Abstract
Microbial diversity of septic tank effluent (STE) and the biomat that is formed as a result of STE infiltration on soil were characterized by 16S rRNA gene sequence analysis. Results indicate that microbial communities are different within control soil, STE, and the biomat and that microbes found in STE are not found in the biomat. The development of a stable soil biomat appears to provide the best on-site water treatment or protection for subsequent groundwater interactions of STE.
On-site wastewater treatment systems serve approximately 22 million homes throughout the United States (39), resulting in ∼15 billion liters of wastewater, typically treated in septic tanks (Fig. 1A), being discharged into the environment daily (8). Septic tank effluent (STE) is normally discharged into subsurface trenches or beds (i.e., soil treatment units [STU]) (40), where many pathogenic bacteria, viruses, and nutrients are removed, transformed, or destroyed.
FIG. 1.
Shown are typical on-site wastewater system components (A), a chamber system with an open infiltrative architecture (B), and a stone-and-pipe system with a gravel-laden infiltrative surface architecture (C). (Adapted with permission from Infiltrator Systems, Inc.)
Within the STU, a complex ecosystem referred to as a biomat evolves over time at and near the soil infiltrative surface in response to STE application (5, 6, 32). This region (1 to 2 cm in thickness) becomes dark in color and has high organic matter accumulation, high water content, and high microbial densities. The biomat may be important from a purification perspective (42); however, excessive soil pore clogging can be detrimental to long-term hydraulic functioning.
Biomat formation and potential soil clogging appear to be dependent on several factors: wastewater composition and loading rate (18, 31, 32, 38), soil characteristics (23), microorganisms (2, 11, 14, 24, 29, 33), temperature (17), and wastewater application method (17). Although it is believed that microorganisms are involved, research examining the microbial community of the biomat is limited and until recently has focused only on using plating techniques to determine bacterial numbers (27, 29). Few researchers have attempted to characterize the microbial community found in the biomat (3, 29, 41). In this study, traditional culturing techniques and a molecular approach (see “Methods” in the supplemental material) were used to explore the diversity of microbes found in domestic STE and the biomat that developed in the STU in a sandy loam soil.
The biomat that developed within an Ascalon sandy loam soil as a result of STE infiltration over a period of 30 months was characterized using 16S rRNA gene sequence analysis and culturing techniques (37). The microbial communities were analyzed by using STE and biomat samples from two different pilot-scale infiltration units for STE; one (MP10) had an open architecture above the soil infiltrative surface (Fig. 1B), and one (MP9) had gravel aggregate sitting on the soil infiltrative surface (Fig. 1C). For use as a negative control, samples of sandy loam soil were also collected from an infiltration unit at the same site that received clean tap water and had an open architecture. Biomat samples were taken at two depth intervals, 0.0 to 0.5 cm and 0.5 to 1.0 cm from test units MP10 and MP9, and the control sample was taken from 0.0 to 0.5 cm. At the time of sampling, the three infiltration units had received STE or tap water for 30 months.
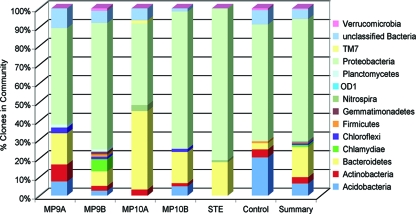
A total of 447 bacterial 16S rRNA gene sequences were generated from samples of the STE and the biomat (two STE units and a tap water unit). This number likely represents a great undersampling of these environments; however, these sequences are sufficient to provide insight into the microbial community phylotype composition. Although “universal” (i.e., all three phylogenetic domains amplified) PCR primers 515F and 1391R (19) were used, only organisms from the bacterial domain were detected. As in a previous study of a biomat (3) and other studies of soil microbial communities (9, 28), Proteobacteria sequences dominated the biomat samples, but Bacteroidetes and Acidobacteria sequences were also found in significant abundance (Fig. 2). The control community was comprised mainly of sequences from Proteobacteria, Acidobacteria, and Actinobacteria, with a distribution similar to that of the biomat. However, only 31% of sequences at the family level were shared between the 0.0- to 0.5-cm-depth samples of each biomat and the control. Furthermore, 95% of the operational taxonomic units (OTUs) found in the control sample were not found in any other sample analyzed in this study.
FIG. 2.
Bacterial phylum diversity based on rRNA gene clone libraries and the Ribosomal Database Project classification (7). MP9A, gravel-laden architecture test unit (0.5 cm); MP9B, gravel-laden architecture test unit (1.0 cm); MP10A, open architecture test unit (0.5 cm); MP10B, open architecture test unit (1.0 cm); STE; Control, negative control infiltrative surface architecture; and Summary, a compilation of the sequences of all clones used in this study and examined in all environments. Percentages are based on the total number of clones within each sample.
The lack of shared sequences suggests that the nutrient-rich STE fosters the growth of a different soil microbial community. The composition of the effluent applied (clean water versus STE) appears to lead to distinct microbial communities, based on the relatively low percentage of shared OTUs. Additionally, scanning electron micrographs of both biomat and control soil samples were strikingly different, supporting the differences observed by molecular comparative methods (see Fig. S1 in the supplemental material).
The STE library was also dominated by proteobacteria (81%), but the class distribution differed considerably from both the control and biomat (see Fig. S2 in the supplemental material). Epsilonproteobacteria (from the genera Sulfurospirillum, Arcobacter, and Sulfurimonas) species represented the dominant subphyla in the STE sample but were absent from all other samples. None of the OTUs found in STE were found in the biomat samples, suggesting that the microbial community of the biomat originates in the soil rather than from the influx of organisms entering the soil in STE. This also may indicate limited survival or the possibility that they are outcompeted by indigenous soil microbes. Species of both the Sulfurospirillum and Sulfurimonas genera have also been found in aquatic environments (13, 16) and contaminated sediments (22). Sequences of Arcobacter spp., aerotolerant gram-negative spiral-shaped bacteria, were the most dominant found in STE. Arcobacter spp. have been found in mangrove sediments (21) and activated sludge (34) and have been associated with human and animal diarrhea (1, 20, 36). The lack of Arcobacter spp. in any of the biomat samples indicates a rapid die-off or predation at the soil infiltrative surface or the possibility that they are vastly outnumbered in this environment and were not detected by the sequencing performed.
There was far less overall phylogenetic diversity in STE than in the biomat samples (see Table S1 in the supplemental material). The phylum distribution in the STE is similar to that of the human gut, which contains some of the highest cell densities of any ecosystem (43); however, diversity at the phylum level is among the lowest (15). Studies have shown that the human gut and feces are dominated by Bacteroidetes (30%) and Firmicutes (30%) species (4, 10). The lack of sequences grouping with Firmicutes spp. in this study suggests that they may not survive the septic tank environment. Not surprisingly, Escherichia coli sequences were not detected in this study, as these bacteria represent less than 0.1% of the species found in the colon (10, 35).
The amount of fecal coliforms and E. coli bacteria was reduced by a minimum of 90% and 85%, respectively, from the STE to the biomat. Similar to results of previous studies (12, 25, 26, 30), culturable heterotrophic bacteria, fecal coliforms, and E. coli bacteria decreased with the increase in depth (1 to 10 cm) in both of the STE test units. The number of heterotrophic bacteria was 3 to 5 orders of magnitude greater in all samples than counts for fecal coliforms and E. coli. At all depths, the number of fecal coliforms and E. coli were greater in test unit MP10 than in MP9; this may be due in part to differences between open versus gravel-laden infiltration surfaces, with the gravel architecture of MP9 being more conducive to the formation of a water-treating biomat.
Further analysis may determine the roles that microbes play in the treatment of organic and inorganic pollutants in the biomat. Ultimately, understanding the microbial community will allow engineers to implement on-site systems that control the rate of biomat formation and thus keep clogging and subsequent failure to a minimum. This improved design and operation would help to protect the underlying groundwater.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to the GenBank database and assigned the accession numbers EU403601 to EU404047. Clone names designate the community (MP9A, test unit 9, 0.5 cm; MP9B, test unit 9, 1.0 cm; MP10A, test unit 10, 0.5 cm; MP10B, test unit 10, 1.0 cm; STEU-STE, CTRL-negative control test unit, 0.5 cm; plus the clone number [e.g., MP9A1]).
Supplementary Material
Acknowledgments
We thank Jim McKinley for assistance with sample collection, the Pace lab at the University of Colorado for sequencing assistance, and Robert Findlay and Jen Mosher at the University of Alabama for analysis of biomass. We also thank Kathryn Lowe for technical guidance and Charles Pepe-Ranney for lab support.
This work was sponsored and supported by the Colorado School of Mines and by Infiltrator Systems, Inc.
Footnotes
Published ahead of print on 20 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abdelbaqi, K., A. Buissonnière, V. Prouzet-Mauleon, J. Gresser, I. Wesley, F. Mégraud, and A. Ménard. 2007. Development of a real-time fluorescence resonance energy transfer PCR to detect Arcobacter species. J. Clin. Microbiol. 45:3015-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, L. E. 1947. Effect of microorganisms on permeability of soil under prolonged submergence. Soil Sci. Soc. 63:439-450. [Google Scholar]
- 3.Amador, J. A., D. A. Potts, M. C. Savin, P. Tomlinson, J. H. Gorres, and E. L. Nicosia. 2006. Mesocosm-scale evaluation of faunal and microbial communities of aerated and unaerated leachfield soil. J. Environ. Qual. 35:1160-1169. [DOI] [PubMed] [Google Scholar]
- 4.Bäckhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 5.Beach, D. N., J. E. McCray, K. S. Lowe, and R. L. Siegrist. 2005. Temporal changes in hydraulic conductivity of sand porous media biofilters during wastewater infiltration due to biomat formation. J. Hydrol. 311:230-243. [Google Scholar]
- 6.Bouma, J. 1975. Unsaturated flow during soil treatment of septic tank effluent. J. Environ. Eng. 101:967-981. [Google Scholar]
- 7.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conn, K. E., L. B. Barber, G. K. Brown, and R. L. Siegrist. 2006. Occurrence and fate of organic contaminants during onsite wastewater treatment. Environ. Sci. Technol. 40:7358-7366. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankenberger, W. T., F. R. Troeh, and L. C. Dumenil. 1979. Bacterial effects on hydraulic conductivity of soils. Soil Sci. Soc. Am. J. 43:333-338. [Google Scholar]
- 12.Garcia-Pichel, F., S. L. Johnson, D. Youngkin, and J. Belnap. 2003. Small-scale vertical distribution of bacterial biomass and diversity in biological soil crusts from arid lands in the Colorado plateau. Microb. Ecol. 46:312-321. [DOI] [PubMed] [Google Scholar]
- 13.Grote, J., M. Labrenz, B. Pfeiffer, G. Jost, and K. Jurgens. 2007. Quantitative distributions of Epsilonproteobacteria and a Sulfurimonas subgroup in pelagic redoxclines of the central Baltic Sea. Appl. Environ. Microbiol. 73:7155-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, R. P., and D. Swartzendruber. 1962. Flow-associated reduction in the hydraulic conductivity of quartz sand. Soil Sci. Soc. Proc. 26:6-10. [Google Scholar]
- 15.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, A., and K. Finster. 2005. Isolation and characterization of Sulfurospirillum carboxydovorans sp. nov., a new microaerophilic carbon monoxide oxidizing epsilon Proteobacterium. Antonie van Leeuwenhoek 87:339-353. [DOI] [PubMed] [Google Scholar]
- 17.Kristiansen, R. 1981. Sand-filter trenches for purification of septic tank effluent. I. The clogging mechanism and soil physical environment. J. Environ. Qual. 10:361-364. [Google Scholar]
- 18.Laak, R. 1970. Influence of domestic wastewater pretreatment on soil clogging. J. Water Pollut. Control Fed. 42:1495-1500. [Google Scholar]
- 19.Lane, D. J. 1991. Nucleic acid techniques in bacterial systematics. Wiley, New York, NY.
- 20.Lerner, J., V. Brumberger, and V. Preac-Mursic. 1994. Severe diarrhea associated with Arcobacter butzleri. Eur. J. Clin. Microbiol. Infect. Dis. 13:660-662. [DOI] [PubMed] [Google Scholar]
- 21.Liao, P. C., B. H. Huang, and S. Huang. 2007. Microbial community composition of the Danshui river estuary of Northern Taiwan and the practicality of the phylogenetic method in microbial barcoding. Microb. Ecol. 54:497-507. [DOI] [PubMed] [Google Scholar]
- 22.Luijten, M. L., J. de Weert, H. Smidt, H. T. Boschker, W. M. de Vos, G. Schraa, and A. J. Stams. 2003. Description of Sulfurospirillum halorespirans sp. nov., an anaerobic, tetrachloroethene-respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int. J. Syst. Evol. Microbiol. 53:787-793. [DOI] [PubMed] [Google Scholar]
- 23.Magdoff, F. R., and J. Bouma. 1974. The development of soil clogging in sands leached with septic tank effluent, p. 37-47. In Proceedings of the National Home Sewage Disposal Symposium. American Society of Agricultural Engineers, Chicago, IL.
- 24.McCalla, T. M. 1950. Studies of the effect of microorganisms on rate of percolation of water though soil. Soil Sci. Soc. Proc. 15:182-186. [Google Scholar]
- 25.Pederson, K. 1993. The deep subterranean biosphere. Earth Sci. Rev. 34:243-260. [Google Scholar]
- 26.Pell, M., and H. Ljunggren. 1984. Reduction of organic matter with focus on the clogging phenomenon, p. 311-327. In Proceedings of the International Conference on New Technology for Wastewater Treatment and Sewerage in Rural and Suburb Areas. Tampere University of Technology, Hanasaari, Finland.
- 27.Pell, M., F. Nyberg, and H. Ljunggren. 1990. Microbial numbers and activity during infiltration of septic-tank effluent in a subsurface sand filter. Water Res. 24:1347-1354. [Google Scholar]
- 28.Roesch, L. F., R. R. Fulthorpe, A. Riva, G. Casella, A. K. Hadwin, A. D. Kent, S. H. Daroub, F. A. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ronner, A. B., and A. C. L. Wong. 1994. Microbial clogging of wastewater infiltration systems, p. 559-562. In Proceedings of the 7th National Symposium on Individual and Small Community Sewage Systems. American Society of Agricultural Engineers, Atlanta, GA.
- 30.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 31.Siegrist, R. L. 1985. Soil clogging effects of effluent composition and loading rate, p. 249-256. In Proceedings of the 5th Northwest Onsite Wastewater Treatment Short Course. University of Washington, Seattle.
- 32.Siegrist, R. L., and W. C. Boyle. 1987. Wastewater-induced soil clogging development. J. Environ. Eng. 113:550-566. [Google Scholar]
- 33.Siegrist, R. L., R. Smed-Hildmann, Z. K. Filip, and P. D. Jenssen. 1991. Humic substance formation during wastewater infiltration, p. 223-232. In Proceedings of the 6th National Symposium on Individual and Small Community Sewage Systems. American Society of Agricultural Engineers, St. Joseph, MI.
- 34.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K. H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor, D. N., J. A. Kiehlbauch, W. Tee, C. Pitarangsi, and P. Echeverria. 1991. Isolation of group 2 aerotolerant Campylobacter species from Thai children with diarrhea. J. Infect. Dis. 163:1062-1067. [DOI] [PubMed] [Google Scholar]
- 37.Tomaras, J. M. B., J. R. Spear, J. W. McKinley, and R. L. Siegrist. 2006. The microbial characteristics of the wastewater-induced soil biozone, p. 28-30. In Proceedings of the 15th Annual National Onsite Wastewater Recycling Association (NOWRA) Technical Education Conference and Exposition. NOWRA, Denver, CO.
- 38.Tyler, J. E., J. C. Converse, and J. R. Keys. 1995. Soil acceptance of wastewater affected by wastewater quality, p. 96-109. In Proceedings of the 8th Northwest Onsite Wastewater Treatment Short Course and Equipment Exhibition. University of Washington, Seattle.
- 39.U.S. Census Bureau. 2002. American housing survey for the United States: 2001. Current housing reports, series H150/01. U.S. Government Printing Office, U.S. Census Bureau, Washington, DC. http://www.census.gov/hhes/www/housing/ahs/ash01_2000wts/tab14.html.
- 40.U.S. Environmental Protection Agency. 2002. Homeowner's guide to septic systems. EPA-832-B-02-005. Office of Water, U.S. Environmental Protection Agency, Washington, DC.
- 41.Van Cuyk, S. 2003. Fate of virus during wastewater renovation in soil porous media biofilters. Ph.D. thesis. Colorado School of Mines, Golden, CO.
- 42.Van Cuyk, S., R. L. Siegrist, K. Lowe, and R. W. Harvey. 2004. Evaluating microbial purification during soil treatment of wastewater with multicomponent tracer and surrogate tests. J. Environ. Qual. 33:316-329. [DOI] [PubMed] [Google Scholar]
- 43.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.