Abstract
Polyamine biosynthesis is a drug target for the treatment of African sleeping sickness; however, mechanisms regulating the pathway in Trypanosoma brucei are not well understood. Recently, we showed that RNA interference (RNAi)-mediated gene silencing or the inhibition of S-adenosylmethionine decarboxylase (AdoMetDC) led to the upregulation of the AdoMetDC activator, prozyme, and ornithine decarboxylase (ODC) proteins. To determine if this regulatory response is specific to AdoMetDC, we studied the effects of the RNAi-induced silencing of the spermidine synthase (SpdSyn) and ODC genes in bloodstream form T. brucei. The knockdown of either gene product led to the depletion of the polyamine and trypanothione pools and to cell death. Decarboxylated AdoMet levels were elevated, while AdoMet was not affected. There was no significant effect on the protein levels of other polyamine pathway enzymes. The treatment of parasites with the ODC inhibitor α-difluoromethylornithine gave similar results to those observed for ODC knockdown. Thus, the cellular response to the loss of AdoMetDC activity is distinctive, suggesting that AdoMetDC activity controls the expression levels of the other spermidine biosynthetic enzymes. RNAi-mediated cell death occurred more rapidly for ODC than for SpdSyn. Further, the ODC RNAi cells were rescued by putrescine, but not spermidine, suggesting that the depletion of both putrescine and spermidine is more detrimental than the depletion of spermidine alone. This finding may contribute to the effectiveness of ODC as a target for the treatment of African sleeping sickness, thus providing important insight into the mechanism of action of a key antitrypanosomal agent.
Human African trypanosomiasis (HAT) is a severe disease that is endemic in central Africa and is caused by the protozoan parasite Trypanosoma brucei. The drugs currently available to treat HAT are limited by toxicity and difficult dosing schemes (6). There is an unprecedented need to develop a new class of safe and effective drugs for HAT treatment. The polyamine biosynthesis pathway has been extensively studied as a promising drug target for the chemotherapeutic intervention of HAT (17). α-Difluoromethylornithine (DFMO), a suicide inhibitor of ornithine decarboxylase (ODC), is the safest and most effective drug for the treatment of late-stage HAT caused by T. brucei gambiense, and it is the only antitrypanosomal drug with a known mechanism of action (2, 4, 6).
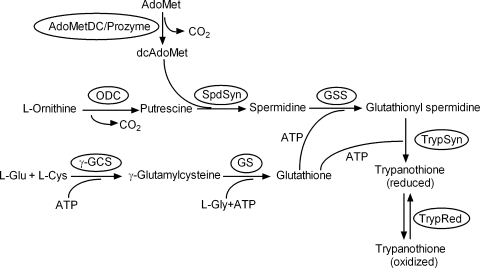
Polyamines are essential for both eukaryotic and prokaryotic cell growth and differentiation (8, 29). In eukaryotic cells, the polyamines putrescine and spermidine are synthesized from l-ornithine and S-adenosylmethionine (AdoMet) (Fig. 1). ODC catalyzes the first committed step, producing putrescine, while S-adenosylmethionine decarboxylase (AdoMetDC) generates decarboxylated S-adenosylmethionine (dcAdoMet), which serves as the aminopropyl group donor for spermidine synthesis. In the trypanosomatids, spermidine is conjugated to two molecules of glutathione to form trypanothione, which is required to maintain cellular redox balance (12). Thus, polyamines play a unique role in these protozoan pathogens.
FIG. 1.
Polyamine and trypanothione biosynthetic pathways.
Both genetic and chemical data have demonstrated that the polyamine and trypanothione biosynthetic pathways are essential in T. brucei. Gene knockout or RNA interference (RNAi) studies in T. brucei have shown that the polyamine and trypanothione biosynthetic enzymes are required for parasite growth (1, 19, 23, 24, 35, 40). In addition to DFMO, inhibitors of AdoMetDC have also been reported to have antitrypanosomal activity (3, 5, 7), further demonstrating that polyamine metabolism is of clear medical and pharmacological importance in these parasites. DFMO treatment depletes the cells of putrescine and trypanothione, while leading to a partial depletion of spermidine (11). The loss of these polyamine pools has been clearly implicated in the mechanism of action of DFMO killing. Additionally, DFMO has been reported to elevate AdoMet levels in some T. brucei isolates, and this response has been put forward as a potential mechanism accounting for species differences in DFMO sensitivity (41).
Eukaryotic cells tightly regulate the polyamine biosynthetic pathway to maintain polyamine homeostasis (10, 27-29). Excess polyamine biosynthesis is associated with tumor promotion in mammals, while a deficiency of polyamines leads to cell growth arrest. Polyamine synthesis, degradation, and transport have each been shown to be under regulatory control. The key rate-limiting enzymes ODC and AdoMetDC are regulated at the levels of transcription, translation, and posttranslational modification (8, 9, 28, 34). ODC is further regulated by a protein inhibitor termed antizyme, which regulates its rate of degradation in response to polyamine concentration (10). Finally, antizyme levels are modulated by translational frame shifting and by binding to antizyme inhibitor (25).
In contrast to this wealth of regulatory control in mammalian cells, an understanding of the mechanisms that regulate the polyamine biosynthesis pathway in trypanosomes is only just emerging. The regulatory mechanisms found in mammalian cells do not appear to operate in T. brucei, and they appear to have evolved novel control mechanisms. There is no evidence for the regulation of ODC and AdoMetDC protein turnover (15, 30, 40). An antizyme homolog has not been found in the T. brucei genome, and the AdoMetDC gene does not appear to carry an upstream open reading frame that could regulate translation. Instead, we recently discovered a unique mechanism for the regulation of AdoMetDC in trypanosomatids. T. brucei AdoMetDC is activated 1,200-fold by forming a heterodimer with an inactive homolog termed prozyme, which is found only in the trypanosomatids (39). Gene silencing by RNAi or the chemical inhibition of AdoMetDC led to 25-fold and 7-fold increases in the protein levels of prozyme and ODC, respectively, with the data suggesting that the expression of these enzymes is translationally regulated (40).
To determine if additional enzymes in the polyamine biosynthetic pathway are also involved in the regulatory response, we studied the effect of the RNAi-mediated silencing of the spermidine synthase (SpdSyn) and ODC genes in bloodstream form (BSF) T. brucei cells. The effects of DFMO treatment were studied as a comparison. The knockdown of ODC or SpdSyn led to polyamine and trypanothione depletion and to cell growth arrest. However, neither ODC nor SpdSyn knockdown, nor treatment with DFMO, resulted in any significant effects on the expression levels of other polyamine pathway enzymes. These data demonstrate that the regulatory response observed upon the depletion of AdoMetDC activity is unique and suggest that AdoMetDC is the central player in the control of pathway flux. Finally, in comparison to other pathway enzymes, the knockdown of ODC by RNAi led to a rapid cell death that could not be rescued by spermidine, suggesting alternative roles for putrescine or the possibility that mechanisms other than polyamine depletion contribute to cell death. These studies contribute to our understanding of the regulatory control points in the polyamine biosynthetic pathway, providing important insight into the enzymes in the pathway that are best targeted for drug discovery projects.
MATERIALS AND METHODS
Materials.
All reagents were purchased from Sigma-Aldrich unless otherwise indicated.
Trypanosome cultures.
BSF trypanosomes were cultured in HMI-9 medium with 10% fetal bovine serum at 37°C under 5% CO2 (18). For the spermidine rescue experiments, chicken serum was substituted for bovine serum to avoid polyamine oxidase-driven toxicity (31). Cells were maintained in the mid-log phase (105 to 106 cells/ml) and selected in medium containing the appropriate antibiotics (15 μg/ml of G418, 50 μg/ml of hygromycin, and 2.5 μg/ml of phleomycin). Cell densities were determined by counting with a hemocytometer (Brightline; Fisher). Growth curves were plotted as time versus total cell numbers, calculated as the product of the cell density and the total dilution factors (38).
SpdSyn RNAi and ODC RNAi constructs.
The SpdSyn (EC 2.5.1.16) and ODC (EC 4.1.1.17) RNAi constructs were generated as described previously (38). Briefly, a 549-bp fragment (corresponding to coding nucleotides 108 to 656) from the T. brucei SpdSyn gene or a 420-bp fragment (corresponding to coding nucleotides 894 to 1313) from the T. brucei ODC gene was amplified by PCR using gene-specific primers and ligated into the pJM326 vector in the forward direction and into the pLEW100 vector in the reverse direction. The HindIII and XbaI fragments from the resulting pJM326 construct, containing the gene fragment plus a stuffer region, were ligated into the pLEW100 vector containing the XbaI-MluI gene fragment. The final constructs have two copies of the target gene in opposite directions separated by a stuffer region. The primers were as follows: for SpdSyn, HindIII-TCAAGCTTAACAAAGTTCCAGCACCTGTC, NheI-AAGCTAGCATAAAGTCG GCCATCCCTTC, MluI-AAACGCGTAACAAAGTTCCAGCACCTGTC, and XbaI-CCTCTAGAATAAAGTCGGCCATCCCTTC, and for ODC, HindIII-AAGCTTCTACGTTGCTTCAGCTTTCAC, NheI-GCTAGCCGGACAACATGGTCTGGTAG, MluI-ACGCGTCTACGTTGCTTCAGCTTTCAC, and XbaI-TCTAGACGGACAACATGGTCTGGTAG.
Transfection of T. brucei parasites.
90-13 BSF cells (a gift from George Cross) were grown to mid-log phase (∼106 to ∼107 cells/ml). DNA (100 μg of linearized construct) was electroporated into cells (∼5 × 106 to ∼8 × 106) at room temperature by a single pulse on the Bio-Rad gene pulser electroporation system as described previously (19). Cells were suspended into HMI-9 medium (24 ml) plus G418 (15 μg/ml) and hygromycin (50 μg/ml), dispensed into a 24-well plate at 1 ml/well, and incubated for 6 h at 37°C before phleomycin (5 μg/ml) was added to select for plasmid integration into the rRNA locus. RNAi targeting SpdSyn or ODC message was induced by the addition of tetracycline (Tet; 1 μg/ml) into the culture every 24 h. Spermidine (100 μM) or putrescine (500 μM) was included in the medium as a rescue agent where indicated.
Northern analysis.
mRNA was prepared from at least 1 × 108 cells, separated on 1% denaturing agarose gels (1 μg/lane), and transferred to a positively charged nylon membrane (BrightStar-Plus; Ambion). Northern blot analysis was performed as recommended by the manufacturer using a Micro Poly(A) Purist kit (Ambion) as previously described (40).
Western analysis.
Protein levels were evaluated by Western blotting as described previously (19, 40). Briefly, log-phase cells (1 × 108) were resuspended in lysis buffer (50 mM HEPES [pH 8.0], 100 mM NaCl, 5 mM 2-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 2 μg/ml antipain, 10 μg/ml benzamidine, 1 μg/ml pepstatin, and 1 μg/ml chymostatin) and lysed by three freeze-thaw cycles. The supernatant was collected after centrifugation, and the total protein was separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (15 μg for SpdSyn, trypanothione synthetase [TrypSyn], trypanothione reductase [TrypRed], and tubulin and 30 μg for ODC, AdoMetDC, prozyme, and γ-glutamylcysteine synthetase [γ-GCS]), and transferred to a polyvinylidene difluoride membrane (Hybond-P; Amersham Biosciences/GE Healthcare). Membranes were probed with rabbit polyclonal antibodies raised against T. brucei ODC, AdoMetDC (EC 4.1.1.50), prozyme, and γ-GCS (EC 6.3.2.2); rabbit polyclonal antibody raised against Leishmania donovani SpdSyn; and rat polyclonal antibodies raised against T. brucei TrypSyn (EC 6.3.1.9) and TrypRed (EC 1.8.1.12), using antibody dilutions and conditions as previously described (19, 40), followed by visualization with ECL Western blotting detection reagents (Amersham Biosciences/GE Healthcare). Controls for the ODC and the γ-GCS antibodies were also run on gels; these included recombinant ODC and procyclic 29-13 cell lysates in which higher levels of γ-GCS expression are observed.
Quantification of intracellular polyamine pools.
T. brucei cells (1 × 107) were collected after centrifugation, washed twice in phosphate-buffered saline (PBS; 20 ml), resuspended in buffer (100 mM MOPS [morpholinepropanesulfonic acid] buffer [pH 8.0], 50 mM NaCl, 2 mM EDTA, and 20 mM MgCl2), and lysed by three freeze-thaw cycles. The protein was precipitated with trichloroacetic acid (9.2% final concentration), and the clear supernatants were collected after centrifugation (13,500 rpm for 3 min). Aliquots (5 μl) were labeled with AccQ-tag reagent (Waters) and analyzed by high-performance liquid chromatography (HPLC) using the previously described system and gradients (26, 40).
Quantification of intracellular thiol pools.
Log-phase cells (1 × 108) were harvested, washed twice in PBS (20 ml), and resuspended in buffer (20 mM HEPPS [pH 8.0], 2 mM diethylenetriaminepentaacetic acid) plus monobromobimane (final concentration of 5 mM) (Invitrogen). After three freeze-thaw cycles, the lysates were heated at 70°C for 3 min and cooled on ice. The protein was precipitated by the addition of methanesulphonic acid (2 M, pH 1.6), followed by incubation (on ice for 30 min) and centrifugation at 13,500 rpm for 5 min. The resulting supernatant was analyzed by HPLC using the previously described system and gradients (11, 26, 40). Reduced glutathionyl spermidine (GSH-Spd) and reduced trypanothione [T(SH)2] standards for the standard curves were prepared by incubation with tris(2-carboxyethyl)phosphine (2 mM) at room temperature for 30 min.
Quantification of intracellular AdoMet and dcAdoMet.
Cells (1 × 108) were harvested, washed twice in PBS (20 ml), and lysed in 20% trichloroacetic acid at 4°C overnight. After centrifugation (13,000 × g for 10 min at 4°C) of the cells, the supernatant was collected. Precolumn derivatization was carried out to form the fluorescent 1,N6-etheno derivatives of the adenine moiety of AdoMet and dcAdoMet (37). Samples were derivatized by the addition of chloroacetaldehyde (final concentration of 2 M) and sodium acetate (0.44 M final concentration) followed by incubation at pH 3.0 at 40°C for 7 h. Standards of AdoMet (Sigma) and dcAdoMet (kindly provided by Akira Shirahata) were derivatized by the same method. The derivatized samples were then analyzed by HPLC on an Ultrasphere IP 4.6- by 250-mm column (Beckman) using a mobile phase containing medium A (0.1 M NaH2PO4, 8 mM 1-octanesulfonic acid, 0.1 mM EDTA, 2% [vol/vol] acetonitrile [pH 3.65]) and medium B (0.2 M NaH2PO4, 8 mM 1-octanesulfonic acid, 26% [vol/vol] acetonitrile [pH 4.30]) using the following gradient: 25% to 60% medium B in 45 min, followed by 60% to 100% medium B in 1 min and a 100% medium B wash for 8 min before regenerating the starting condition. The etheno derivatives of AdoMet and dcAdoMet were detected with λEx at 270 nm and λEm at 410 nm and the peak areas measured. The concentrations were then determined from a standard curve of concentration versus peak area that was created using known concentrations of the etheno derivatives of AdoMet and dcAdoMet.
Gene accession numbers.
The GeneDB accession numbers are as follows: ODC, Tb11.01.5300; SpdSyn, Tb09.v1.0380; AdoMetDC, Tb927.6.4410, Tb927.6.4460; prozyme, Tb927.6.4470; γ-GCS, Tb10.389.1360; TrypSyn, Tb927.2.4370; and TrypRed, Tb10.406.0520.
RESULTS
The silencing of SpdSyn or ODC genes by RNAi leads to the death of T. brucei BSF cells.
To test the effects of reduced SpdSyn or ODC expression on BSF T. brucei parasites, we generated stable cell lines with an inducible RNAi construct targeting each gene. These lines contain a Tet-inducible stem-loop vector with portions of the SpdSyn (549 bp) or ODC (420 bp) coding regions cloned in opposite orientations around a spacer sequence and integrated into the rRNA gene locus of T. brucei BSF 90-13 cells. The addition of Tet leads to the production of a double-stranded stem-loop RNA targeting either SpdSyn or ODC mRNA for degradation. Northern and Western blot analyses indicated that SpdSyn mRNA (Fig. 2A, inset, S panel) and protein (Fig. 2B) were reduced >90% by day 2 after induction of the SpdSyn RNAi line with Tet, while the induction of ODC RNAi led to a 90% depletion of ODC mRNA (Fig. 3A, inset, O panel) and protein (Fig. 3B) within 24 h of induction. Tubulin mRNA (Fig. 2A and 3A, insets, T panels) and protein levels (Fig. 2B and 3B) were evaluated as a loading control.
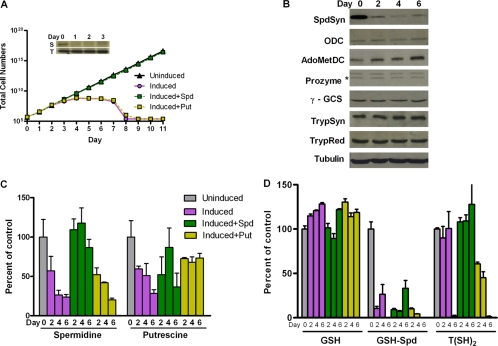
FIG. 2.
Effects of the RNAi-mediated silencing of the SpdSyn gene. (A) Cell growth curves. Total cell numbers calculated as the product of cell density and the total dilution factor are plotted versus time (days) post-Tet induction (1 μg/ml) in the presence or absence of spermidine (Spd; 0.1 mM) or putrescine (Put; 0.5 mM). The inset shows Northern analysis for SpdSyn (S panel) and the tubulin control (T panel). Data shown are the averages of the results from two experiments. (B) Western blot analysis of the polyamine and trypanothione biosynthetic pathway enzymes. (C) Intracellular polyamine levels. (D) Intracellular thiol levels. Uninduced SpdSyn RNAi cells are included as a control. Errors represent the standard errors of the means of the results from three independent biological replicates.
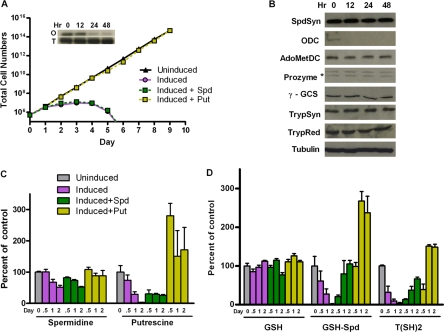
FIG. 3.
Effects of the RNAi-mediated silencing of the ODC gene. (A) Cell growth curves. Total cell numbers calculated as the product of cell density and the total dilution factor are plotted versus time (days) post-Tet induction (1 μg/ml) in the presence or absence of spermidine (Spd; 0.1 mM) or putrescine (Put; 0.5 mM). The inset shows Northern analysis for ODC (O panel) and the tubulin control (T panel). Data shown are the averages of the results from two experiments. (B) Western blot analysis of the polyamine and trypanothione biosynthetic pathway enzymes. (C) Intracellular polyamine levels. (D) Intracellular thiol levels. Uninduced ODC RNAi cells are included as a control. Errors represent the standard errors of the means of the results from three independent biological replicates.
The mRNA knockdown of SpdSyn or ODC led to cell growth arrest and to cell death after several cell cycles, similarly to previous reports (24, 35, 38). SpdSyn RNAi cells induced with Tet grew slowly from day 3 and began to die after day 7 (Fig. 2A). The addition of exogenous spermidine (0.1 mM) to the Tet-induced SpdSyn RNAi cells restored normal growth relative to the uninduced cells, while the addition of exogenous putrescine (0.5 mM) did not. The RNAi-mediated silencing of the ODC gene led to more rapid growth arrest, which occurred by day 2, followed by cell death by day 4 (Fig. 3A). The Tet-induced ODC RNAi line was rescued by the addition of putrescine (0.5 mM) but not by spermidine (0.1 mM).
The demonstration that exogenous spermidine rescues the growth defect caused by the RNAi-mediated silencing of the SpdSyn gene differs from previous reports that found spermidine to be toxic to the cells and unable to rescue the effects of SpdSyn knockdown by RNAi (35). This difference can be attributed to the fact that in the prior report, bovine serum was used to culture the parasites, whereas we used chicken serum. Bovine serum contains polyamine oxidases that utilize spermidine as a substrate and generate hydrogen peroxide, which is toxic to the cells (31, 32). Chicken serum does not have these oxidases, and spermidine at the concentrations used is not toxic to BSF parasites under these conditions.
Effects of SpdSyn or ODC RNAi-mediated gene silencing on the expression of polyamine and trypanothione biosynthetic enzymes.
To examine the possibility of cross-regulation between polyamine and trypanothione biosynthesis, the protein levels of the other enzymes in both pathways were evaluated by Western blotting after Tet induction of SpdSyn or ODC RNAi. The SpdSyn RNAi cells were followed for 6 days after induction, and a modest increase (two- to threefold) was observed in the steady-state levels of the AdoMetDC protein while the expression levels of the remaining enzymes were unchanged (Fig. 2B). The ODC RNAi cells were followed for only 48 h after induction because of the rapid cell death phenotype. Over this time course, the depletion of ODC did not affect the steady-state protein levels of other enzymes in the pathway (Fig. 3B). As a control, the protein expression levels were also examined in untransformed 90-13 cells with or without the addition of Tet, demonstrating that Tet itself did not affect the expression profile of pathway enzymes (data not shown).
SpdSyn or ODC RNAi-mediated gene silencing leads to reduced polyamine and trypanothione levels.
The effects of SpdSyn and ODC knockdown on intracellular polyamine and reduced thiol pools were examined by HPLC analysis (Table 1 and Fig. 2 and 3). After 6 days of Tet induction, the spermidine levels in SpdSyn RNAi cells decreased to 20% of the uninduced control (Fig. 2C), while both the GSH-Spd and T(SH)2 levels were depleted to <5% of those of the controls (Fig. 2D). The spermidine and GSH-Spd levels began to drop after 2 days of induction, while the depletion of T(SH)2 did not occur until 6 days after induction. Exogenous spermidine (0.1 mM) partially restored the spermidine and GSH-Spd levels, while fully restoring the T(SH)2 levels in these cells. In contrast, putrescine (0.5 mM) was unable to replenish the spermidine, GSH-Spd, or T(SH)2 pool, a result which correlates with the observation that cell growth arrest was rescued by spermidine but not putrescine. The silencing of the ODC gene caused a near complete depletion of putrescine, a 50% reduction in spermidine (Fig. 3C), and a > 90% reduction in GSH-Spd and T(SH)2 levels (Fig. 3D) 48 h after induction. Exogenous putrescine (0.5 mM) completely restored both the polyamine and thiol pools. In contrast, while the addition of exogenous spermidine (0.1 mM) restored the GSH-Spd and T(SH)2 pools to >70% of the control levels, the spermidine and putrescine levels remained low at only 50% and 24%, respectively, of the levels in the uninduced controls. The cells were not viable under these conditions. GSH levels remained similar to wild-type levels in all of the experimental systems.
TABLE 1.
The effects of genetic knockdown of polyamine biosynthetic enzymes on polyamine levels and the biosynthetic enzymes in blood form T. brucei
| Genetic knockdown/chemical inhibitiona | Cell response (% of the wild-type level)b
|
||||||
|---|---|---|---|---|---|---|---|
| Putrescine | Spermidine | GSH-Spd | T(SH)2 | AdoMet | dcAdoMet | Pathway enzymes | |
| ODC | 0 | 50 | <5 | <5 | 100 | 400 | nc |
| DFMO treatment | 0 | 40 | 20 | 40 | 100 | 4,000 | nc |
| SpdSyn | 30 | 20 | <5 | <5 | 100 | 18,000 | nc |
| AdoMetDCc | 700 | 40 | <5 | <5 | ND | ND | 2,500, prozyme; 700, ODC |
| Prozymec | 1,000 | <5 | <5 | <5 | ND | ND | 500, ODC |
For ODC, SpdSyn, and AdoMetDC, the results are from RNAi-mediated gene silencing, while for prozyme, the results are taken from a gene knockout cell line expressing a Tet-inducible regulated copy of the gene. For ODC, data represent the 48-h time point after Tet induction; for SpdSyn and AdoMetDC, data represent the 6-day time point after Tet induction; and for prozyme, data represent the 4-day time point after Tet withdrawal.
ND, not determined; nc, no significant change.
Data taken from reference 40.
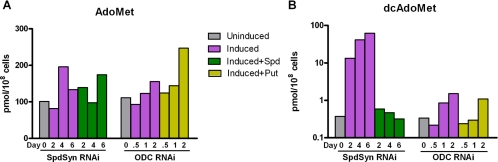
Decarboxylated AdoMet but not AdoMet levels are increased upon SpdSyn or ODC RNAi-mediated gene silencing.
To explore additional mechanisms that contribute to cell death beyond polyamine and T(SH)2 depletion, we evaluated the effects of SpdSyn and ODC RNAi-mediated gene silencing on AdoMet and dcAdoMet levels (Table 1 and Fig. 4). Intracellular dcAdoMet levels increased >100-fold upon Tet induction of SpdSyn RNAi, while the addition of exogenous spermidine (0.1 mM) restored the wild-type levels. The induction of ODC RNAi by Tet led to a small (fourfold) increase in dcAdoMet levels. The intracellular AdoMet levels were not significantly affected by the knockdown of either ODC or SpdSyn.
FIG. 4.
Effect of the RNAi-mediated silencing of the SpdSyn gene (A) or the ODC gene (B) on AdoMet and dcAdoMet levels. SpdSyn RNAi and ODC RNAi cells were induced with Tet (1 μg/ml) in the presence or absence of spermidine (Spd; 0.1 mM) or putrescine (Put; 0.5 mM). SpdSyn RNAi cells (1 × 108) were harvested after 0, 2, 4, and 6 days of induction; ODC RNAi cells (1 × 108) were harvested after 0, 12, 24, and 48 h of induction. After acid extraction from the cell lysates followed by derivatization, AdoMet and dcAdoMet levels were determined by HPLC analysis. Uninduced SpdSyn RNAi and ODC RNAi cells were included as a control. Data shown are the averages of the results from two independent experiments.
Comparative effects of DFMO on polyamine and trypanothione biosynthesis.
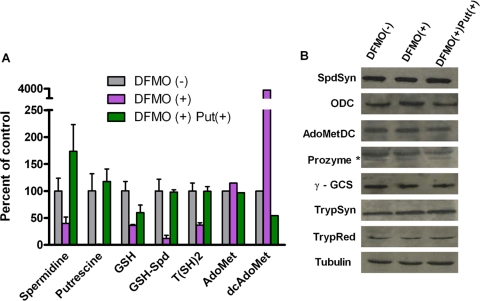
The growth effect of DFMO on 90-13 BSF parasites was evaluated by generating a dose-response curve (50% effective concentration, 25 μM) to determine the level of DFMO to be used in the subsequent experiments. The cells were next treated with DFMO at the 50% effective concentration (12.5 μM) for 3 days in the presence or in the absence of putrescine (0.5 mM). The effects of DFMO on the polyamine, trypanothione, AdoMet, and dcAdoMet pools were examined by HPLC analysis (Table 1 and Fig. 5A), and the protein levels of the polyamine pathway enzymes were evaluated by Western blotting (Fig. 5B). As previously reported (11), DFMO treatment led to a complete depletion of putrescine, whereas the spermidine, GSH-Spd, and T(SH)2 levels were decreased to 20 to 40% of that of the DFMO (−) control. The addition of exogenous putrescine restored the spermidine, putrescine, GSH-Spd, and T(SH)2 levels. The GSH level was also reduced to 40% of that of the control cells but was restored to 60% of that of the untreated control by the addition of putrescine. The effects on GSH levels have not previously been reported, and these data suggest that GSH may become oxidized due to the accumulation of oxidized trypanothione. The dcAdoMet levels increased by ∼40-fold in DFMO-treated T. brucei cells, and this effect was reversed by the addition of putrescine. No significant change in the AdoMet level was observed. Finally, DFMO treatment led to an approximately twofold increase in the ODC protein level, while the steady-state protein levels of SpdSyn, AdoMetDC, γ-GCS, prozyme, TrypSyn, and TrypRed were unaffected.
FIG. 5.
Effects of DFMO on the polyamine and trypanothione biosynthesis. 90-13 cells were treated with DFMO (12.5 μM for 3 days) in the presence (+) or absence (−) of putrescine (Put; 0.5 mM). (A) Polyamines, thiols, AdoMet, and dcAdoMet were quantitated by HPLC analysis. Errors represent the standard errors of the means of the results from three independent biological replicates, except with AdoMet and dcAdoMet, where n = 2. (B) Western blot analysis of the polyamine and trypanothione biosynthetic enzymes.
DISCUSSION
Polyamine metabolism is tightly regulated in eukaryotic cells to maintain a balance of polyamine biosynthesis, degradation, and transport; however, the previously described mechanisms do not appear to operate in trypanosomes. We have now completed a systematic analysis of the effects of the genetic knockdown of the four proteins involved in spermidine biosynthesis in BSF T. brucei (summarized in Table 1). This experimental data set provides important insight into polyamine regulation in T. brucei, and it is a valuable tool for further analysis of pathway flux, including the use of mathematical modeling, as was recently described for the mammalian pathway (33).
ODC, SpdSyn, and AdoMetDC (40) were evaluated by RNAi-mediated gene silencing, while prozyme was evaluated using a regulated knockout strategy (40). The knockdown of each of these proteins leads to cell growth arrest, correlating with a near-complete depletion of T(SH)2 and GSH-Spd. The effects on the polyamines putrescine and spermidine are, however, specific to the point where the pathway was perturbed. Putrescine is only depleted by ODC knockdown or inhibition, while putrescine was, in contrast, elevated only by AdoMetDC knockdown or inhibition (either chemical or by the loss of the activator prozyme). Interestingly, while T(SH)2 is completely depleted by the knockdown of any one of the pathway enzymes, spermidine pools are retained at 20 to 50% of the wild-type levels in all cell lines except for the prozyme knockout cell line. The ODC and SpdSyn knockdown data provide further support for the hypothesis that the catabolism of T(SH)2 may contribute to spermidine homeostasis under the stress of reduced synthesis from putrescine (40). Trypanothione synthase contains both a synthetic domain and a catabolic domain, though the exact role of the latter in T(SH)2 metabolism has not been determined (13).
The data from the genetic knockdown of the pathway enzymes demonstrate that AdoMetDC is the key regulatory control point for polyamine biosynthesis in T. brucei, as it is the only enzyme for which perturbation led to changes in the expression of other pathway enzymes. The finding that AdoMetDC activity levels are correlated to ODC and prozyme expression also suggests that the rate-limiting steps in polyamine biosynthesis are likely to be catalyzed by ODC and AdoMetDC. In mammalian cells, spermidine concentration controls protein expression levels of ODC, AdoMetDC, and the ODC protein inhibitor antizyme by affecting translation (10, 27-29). In T. brucei, polyamine pool analysis of the genetic knockdown cell lines suggests that a different mechanism must exist to control prozyme and ODC protein expression. While the data support a translational control mechanism (40), fluctuations in spermidine concentration do not appear to play a regulatory role. Spermidine pools are similarly decreased by AdoMetDC, ODC, and SpdSyn knockdown, yet prozyme and ODC protein levels increase upon the knockdown of AdoMetDC but not upon the knockdown of ODC and SpdSyn. The only unique difference in the polyamine levels between these lines is the elevation of putrescine observed upon AdoMetDC knockdown or inhibition. However, this effect is most likely a consequence of the loss of the downstream metabolic pathway for putrescine utilization in spermidine biosynthesis. Thus, the regulatory switch that controls ODC and prozyme translation has yet to be identified.
An analysis of the ODC RNAi line suggests that additional cell-killing mechanisms beyond the depletion of the spermidine and T(SH)2 pools contribute to cell death when ODC protein is inhibited or depleted. ODC RNAi-mediated gene silencing led to more rapid cell death than was observed for SpdSyn or AdoMetDC knockdown (within 48 h versus 4 to 6 days) (40). Further cell growth could only be rescued by the addition of putrescine but not by spermidine, despite the finding that GSH-Spd and T(SH)2 pools were restored by the addition of either amine. These results are in contrast to results reported for L. donovani for which either putrescine or spermidine rescued the genetic knockout of ODC (20).
One plausible hypothesis for the finding that ODC knockdown is not rescued by spermidine was that elevated AdoMet and dcAdoMet levels contribute to cell death when ODC is inhibited or depleted from the cell. It was previously reported that DFMO treatment of sensitive T. brucei rhodesiense clinical isolates, but not refractory strains, led to a >60-fold elevation in AdoMet and dcAdoMet levels and consequently to a significant increase of the methylation index that was postulated to have a role in cell killing (41). Similar changes were observed in the lab strain of T. brucei brucei (EATRO 110). However, in our experimental system, while dcAdoMet levels are increased fourfold by the RNAi-mediated knockdown of ODC, and 40-fold by DFMO treatment, we observe no effect on the AdoMet levels in these cells. This alternative response may reflect heterogeneity between T. brucei brucei laboratory strains. Indeed, a recent study suggests that T. brucei rhodesiense field isolates show significant genetic diversity and polymorphisms throughout the genome (16).
The increase in dcAdoMet levels after the knockdown of either ODC or SpdSyn is consistent with the fact that the cells can no longer utilize this metabolite for the synthesis of spermidine. These data further suggest that AdoMetDC levels do not decrease to compensate for this build-up. In contrast, the data suggest that AdoMet synthetase may be a potential control point and that the levels of this enzyme may decrease in response to the buildup of dcAdoMet, thus allowing AdoMet levels to remain constant.
While elevated dcAdoMet levels were observed after ODC gene silencing, the high dcAdoMet levels do not appear to play a role in spermidine-independent growth arrest, as dcAdoMet levels were similarly increased by the RNAi-mediated gene silencing of SpdSyn. It remains possible that putrescine plays a novel role in T. brucei beyond serving as a precursor for spermidine biosynthesis. Putrescine has been shown to be an important substrate for transglutaminase in plants (14), and solute transporters (antiporter or symporter) that utilize putrescine have been described for Escherichia coli cells and colon epithelial cells (21, 22, 36).
Alternatively, these data may suggest that an irreversible event caused by polyamine or trypanothione depletion occurs before sufficient spermidine is incorporated to protect the cells. In support, spermidine is poorly taken up by T. brucei BSF cells, and it is unable to rescue the growth arrest caused by the knockout of prozyme, which also occurs with a similarly rapid time course as observed for ODC RNAi-mediated knockdown (40). In contrast, spermidine is able to rescue the growth effects caused by the RNAi-mediated knockdowns of AdoMetDC and SpdSyn, which are both slower events providing more time to incorporate sufficient spermidine into the cells to protect them from cell death.
Finally, our data provide important mechanistic insight into the effectiveness of DFMO as an antitrypanosomal agent. The knockdown of each of the spermidine biosynthetic enzymes (ODC, SpdSyn, and AdoMetDC) and the regulator prozyme leads to cell death, and in no case have we observed a growth reversal phenotype leading to cell escape of the genetic block. Escape of the genetic block has been observed in T. brucei after the RNAi-mediated gene silencing of TrypSyn or after the regulated knockout of TrypRed (1, 23). In contrast, the loss of ODC and prozyme proteins by genetic knockdown strategies leads to very rapid cell killing, and the spermidine rescue data suggest that an irreversible event occurs early in the time course after gene expression is silenced, showing that the parasite is highly sensitive to polyamine depletion. Thus, the data suggest that the inhibition of the early, rate-limiting enzymes in the pathway (ODC and AdoMetDC/prozyme) provides a more effective strategy to inhibit polyamine/trypanothione biosynthesis and parasite growth than the inhibition of enzymes later in the pathway (e.g., TrypSyn and TrypRed). These data provide an explanation as to why, despite the relatively low potency of DFMO on the parasite, DFMO is an effective antitrypanosomal agent and ODC is a particularly effective target in this organism.
Acknowledgments
We thank Buddy Ullman for providing antibody to spermidine synthase and Alan Fairlamb for providing antibodies to trypanothione synthetase and trypanothione reductase. We are grateful to Paul England for the pLEW100 and pJM326 vectors and to George Cross for the 90-13 cells. We thank Tony Michael for critical reading of the manuscript.
This work was supported by the National Institutes of Health (R01 AI34432) (to M.A.P.) and the Welch Foundation (I-1257) (to M.A.P.).
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Ariyanayagam, M. R., S. L. Oza, M. L. S. Guther, and A. H. Fairlamb. 2005. Phenotypic analysis of trypanothione synthetase knockdown in the African trypanosome. Biochem. J. 391425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacchi, C., H. Nathan, S. Hunter, P. McCann, and A. Sjoerdsma. 1980. Polyamine metabolism: a potential therapeutic target in trypanosomes. Science 210332-334. [DOI] [PubMed] [Google Scholar]
- 3.Bacchi, C. J., R. Brun, S. L. Croft, K. Alicea, and Y. Buhler. 1996. In vivo trypanocidal activities of new S-adenosylmethionine decarboxylase inhibitors. Antimicrob. Agents Chemother. 401448-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacchi, C. J., J. Garofalo, D. Mockenhaupt, P. P. McCann, K. A. Diekema, A. E. Pegg, H. C. Nathan, E. A. Mullaney, L. Chunosoff, A. Sjoerdsma, and S. H. Hutner. 1983. In vivo effects of alpha-DL-difluoromethylornithine on the metabolism and morphology of Trypanosoma brucei brucei. Mol. Biochem. Parasitol. 7209-225. [DOI] [PubMed] [Google Scholar]
- 5.Bacchi, C. J., H. C. Nathan, N. Yarlett, B. Goldberg, P. P. McCann, A. J. Bitonti, and A. Sjoerdsma. 1992. Cure of murine Trypanosoma brucei rhodesiense infections with an S-adenosylmethionine decarboxylase inhibitor. Antimicrob. Agents Chemother. 362736-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett, M. P., D. W. Boykin, R. Brun, and R. R. Tidwell. 2007. Human African trypanosomiasis: pharmacological re-engagement with a neglected disease. Br. J. Pharmacol. 1521155-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bitonti, A. J., T. L. Byers, T. L. Bush, P. J. Casara, C. J. Bacchi, A. B. Clarkson, Jr., P. P. McCann, and A. Sjoerdsma. 1990. Cure of Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense infections in mice with an irreversible inhibitor of S-adenosylmethionine decarboxylase. Antimicrob. Agents Chemother. 341485-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casero, R. A., Jr., and L. J. Marton. 2007. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 6373-390. [DOI] [PubMed] [Google Scholar]
- 9.Childs, A. C., D. J. Mehta, and E. W. Gerner. 2003. Polyamine-dependent gene expression. Cell. Mol. Life Sci. 601394-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffino, P. 2001. Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol. 2188-194. [DOI] [PubMed] [Google Scholar]
- 11.Fairlamb, A. H., G. B. Henderson, C. J. Bacchi, and A. Cerami. 1987. In vivo effects of difluoromethylornithine on trypanothione and polyamine levels in bloodstream forms of T. brucei. Mol. Biochem. Parasitol. 24185-191. [DOI] [PubMed] [Google Scholar]
- 12.Fries, D. S., and A. H. Fairlamb. 2003. Antiprotozoal agents, p. 1033-1087. In D. Abraham (ed.), Burger's medicinal chemistry and drug discovery, 6th ed., vol. 5. John Wiley & Sons, Inc., New York, NY. [Google Scholar]
- 13.Fyfe, P. K., S. L. Oza, A. H. Fairlamb, and W. N. Hunter. 2008. Leishmania trypanothione synthetase-amidase structure reveals a basis for regulation of conflicting synthetic and hydrolytic activities. J. Biol. Chem. 28317672-17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galston, A. W., and R. K. Sawhney. 1990. Polyamines in plant physiology. Plant Physiol. 94406-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghoda, L., M. A. Phillips, K. Bass, C. C. Wang, and P. Coffino. 1990. Trypanosome ornithine decarboxylase is stable because it lacks sequences found in the carboxyl terminus of the mouse enzyme which target the latter for intracellular degradation. J. Biol. Chem. 26511823-11826. [PubMed] [Google Scholar]
- 16.Gibson, W. 2007. Resolution of the species problem in African trypanosomes. Int. J. Parasitol. 37829-838. [DOI] [PubMed] [Google Scholar]
- 17.Heby, O., S. C. Roberts, and B. Ullman. 2003. Polyamine biosynthetic enzymes as drug targets in parasitic protozoa. Biochem. Soc. Trans. 31415-419. [DOI] [PubMed] [Google Scholar]
- 18.Hirumi, H., and K. Hirumi. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75985-989. [PubMed] [Google Scholar]
- 19.Huynh, T. T., V. T. Huynh, M. A. Harmon, and M. A. Phillips. 2003. Gene knockdown of γ-glutamylcysteine synthetase by RNAi in the parasitic protozoa Trypanosoma brucei demonstrates that it is an essential enzyme. J. Biol. Chem. 27839794-39800. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, Y., S. C. Roberts, A. Jardin, N. S. Carter, S. Shih, M. R. Ariyanayagam, A. H. Fairlamb, and B. Ullman. 1999. Ornithine decarboxylase gene deletion mutants of Leishmania donovani. J. Biol. Chem. 2743781-3788. [DOI] [PubMed] [Google Scholar]
- 21.Kashiwagi, K., S. Miyamoto, F. Suzuki, H. Kobayashi, and K. Igarashi. 1992. Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 894529-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashiwagi, K., R. Pistocchi, S. Shibuya, S. Sugiyama, K. Morikawa, and K. Igarashi. 1996. Spermidine-preferential uptake system in Escherichia coli. Identification of amino acids involved in polyamine binding in PotD protein. J. Biol. Chem. 27112205-12208. [DOI] [PubMed] [Google Scholar]
- 23.Krieger, S., W. Schwarz, M. R. Ariyanayagam, A. H. Fairlamb, R. L. Krauth-Siegel, and C. Clayton. 2000. Trypanosomes lacking trypanothione reductase are avirulent and show increased sensitivity to oxidative stress. Mol. Microbiol. 35542-552. [DOI] [PubMed] [Google Scholar]
- 24.Li, F., S. Hua, C. C. Wang, and K. Gottesdiener. 1998. Trypanosoma brucei brucei: characterization of an ODC null bloodstream form mutant and the action of alpha-difluoromethylornithine. Exp. Parasitol. 88255-257. [DOI] [PubMed] [Google Scholar]
- 25.Mangold, U. 2006. Antizyme inhibitor: mysterious modulator of cell proliferation. Cell. Mol. Life Sci. 632095-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osterman, A. L., H. B. Brooks, L. K. Jackson, J. J. Abbott, and M. A. Phillips. 1999. Lys-69 plays a key role in catalysis by T. brucei ornithine decarboxylase through acceleration of the substrate binding, decarboxylation and product release steps. Biochemistry 3811814-11826. [DOI] [PubMed] [Google Scholar]
- 27.Pegg, A. E., H. Xiong, D. J. Feith, and L. M. Shantz. 1998. S-adenosylmethionine decarboxylase: structure, function and regulation by polyamines. Biochem. Soc. Trans. 26580-586. [DOI] [PubMed] [Google Scholar]
- 28.Pegg, A. E. 2006. Regulation of ornithine decarboxylase. J. Biol. Chem. 28114529-14532. [DOI] [PubMed] [Google Scholar]
- 29.Pegg, A. E., and D. J. Feith. 2007. Polyamines and neoplastic growth. Biochem. Soc. Trans. 35295-299. [DOI] [PubMed] [Google Scholar]
- 30.Phillips, M. A., P. Coffino, and C. C. Wang. 1987. Cloning and sequencing of the ornithine decarboxylase gene from Trypanosoma brucei. Implications for enzyme turnover and selective α-difluoromethylornithine inhibition. J. Biol. Chem. 2628721-8727. [PubMed] [Google Scholar]
- 31.Roberts, S., J. Scott, J. Gasteier, Y. Jiang, B. Brooks, A. Jardim, N. Carter, O. Heby, and B. Ullman. 2002. S-adenosylmethionine decarboxylase from Leishmania donovani: molecular, genetic and biochemical characterization of null mutants and overproducers. J. Biol. Chem. 2775902-5909. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, S. C., Y. Jiang, A. Jardim, N. S. Carter, O. Heby, and B. Ullman. 2001. Genetic analysis of spermidine synthase from Leishmania donovani. Mol. Biochem. Parasitol. 115217-226. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Caso, C., R. Montanez, M. Cascante, F. Sanchez-Jimenez, and M. A. Medina. 2006. Mathematical modeling of polyamine metabolism in mammals. J. Biol. Chem. 28121799-21812. [DOI] [PubMed] [Google Scholar]
- 34.Ruan, H., L. M. Shantz, A. E. Pegg, and D. R. Morris. 1996. The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J. Biol. Chem. 27129576-29582. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, M. C., H. Kaur, B. Blessington, J. M. Kelly, and S. R. Wilkinson. 2008. Validation of spermidine synthase as a drug target in African trypanosomes. Biochem. J. 409563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uemura, T., H. F. Yerushalmi, G. Tsaprailis, D. E. Stringer, K. E. Pastorian, L. Hawel III, C. V. Byus, and E. W. Gerner. 2008. Identification and characterization of a diamine exporter in colon epithelial cells. J. Biol. Chem. 28326428-26435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner, J., Y. Hirth, N. Claverie, and C. Danzin. 1986. A sensitive high-performance liquid chromatographic procedure with fluorometric detection for the analysis of decarboxylated S-adenosylmethionine and analogs in urine samples. Anal. Biochem. 154604-617. [DOI] [PubMed] [Google Scholar]
- 38.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 27540174-40179. [DOI] [PubMed] [Google Scholar]
- 39.Willert, E. K., R. Fitzpatrick, and M. A. Phillips. 2007. Allosteric regulation of an essential trypanosome polyamine biosynthetic enzyme by a catalytically dead homolog. Proc. Natl. Acad. Sci. USA 1048275-8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willert, E. K., and M. A. Phillips. 2008. Regulated expression of an essential allosteric activator of polyamine biosynthesis in African trypanosomes. PLoS Pathog. 4e1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yarlett, N., and C. J. Bacchi. 1988. Effect of DL-alpha-difluoromethylornithine on methionine cycle intermediates in Trypanosoma brucei brucei. Mol. Biochem. Parasitol. 271-10. [DOI] [PubMed] [Google Scholar]