that category (Table 6). Upregulation was detected for the genes ALS2, ALS4, and UTR2, which encode adhesion molecules and are part of a set of genes shown to be upregulated in filamenting cells (1, 78). Differential regulation was also identified for two transcription factors involved in filamentation, CPH2 and CZF1 although, surprisingly, these genes were downregulated by twofold, and no upregulation of any of the remaining known transcriptional activators of filamentation could be detected. However, we detected a strong upregulation of STE11, the potential MAPKKK in the filamentation MAPK pathway (15), whose downstream effector is the transcriptional regulator Cph1p. Interestingly, the forkhead transcription factor FKH2, as well as the pololike kinase CDC5, were downregulated by twofold; deletion of these genes results in constitutive pseudohyphal growth (9) or elongated buds (4), respectively. The constitutive filamentous growth phenotype of Cdc53p-depleted cells together with the differential regulation of several filamentation-associated genes indicates an important role for Cdc53p in regulating growth morphology in C. albicans (see Discussion).
TABLE 6.
Subclassification of the GO term filamentous growth of the transcriptional profile of Cdc53p-depleted cells
| GO term | Pa | No. of regulated genesb | Background frequencyc | Regulation of KTY31 vs KTY3 | Gene name (open reading frame no.) |
|---|---|---|---|---|---|
| Filamentous growth | 0.82 | 23 | 323 | ||
| Invasive growth (sensu Saccharomyces) | 0.08 | 1 | 15 | Up | ALS4 (orf19.4555) |
| 2 | Down | CDC14 (orf19.4192), GAL10 (orf19.3672) | |||
| Pseudohyphal growth | 0.10 | 2 | 36 | Up | STE11 (orf19.844), ALS4 (orf19.4555) |
| 3 | Down | FKH2 (orf19.5389), ASH1 (orf19.5343), CPH2 (orf19.1187) | |||
| Signal transduction during filamentous growth | 0.43 | 1 | 8 | Down | CDC5 (orf19.6010) |
| Hyphal growth | 0.73 | 3 | 139 | Up | ALS2 (orf19.1097), UTR2 (orf19.1671), ALS4 (orf19.4555) |
| 5 | Down | FKH2 (orf19.5389), MCM1 (orf19.7025), CDC14 (orf19.4192), CPH2 (orf19.1187), SSK1 (orf19.5031) | |||
| Other | 4 | Up | SSU1 (orf19.7313), PHHB (orf19.2079), FGR3 (orf19.3845), FGR43 (orf19.4786) | ||
| 7 | Down | CLB4 (orf19.7186), CZF1 (orf19.3127), orf19.6720, ECM4 (orf19.2613), GAT2 (orf19.4056), CHA1 (orf19.1996), FGR41 (orf19.4910) |
That is, the significance of overrepresentation of listed GO terms comparing the transcriptional profiles of strains KTY31 to KTY3 in the presence of doxycycline.
That is, the number of differentially regulated genes belonging to the listed GO terms.
That is, the number of genes belonging to the listed GO terms on a genome-wide level.
Whereas classification by GO terms is valuable for the determination of Cdc53p-dependent pathways, analysis of individual differentially transcribed genes can also yield important clues in the involvement of Cdc53p activity in different cellular processes (see Table S2 in the supplemental material). The strongest upregulation in Cdc53p-depleted cells was detected for AOX2 (orf19.4773, 195-fold), an alternative oxidase that is involved in the cyanide-resistant respiratory pathway (30). Two further differentially transcribed genes that were not part of our GO term analysis presented above are FAD3 (orf19.4993) and FAD2 (orf19.118), which were upregulated upon Cdc53p depletion by six- and twofold, respectively. These genes encode fatty acid desaturases that are involved in α-linolenic acid and linoleic acid synthesis, respectively, which in turn are major membrane components (49). Furthermore, we detected strong downregulation of the chitinases CHT2 (orf19.3895, 6-fold) and CHT3 (orf19.7586, 17-fold), two important factors in the homeostasis of chitin, a cell wall component that contributes to its mechanical strength and is enriched at septa (44). The differential regulation of these genes, together with transcriptional changes (in both directions) of several more genes, which affect membrane composition, cell wall integrity, and cell-cell interaction (e.g., genes coding for glycosylphosphatidylinositol-anchored proteins), suggests that Cdc53p is a major regulator of fungal growth, development, and survival.
DISCUSSION
In S. cerevisiae, the SCFCdc4 complex is responsible for degrading the cell cycle inhibitor Sic1p (75). Mutants in one of the proteins needed for ubiquitination-dependent degradation of Sic1p, including Cdc53p, display multiple elongated buds and arrest in the G1 phase (24, 43). Our study indicates that repression of CDC53 in C. albicans rather results in cell elongation and subsequent pseudohypha formation (Fig. 3C), the cells probably evading immediate cell cycle arrest by switching to a filamentous growth mode. This is in accordance with the demonstrated ability of C. albicans to form polarized evaginations in all stages of the cell cycle, therefore uncoupling hyphal elongation and cell cycle morphogenesis programs (25). However, the occurrence of a mix of pseudohyphae and real hyphae after 24 h of growth under CDC53 repressing conditions that seem to be defective in cytokinesis and septation, as can be observed by the missing nuclei and septa in/between part of the filaments (Fig. 3C), suggests that finally cell death prevails. This proposition is strengthened by the growth phenotype of single colonies depleted in Cdc53p, which stop growing after 24 h (Fig. 2), as well as the results of the propidium iodide staining, which confirmed an earlier onset of cell death for Cdc53p-depleted cells compared to the controls (Table 2). In addition, we were unsuccessful in creating a homozygous knockout mutant, with both alleles of CDC53 deleted, a further evidence of the essentiality of C. albicans CDC53, similar to S. cerevisiae CDC53 (43). This growth defect of Cdc53p-depleted cells is specific for the cullin moiety of the C. albicans SCFCdc4 complex, since C. albicans homozygous cdc4−/− cells grow as air-protruding filaments (3). The different growth response of C. albicans and S. cerevisiae to Cdc53p depletion, as well as the viability of C. albicans cdc4−/− cells compared to the essentiality of the ScCDC4 gene may be related to the different functions of the cell cycle inhibitor Sic1p of S. cerevisiae and its homologue in C. albicans, Sol1p, since stabilization of Sol1p leads to only a slight delay in G1-S phase progression, contrary to the cell cycle arrest in G1 phase caused by stabilization of Sic1p (3). The finding that C. albicans Cdc53p is essential, coupled to the fact that proteins which are part of SCF-E2 complexes are not (e.g., Cdc4p and Grr1p [3, 39]), suggests that the essentiality of Cdc53p is potentially caused by the combined dysfunction of several/all SCF ligases (synthetic lethal effect), highlighting the importance of Cdc53p in C. albicans cell physiology. However, it is also possible that Cdc53p essentiality results from its interaction with a single essential protein that remains to be identified. The genetic analysis of other components of potential SCF-E2 complexes in C. albicans will help in answering this question.
The identification of the pathway involved in the Cdc53p-dependent filamentation by transcriptional profiling was limited, since no general transcriptional filamentation response could be detected (35, 48, 51). In fact, the transcripts of CPH2 and CZF1, two genes coding for transcriptional activators of filamentous growth (14, 36), were found to be downregulated in Cdc53p-depleted cells (Table 6). On the other hand, we detected a strong upregulation of STE11 encoding a MAPKKK (fivefold), indicating a possible involvement of the MAPK pathway in filamentation via its downstream effector Cph1p. Although no differential regulation for the majority of Cph1p-dependent genes, e.g., HWP1, HYR1, and ECE1 (35), was identified in the CDC53 repressed strain by genome-wide transcriptional profiling, HWP1 could be detected as upregulated by Northern blot analysis using the same RNA preparations as for the microarray hybridization experiment (data not shown). Hence, the highly stringent criteria used for the DNA microarray data analysis could have caused some filamentation-dependent genes to evade detection (HWP1 and HYR1 were upregulated by 1.2- and 1.4-fold, respectively; see Table S2 in the supplemental material). Therefore, the involvement of this pathway in the filamentation response of Cdc53p-depleted cells cannot be excluded. Transcriptional changes of other factors that are consistent with the pseudohyphal growth phenotype of the conditional CDC53 strain are the downregulation of the forkhead transcription factor FKH2 (9), as well as the downregulation of the pololike kinase CDC5 (4) although, apart from downregulation of the histone machinery (Table 3, group VIII), the expression profile of the cdc5 mutant does not correlate with the one obtained upon Cdc53p depletion. Considering these results, it seems less likely that a single transcriptional activation pathway is responsible for the filamentation phenotype of Cdc53p-depleted cells but rather that a combination of different effects caused by the failed degradation and/or activation of several proteins.
A very clear transcriptional response to Cdc53p depletion was obtained with the GO term amino acid metabolic process, similar to the transcriptional profile of amino acid-starved cells. In C. albicans, the amino acid starvation response is directly dependent on the transcriptional regulator Gcn4p, a functional homologue of ScGcn4p (69). Whereas in S. cerevisiae the amino acid starvation response is regulated mainly at the translational level by the eIF2α kinase ScGcn2p (reviewed in references 27 and 28), activation of the amino acid starvation response in C. albicans depends mainly on the transcriptional regulation of the GCN4 gene (68). The activity of ScGcn4p is further regulated at the posttranslational level by proteasomal degradation, mediated by phosphorylation by the Cyclin Pcl5-dependent kinase Pho85 (64) and subsequent ubiquitination by the SCFCdc4-Cdc34p complex (45). Similarly, CaGcn4p, which is produced in rich medium, is rapidly degraded with a half-life of less than 5 min in response to amino acid availability (23, 69). Although it has been shown that degradation of CaGcn4p is also dependent on phosphorylation by the cyclin Pcl5-dependent kinase Pho85 (23), the ubiquitin ligase involved in that process remains to be identified. The transcriptome analysis presented here revealed upregulation of amino acid starvation response genes under Cdc53p-depleted conditions, a finding consistent with an increase in Gcn4p activity. Since the fluorescence signal intensities indicate high abundance of the GCN4 transcript in rich medium without a change in transcript levels between the wild-type and the Cdc53p-depleted cells, a potential increase in Gcn4p activity would appear to be mainly regulated at the protein degradation level, suggesting the involvement of a Cdc53p-containing SCF complex in the degradation of Gcn4p in C. albicans. Interestingly, it has been shown that amino acid starvation also induces a filamentation response in C. albicans (54, 69), with Gcn4p stimulating pseudohyphal growth in an Efg1- and partially Ras1p-dependent fashion, but independent of Cph1p, the downstream effector of the MAPK pathway (69). Therefore, the observed filamentation of C. albicans cells depleted of Cdc53p could be mediated at least in part by nondegraded Gcn4p.
A similar Gcn4p-dependent response to mutations in the SCF-E2 complex components CDC53 and CDC34 in S. cerevisiae was identified in a genome-wide transcriptional analysis carried out by Varelas et al. (72), with the GO term amino acid metabolism identified in their study, a daughter term of the GO terms amino acid and derivative metabolic process and organic acid metabolic process identified in our study (Table 3, cluster I). These authors also detected transcriptional changes of genes involved in sulfur utilization, which they attributed to alterations of the transcription factor Met4p in the S. cerevisiae cdc53-1 and cdc34-2 mutants (72). Likewise, downregulation of CDC53 in C. albicans resulted in differential regulation of 7 of 12 sulfur utilization genes (P = 4.68E−06), a daughter GO term of the sulfur metabolic process (Table 3, cluster I), possibly due to changes in the availability of the as-yet-uncharacterized potential SCF-target CaMet28p, the homolog of ScMet4p. Regarding the GO term cell wall organization, only minor correlation between the Varelas et al. study (72) and the present study could be detected. Furthermore, the significance of this GO term being affected by Cdc53p depletion in C. albicans (3.33E−01) was described above our cutoff level, rendering it nonsignificant. However, we detected differential regulation of genes in C. albicans, which affect membrane composition and cell wall integrity, including glucan 1,3-β-glucosidases (ENG1 [orf19.3066], XOG1 [orf19.2990], SCW11 [orf19.3893], and EXG2 [orf19.2952]; P value of 2.2E−03) and the two chitinases CHT2 (orf19.3895) and CHT3 (orf19.7586), suggesting that Cdc53p depletion affects certain aspects of membrane rearrangements in C. albicans. Concerning the remaining GO terms vitamin metabolism, sodium transport, cell-cell adhesion, sporulation, purine base metabolism, heavy metal ion homeostasis, and signal transduction of mating signals as reported in the Varelas et al. study (72), no similar response could be detected in our study. Finally, the identification of the following categories unique to Cdc53p depletion in C. albicans, namely, transport, response to stimulus, respiratory chain complex IV assembly, ethanol metabolic process, cell separation during cytokinesis, fermentation, and chromatin assembly or disassembly (Table 3), indicates that depletion and/or mutation of the cullin subunit of the SCF complex affects different sets of genes or pathways in C. albicans compared to S. cerevisiae.
Of major importance for its survival in the host and its pathogenicity is the ability of C. albicans to acquire iron from the iron-limiting host environment (55). C. albicans uses for that purpose at least three independent high-affinity iron uptake systems: a high-affinity iron permease, an iron-siderophore uptake system, and a heme uptake system (reviewed in reference 67). Interestingly, we found that the second most important GO term that was significantly affected in the transcriptome analysis of Cdc53p-depleted cells (transport) included several genes involved in copper-dependent reductive iron transport across the membrane (Table 5). These include the externally directed ferric reductases CFL2 and CFL5, which in analogy to S. cerevisiae (reviewed in reference 67) are coupled to a ferrous iron transporter complex that consists of a plasma membrane multicopper oxidase (FET3 or FET34) and a high-affinity iron permease (FTR1 or FTR2). Furthermore, activity of the multicopper ferroxidase requires efficient copper uptake by the cells (2, 32, 77) involving the copper transporter Ctr1p (42), and in analogy to S. cerevisiae, the metallochaperone Atx1p and the P-type ATPase Crp1p, with the latter being a homologue of ScCcc2p (reviewed in reference 62). The exclusive upregulation of all of the above-mentioned genes indicates that Cdc53p negatively regulates the copper-dependent reductive iron transport under iron-replete conditions.
In C. albicans, the reductive iron transport system is homeostatically regulated in response to available iron (34). Known regulators include Sfu1p, the homologue of the transcriptional repressor Fep1p in the filamentous fungi Schizosaccharomyces pombe (34, 53) and the pH-responsive transcriptional activator Rim101p (10, 17, 18). However, no general Sfu1p or Rim101p transcriptional response could be observed in the transcriptional profile of Cdc53p-depleted cells, rendering both factors unlikely to elicit the observed Cdc53p-dependent upregulation of the high-affinity iron transport genes. Interestingly, a novel transcriptional activation system for a ferric ion reductase in C. albicans has been recently reported that is similar to the respiratory CCAAT-binding factor regulatory system in S. cerevisiae (5, 31). Further studies should provide interesting information as to its contribution to the iron-homeostasis regulon, as well as its potential regulation by ubiquitination.
Supplementary Material
Acknowledgments
We are grateful to Jean-Sébastien Deneault from the BRI DNA microarray laboratory for performing the microarray experiments and to Christian Charbonneau from the IRIC Bio-Imaging Platform for help with the microscopy analyses. We also want to acknowledge the Candida Genome Database, which is funded by the National Institute of Dental and Craniofacial Research at the U.S. National Institutes of Health. We thank Malcolm Whiteway for critical reading of the manuscript.
This study was supported by the Canadian Institutes of Health Research (CIHR) Team Grant on Fungal Pathogenesis (CTP-79843). The Institute for Research in Immunology and Cancer is supported in part by the Canadian Center of Excellence in Commercialization and Research, the Canadian Foundation for Innovation, and the Fonds de Recherche en Santé du Québec.
Footnotes
Published ahead of print on 6 March 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alberti-Segui, C., A. J. Morales, H. Xing, M. M. Kessler, D. A. Willins, K. G. Weinstock, G. Cottarel, K. Fechtel, and B. Rogers. 2004. Identification of potential cell-surface proteins in Candida albicans and investigation of the role of a putative cell-surface glycosidase in adhesion and virulence. Yeast 21285-302. [DOI] [PubMed] [Google Scholar]
- 2.Askwith, C., D. Eide, A. Van Ho, P. S. Bernard, L. Li, S. Davis-Kaplan, D. M. Sipe, and J. Kaplan. 1994. The FET3 gene of Saccharomyces cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76403-410. [DOI] [PubMed] [Google Scholar]
- 3.Atir-Lande, A., T. Gildor, and D. Kornitzer. 2005. Role for the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis. Mol. Biol. Cell 162772-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachewich, C., D. Y. Thomas, and M. Whiteway. 2003. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 142163-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baek, Y. U., M. Li, and D. A. Davis. 2008. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot. Cell 71168-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee, M., D. S. Thompson, A. Lazzell, P. L. Carlisle, C. Pierce, C. Monteagudo, J. L. Lopez-Ribot, and D. Kadosh. 2008. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 191354-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassilana, M., and R. A. Arkowitz. 2006. Rac1 and Cdc42 have different roles in Candida albicans development. Eukaryot. Cell 5321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassilana, M., J. Blyth, and R. A. Arkowitz. 2003. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot. Cell 29-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bensen, E. S., S. G. Filler, and J. Berman. 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bensen, E. S., S. J. Martin, M. Li, J. Berman, and D. A. Davis. 2004. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 541335-1351. [DOI] [PubMed] [Google Scholar]
- 11.Biswas, S., P. Van Dijck, and A. Datta. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71348-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277105-109. [DOI] [PubMed] [Google Scholar]
- 13.Braun, B. R., M. van Het Hoog, C. d'Enfert, M. Martchenko, J. Dungan, A. Kuo, D. O. Inglis, M. A. Uhl, H. Hogues, M. Berriman, M. Lorenz, A. Levitin, U. Oberholzer, C. Bachewich, D. Harcus, A. Marcil, D. Dignard, T. Iouk, R. Zito, L. Frangeul, F. Tekaia, K. Rutherford, E. Wang, C. A. Munro, S. Bates, N. A. Gow, L. L. Hoyer, G. Köhler, J. Morschhäuser, G. Newport, S. Znaidi, M. Raymond, B. Turcotte, G. Sherlock, M. Costanzo, J. Ihmels, J. Berman, D. Sanglard, N. Agabian, A. P. Mitchell, A. D. Johnson, M. Whiteway, and A. Nantel. 2005. A human-curated annotation of the Candida albicans genome. PLoS. Genet. 136-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34651-662. [DOI] [PubMed] [Google Scholar]
- 15.Cheetham, J., D. A. Smith, A. da Silva Dantas, K. S. Doris, M. J. Patterson, C. R. Bruce, and J. Quinn. 2007. A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol. Biol. Cell 184603-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciechanover, A., and K. Iwai. 2004. The ubiquitin system: from basic mechanisms to the patient bed. IUBMB Life 56193-201. [DOI] [PubMed] [Google Scholar]
- 17.Davis, D. 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 441-7. [DOI] [PubMed] [Google Scholar]
- 18.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deere, D., J. Shen, G. Vesey, P. Bell, P. Bissinger, and D. Veal. 1998. Flow cytometry and cell sorting for yeast viability assessment and cell selection. Yeast 14147-160. [DOI] [PubMed] [Google Scholar]
- 20.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst, J. F. 2000. Transcription factors in Candida albicans: environmental control of morphogenesis. Microbiology 1461763-1774. [DOI] [PubMed] [Google Scholar]
- 22.Feinberg, A. P., and B. Vogelstein. 1984. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137266-267. [DOI] [PubMed] [Google Scholar]
- 23.Gildor, T., R. Shemer, A. Atir-Lande, and D. Kornitzer. 2005. Coevolution of cyclin Pcl5 and its substrate Gcn4. Eukaryot. Cell 4310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goebl, M. G., J. Yochem, S. Jentsch, J. P. McGrath, A. Varshavsky, and B. Byers. 1988. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science 2411331-1335. [DOI] [PubMed] [Google Scholar]
- 25.Hazan, I., M. Sepulveda-Becerra, and H. Liu. 2002. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol. Biol. Cell 13134-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol. 2195-201. [DOI] [PubMed] [Google Scholar]
- 27.Hinnebusch, A. G. 1988. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 52248-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 122-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hope, H., S. Bogliolo, R. A. Arkowitz, and M. Bassilana. 2008. Activation of Rac1 by the guanine nucleotide exchange factor Dck1 is required for invasive filamentous growth in the pathogen Candida albicans. Mol. Biol. Cell 193638-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huh, W. K., and S. O. Kang. 2001. Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem. J. 356595-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, D. C., K. E. Cano, E. C. Kroger, and D. S. McNabb. 2005. Novel regulatory function for the CCAAT-binding factor in Candida albicans. Eukaryot. Cell 41662-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight, S. A., E. Lesuisse, R. Stearman, R. D. Klausner, and A. Dancis. 2002. Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology 14829-40. [DOI] [PubMed] [Google Scholar]
- 33.Kornitzer, D., B. Raboy, R. G. Kulka, and G. R. Fink. 1994. Regulated degradation of the transcription factor Gcn4. EMBO. J. 136021-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan, C. Y., G. Rodarte, L. A. Murillo, T. Jones, R. W. Davis, J. Dungan, G. Newport, and N. Agabian. 2004. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 531451-1469. [DOI] [PubMed] [Google Scholar]
- 35.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 27648988-48996. [DOI] [PubMed] [Google Scholar]
- 36.Lane, S., S. Zhou, T. Pan, Q. Dai, and H. Liu. 2001. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol. Cell. Biol. 216418-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, W. C., M. Lee, J. W. Jung, K. P. Kim, and D. Kim. 2008. SCUD: Saccharomyces cerevisiae ubiquitination database. BMC. Genomics 9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leng, P., P. E. Sudbery, and A. J. Brown. 2000. Rad6p represses yeast-hypha morphogenesis in the human fungal pathogen Candida albicans. Mol. Microbiol. 351264-1275. [DOI] [PubMed] [Google Scholar]
- 39.Li, W. J., Y. M. Wang, X. D. Zheng, Q. M. Shi, T. T. Zhang, C. Bai, D. Li, J. L. Sang, and Y. Wang. 2006. The F-box protein Grr1 regulates the stability of Ccn1, Cln3 and Hof1 and cell morphogenesis in Candida albicans. Mol. Microbiol. 62212-226. [DOI] [PubMed] [Google Scholar]
- 40.Liu, H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4728-735. [DOI] [PubMed] [Google Scholar]
- 41.Lo, H. J., J. R. Köhler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90939-949. [DOI] [PubMed] [Google Scholar]
- 42.Marvin, M. E., P. H. Williams, and A. M. Cashmore. 2003. The Candida albicans CTR1 gene encodes a functional copper transporter. Microbiology 1491461-1474. [DOI] [PubMed] [Google Scholar]
- 43.Mathias, N., S. L. Johnson, M. Winey, A. E. Adams, L. Goetsch, J. R. Pringle, B. Byers, and M. G. Goebl. 1996. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol. Cell. Biol. 166634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCreath, K. J., C. A. Specht, and P. W. Robbins. 1995. Molecular cloning and characterization of chitinase genes from Candida albicans. Proc. Natl. Acad. Sci. USA 922544-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meimoun, A., T. Holtzman, Z. Weissman, H. J. McBride, D. J. Stillman, G. R. Fink, and D. Kornitzer. 2000. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCFCDC4 ubiquitin-ligase complex. Mol. Biol. Cell 11915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michel, S., S. Ushinsky, B. Klebl, E. Leberer, D. Thomas, M. Whiteway, and J. Morschhäuser. 2002. Generation of conditional lethal Candida albicans mutants by inducible deletion of essential genes. Mol. Microbiol. 46269-280. [DOI] [PubMed] [Google Scholar]
- 47.Monge, R. A., E. Roman, C. Nombela, and J. Pla. 2006. The MAP kinase signal transduction network in Candida albicans. Microbiology 152905-912. [DOI] [PubMed] [Google Scholar]
- 48.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO. J. 204742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murayama, S. Y., Y. Negishi, T. Umeyama, A. Kaneko, T. Oura, M. Niimi, K. Ubukata, and S. Kajiwara. 2006. Construction and functional analysis of fatty acid desaturase gene disruptants in Candida albicans. Microbiology 1521551-1558. [DOI] [PubMed] [Google Scholar]
- 50.Nakayama, H., T. Mio, S. Nagahashi, M. Kokado, M. Arisawa, and Y. Aoki. 2000. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect. Immun. 686712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 133452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nantel, A., T. Rigby, H. Hogues, and M. Whiteway. 2007. Microarrays for studying pathology in Candida albicans, p. 181-209. In K. Kavanaugh (ed.), Medical mycology: cellular and molecular techniques. Wiley Press, Hoboken, NJ.
- 53.Pelletier, B., A. Mercier, M. Durand, C. Peter, M. Jbel, J. Beaudoin, and S. Labbe. 2007. Expression of Candida albicans Sfu1 in fission yeast complements the loss of the iron-regulatory transcription factor Fep1 and requires Tup co-repressors. Yeast 24883-900. [DOI] [PubMed] [Google Scholar]
- 54.Pereira, S. A., and G. P. Livi. 1995. A GCN-like response in Candida albicans. Cell. Biol. Int. 1965-69. [DOI] [PubMed] [Google Scholar]
- 55.Ramanan, N., and Y. Wang. 2000. A high-affinity iron permease essential for Candida albicans virulence. Science 2881062-1064. [DOI] [PubMed] [Google Scholar]
- 56.Reuss, O., A. Vik, R. Kolter, and J. Morschhauser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341119-127. [DOI] [PubMed] [Google Scholar]
- 57.Richardson, M. D., and D. W. Warnock. 1997. Fungal infection: diagnosis and management. Blackwell Sciences, Oxford, England.
- 58.Roemer, T., B. Jiang, J. Davison, T. Ketela, K. Veillette, A. Breton, F. Tandia, A. Linteau, S. Sillaots, C. Marta, N. Martel, S. Veronneau, S. Lemieux, S. Kauffman, J. Becker, R. Storms, C. Boone, and H. Bussey. 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 50167-181. [DOI] [PubMed] [Google Scholar]
- 59.Roig, P., and D. Gozalbo. 2002. The Candida albicans UBI3 gene encoding a hybrid ubiquitin fusion protein involved in ribosome biogenesis is essential for growth. FEMS. Yeast Res. 225-30. [DOI] [PubMed] [Google Scholar]
- 60.Roig, P., and D. Gozalbo. 2003. Depletion of polyubiquitin encoded by the UBI4 gene confers pleiotropic phenotype to Candida albicans cells. Fungal Genet. Biol. 3970-81. [DOI] [PubMed] [Google Scholar]
- 61.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 62.Rutherford, J. C., and A. J. Bird. 2004. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot. Cell 31-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saidane, S., S. Weber, X. De Deken, G. St-Germain, and M. Raymond. 2006. PDR16-mediated azole resistance in Candida albicans. Mol. Microbiol. 601546-1562. [DOI] [PubMed] [Google Scholar]
- 64.Shemer, R., A. Meimoun, T. Holtzman, and D. Kornitzer. 2002. Regulation of the transcription factor Gcn4 by Pho85 cyclin PCL5. Mol. Cell. Biol. 225395-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91209-219. [DOI] [PubMed] [Google Scholar]
- 66.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends. Microbiol. 12317-324. [DOI] [PubMed] [Google Scholar]
- 67.Sutak, R., E. Lesuisse, J. Tachezy, and D. R. Richardson. 2008. Crusade for iron: iron uptake in unicellular eukaryotes and its significance for virulence. Trends. Microbiol. 16261-268. [DOI] [PubMed] [Google Scholar]
- 68.Tournu, H., G. Tripathi, G. Bertram, S. Macaskill, A. Mavor, L. Walker, F. C. Odds, N. A. Gow, and A. J. Brown. 2005. Global role of the protein kinase Gcn2 in the human pathogen Candida albicans. Eukaryot. Cell 41687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tripathi, G., C. Wiltshire, S. Macaskill, H. Tournu, S. Budge, and A. J. Brown. 2002. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 215448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ushinsky, S. C., D. Harcus, J. Ash, D. Dignard, A. Marcil, J. Morschhäuser, D. Y. Thomas, M. Whiteway, and E. Leberer. 2002. CDC42 is required for polarized growth in human pathogen Candida albicans. Eukaryot. Cell 195-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.VandenBerg, A. L., A. S. Ibrahim, J. E. Edwards, Jr., K. A. Toenjes, and D. I. Johnson. 2004. Cdc42p GTPase regulates the budded-to-hyphal-form transition and expression of hypha-specific transcripts in Candida albicans. Eukaryot. Cell 3724-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varelas, X., D. Stuart, M. J. Ellison, and C. Ptak. 2006. The Cdc34/SCF ubiquitination complex mediates Saccharomyces cerevisiae cell wall integrity. Genetics 1741825-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walther, A., and J. Wendland. 2003. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 42339-343. [DOI] [PubMed] [Google Scholar]
- 74.Whiteway, M., and C. Bachewich. 2007. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61529-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willems, A. R., M. Schwab, and M. Tyers. 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695133-170. [DOI] [PubMed] [Google Scholar]
- 76.Wodicka, L., H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 151359-1367. [DOI] [PubMed] [Google Scholar]
- 77.Yuan, D. S., R. Stearman, A. Dancis, T. Dunn, T. Beeler, and R. D. Klausner. 1995. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc. Natl. Acad. Sci. USA 922632-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao, X., S. H. Oh, K. M. Yeater, and L. L. Hoyer. 2005. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology 1511619-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, R. C. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416703-709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.