Abstract
Populations of the food- and waterborne pathogen Escherichia coli O157:H7 are comprised of two major lineages. Recent studies have shown that specific genotypes within these lineages differ substantially in the frequencies with which they are associated with human clinical disease. While the nucleotide sequences of the genomes of lineage I strains E. coli O157 Sakai and EDL9333 have been determined, much less is known about the genomes of lineage II strains. In this study, suppression subtractive hybridization (SSH) was used to identify genomic features that define lineage II populations. Three SSH experiments were performed, yielding 1,085 genomic fragments consisting of 811 contigs. Bacteriophage sequences were identified in 11.3% of the contigs, 9% showed insertions and 2.3% deletions with respect to E. coli O157:H7 Sakai, and 23.2% did not have significant identity to annotated sequences in GenBank. In order to test for the presence of these novel loci in lineage I and II strains, 27 PCR primer sets were designed based on sequences from these contigs. All but two of these PCR targets were found in the majority (51.9% to 100%) of 27 lineage II strains but in no more than one (<6%) of the 17 lineage I strains. Several of these linage II-related fragments contain insertions/deletions that may play an important role in virulence. These lineage II-related loci were also shown to be useful markers for genotyping of E. coli O157:H7 strains isolated from human and animal sources.
Enterohemorrhagic Escherichia coli is associated with diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome in humans (31). E. coli serotype O157:H7 predominates in epidemics and sporadic cases of enterohemorrhagic E. coli-related infections in the United States, Canada, Japan, and the United Kingdom (12). Cattle are considered the most important reservoir of E. coli O157:H7 (10, 24, 37, 41), and foods contaminated with bovine feces are thought to be the most common source of human infection with this pathogen (27, 33). The two most important virulence factors of the organism are the production of one or more Shiga toxins (Stx) (6, 20, 32) and the ability to attach to and efface microvilli of host intestinal cells (AE). Stx genes are encoded by temperate bacteriophage inserted in the bacterial chromosome, and genes responsible for the AE phenotype are located on the locus of enterocyte effacement (LEE) as well as other pathogenicity islands (4, 17). All E. coli O157:H7 strains also possess a large plasmid which is thought to play a role in virulence (10, 40, 42).
Octamer-based genome scanning (OBGS) was first used to show that E. coli O157 strains from the United States and Australia could be subdivided into two genetically distinct lineages (21, 22, 46). While both E. coli O157:H7 lineages are associated with human disease and are isolated from cattle, there is a bias in the host distribution between the two lineages, with a significantly higher proportion of lineage I strains isolated from humans than lineage II strains. Several recent studies have shown that there are inherent differences in gene content and expression between populations of lineage I and lineage II E. coli O157:H7 strains. Lejeune et al. (26) reported that the antiterminator Q gene of the stx2-converting bacteriophage 933W was found in all nine OBGS lineage I strains examined but in only two of seven lineage II strains, suggesting that there may be lineage-specific differences in toxin production. Dowd and Ishizaki (9) used DNA microarray analysis to examine expression of 610 E. coli O157:H7 genes and showed that lineage I and lineage II E. coli O157:H7 strains have evolved distinct patterns of gene expression which may alter their virulence and their ability to survive in different microenvironments and colonize the intestines of different hosts (9, 28, 38).
The observations of lineage host bias have been supported and extended by studies using a six-locus-based multiplex PCR termed the lineage-specific polymorphism assay (LSPA-6) (46). However, Ziebell et al. (48) have recently shown that not all LSPA-6 types within lineage II are host biased; e.g., LSPA-6 type 211111 isolation rates from humans and cattle were significantly different from those of other lineage II LSPA-6 types. Therefore, a clearer definition is required of not only the differences between lineages but also the differences among clonal groups within lineages.
The genome sequences of two E. coli O157:H7 strains, Sakai and EDL933 (14, 36), have been determined; however, both of these strains are of lineage I, and there are presently no completed and fully annotated genome sequences available for lineage II strains. In our laboratory, comparative studies utilizing suppression subtractive hybridization (SSH) and comparative genomic hybridization revealed numerous potential virulence factors that are conserved in lineage I strains and that are rare or absent in lineage II strains (42, 47). In this study, we have used SSH to identify genomic regions present in E. coli O157:H7 lineage II strains that are absent from lineage I strains. We wished to examine the distribution of these novel gene segments in E. coli O157:H7 strains and gain insight into their origins and functions. We also attempted to identify molecular markers specific to lineage II strains as well as other markers that would be useful in the genetic subtyping or molecular fingerprinting of E. coli O157:H7 strains in population and epidemiological studies (25). This information may be helpful in the identification of genotypes of the organism associated with specific phenotypes of both lesser and greater virulence (29).
MATERIALS AND METHODS
Bacterial strains.
OBGS type strains (93-001, FDA 516-520, and FRIK 523-2001) were previously described by Kim et al. (21). Zap strains were obtained from David Gally at the University of Edinburgh (30). Escherichia coli O157:H7 strains EDL933 (ATCC 700927) and Sakai (RIMD 0509952) were obtained from the American Type Culture Collection (Manassas, VA). The remaining strains were isolated from human infections or cattle in Canada (Table 1). LSPA-6 genotyping of these strains was performed as previously described (48).
TABLE 1.
Escherichia coli O157:H7 strains included in the study (n = 44)
| Strain | LSPA-6 genotype | Source | Country of origina | Phage type |
|---|---|---|---|---|
| ECI1375 | 111111 | Bovine | Canada | 14 |
| ECI1382 | 111111 | Bovine | Canada | 1 |
| ECI563 | 111111 | Bovine | Canada | 14 |
| ECI577 | 111111 | Bovine | Canada | 4 |
| ECI603 | 111111 | Bovine | Canada | 87 |
| ECI653 | 111111 | Bovine | Canada | 32 |
| ECI665 | 111111 | Bovine | Canada | 1 |
| 93001 | 111111 | Human | U.S.A. | 14 |
| ECI309 | 111111 | Human | Canada | 14 |
| ECI320 | 111111 | Human | Canada | 14 |
| ECI485 | 111111 | Human | Canada | 14 |
| EDL933 | 111111 | Human | U.S.A. | 21 |
| FDA 516 | 111111 | Human | U.S.A. | 21 |
| FDA 518 | 111111 | Human | U.S.A. | 21 |
| FDA 520 | 111111 | Human | U.S.A. | 1 |
| FRIK 523 | 111111 | Human | U.S.A. | 34 |
| Sakai | 111111 | Human | Japan | 32 |
| Zap0032 | 211111 | Bovine | Scotland | 8 |
| Zap0054 | 211111 | Bovine | Scotland | 32 |
| ECI504 | 211111 | Human | Canada | 2 |
| ECI882 | 211111 | Human | Canada | 1 |
| Zap0058 | 211111 | Human | Scotland | 87 |
| ECI241 | 212222 | Human | Canada | 74 |
| FRIK 2001 | 212232 | Bovine | U.S.A. | 54 |
| ECI240 | 212232 | Human | Canada | 54 |
| EC19930200 | 222221 | Bovine | Canada | 23 |
| FRIK 1985 | 222232 | Bovine | U.S.A. | 45 |
| EC19920005 | 222222 | Bovine | Canada | 67 |
| EC19920027 | 222222 | Bovine | Canada | 34 |
| EC19920171 | 222222 | Bovine | Canada | 23 |
| EC19970520 | 222222 | Bovine | Canada | 67 |
| EC20011139 | 222222 | Bovine | Canada | 82 |
| EC20030223 | 222222 | Bovine | Canada | 74 |
| EC20030289 | 222222 | Bovine | Canada | 23 |
| ECI1433 | 222222 | Bovine | Canada | 23 |
| ECI564 | 222222 | Bovine | Canada | 23 |
| ECI633 | 222222 | Bovine | Canada | 23 |
| FRIK 1990 | 222222 | Bovine | U.S.A. | 54 |
| FRIK 1999 | 222222 | Bovine | U.S.A. | 23 |
| FRIK 920 | 222222 | Bovine | U.S.A. | 23 |
| ECI306 | 222222 | Human | Canada | 23 |
| ER6554 | 222222 | Human | Canada | 23 |
| ER6666 | 222222 | Human | Canada | 40 |
| ER6816 | 222222 | Human | Canada | 40 |
U.S.A., United States.
Preparation of the SSH DNA library.
Bacterial cultures were grown overnight in brain heart infusion broth (Difco, Becton, Dickinson and Company, Franklin Lakes, NJ) in a 37°C shaker-incubator (200 rpm) and genomic DNA was extracted from harvested cells using the DNeasy tissue kit (Qiagen, Valencia, CA). The purity and concentration of the genomic DNA were assessed using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Rockland, DE).
SSHs were performed using the Clontech PCR-Select bacterial genome subtraction kit (BD Biosciences, Palo Alto, CA). In addition to the RsaI-digested DNA recommended in the Clontech kit, SSHs were also performed on AluI- and HaeIII-digested DNA to increase the diversity of DNA fragments obtained. Advantage polymerase mix (BD Biosciences) was used during the amplification steps. Three sets of SSH experiments were performed. In the first set, E. coli FRIK 920 (LSPA-6 222222) was subtracted with E. coli Sakai (LSPA-6 111111). In the second set, E. coli FRIK 1999 (LSPA-6 222222) was subtracted with E. coli Sakai. In the third set, E. coli FRIK 2001 (LSPA-6 212232) was subtracted with E. coli 93-001 (LSPA-6 111111).
The SSH DNA fragments isolated in these experiments were cloned into the pCR2.1-TOPO plasmid vector, using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) and plated onto LB agar (Difco) containing 50 μg/ml of ampicillin or kanamycin (Sigma-Aldrich Canada, Oakville, ON, Canada). Prior to cloning, the fragments were incubated for 10 min at 72°C in the presence of AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA) to ensure that all DNA fragments possessed the necessary “A” overhang.
DNA sequencing of SSH DNA library.
PCR amplicons were generated directly from the SSH library clones by transferring whole bacterial cells to 20-μl aliquots of PCR master mix containing 1× buffer II (Applied Biosystems), 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphates, 1 U AmpliTaq DNA polymerase (Applied Biosystems), and 0.2 μM each M13 Forward (5′-GTAAAACGACGGCCAG-3′) and M13 Reverse (5′-CAGGAAACAGCTATGAC-3′) PCR primer sets (Invitrogen). An initial 10-min incubation at 94°C performed to lyse the cells was followed by 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C and a final 10-min extension step at 72°C. PCR amplicons were purified by passage through superfine Sephadex G-50 (Sigma) packed into a MultiScreen 96-well filtration plate (Millipore, Billerica, MA) and sequenced with the DYEnamic ET terminator cycle sequencing kit (GE Health Care, Piscataway, NJ) using M13 Forward and M13 Reverse primer sets. Sequencing products were purified by passage through superfine Sephadex G-50 packed into a MultiScreen 96-well filtration plate prior to nucleotide sequencing on a MegaBace 500 capillary sequencer (GE Health Care).
DNA sequence analysis of the SSH DNA library.
Phred base calling of sequence trace data was performed using the Interphace program (CodonCode Corporation, Dedham, MA). Base-called sequences obtained from each set of SSH experiments were analyzed by the SeqMan 5.05 sequence analysis program (DNAStar Inc., Madison, WI). This program removed vector and adaptor sequences and grouped sequences displaying at least 80% sequence homology into contiguous consensus sequences or “contigs.” Searches were performed on contigs using BLASTN and BLASTX (2) from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/).
PCR screening of putative lineage II conserved markers.
Twenty-seven PCR primer sets were designed to detect frequently occurring sequences within the SSH libraries. These primer sets were used to screen 17 E. coli O157:H7 LSPA-6 111111 strains, 17 genotype 222222 strains, five genotype 211111 strains, and five strains of other genotypes (Table 1). Primer Select 5.08 (DNAStar Inc.) was used to design PCR primer sets (Table 2). All PCR assays were performed in duplicate in 20-μl reaction mixture volumes containing 1× buffer II (Applied Biosystems), 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 1 U AmpliTaq DNA polymerase (Applied Biosystems), 0.2 μM primer sets, and 0.5 ng genomic DNA template. All PCRs included an initial 2-min denaturation step at 94°C, followed by 30 cycles of 1 min denaturation at 94°C, 1 min annealing, and 1 min extension at 72°C, and a final 10-min extension at 72°C. The primer sequences and annealing temperatures for each PCR are listed in Table 2. Positive PCR controls included the SSH tester E. coli strains FRIK 920, FRIK 1999, and FRIK 2001 and negative controls included Campylobacter jejuni ATCC 33560 and a no-template blank. E. coli FRIK 920 amplicons obtained from all primer sets were purified using the QIAquick PCR purification kit (Qiagen) and sequenced on an ABI Prism 277 DNA sequencer (Applied Biosystems, Foster City, CA) to verify that they corresponded to the sequences from the original contig assembly.
TABLE 2.
Primer sets used in PCR screening of lineage II conserved markers among E. coli O157:H7 strains
| Class | Primer set | Forward primer | Reverse primer | Temp (°C) |
|---|---|---|---|---|
| B | A02 | 5′-GCTGGCAGACAATGGCGAGTTT-3′ | 5′-GATACGGCTTGCGGGCTGTG-3′ | 59.3 |
| A03 | 5′-GCCACGGCATTAATAACGCTTCTT-3′ | 5′-CCAGGCATCACTACCCGCAAAAA-3′ | 57.4 | |
| A04 | 5′-GGAGACGGGCGCACTGATTG-3′ | 5′-CTCCACGATTTCGGGGCTTATGTA-3′ | 56.2 | |
| A05 | 5′-TGCGGCATCTGGTCATTCTGGTT-3′ | 5′-ACGGTCTGCGCGAGTTTTGTTT-3′ | 57.3 | |
| A09 | 5′-AGGGGGCATATTTCATTGTTGTGT-3′ | 5′-TAATCTCTTTCGCCTGCCCTTCTT-3′ | 56.4 | |
| A10 | 5′-CGAGGCGTGCGGAGAGGA-3′ | 5′-TTCAGCGCCAGTTCGGTTTTC-3′ | 58.8 | |
| A11 | 5′-AAAACCGTATGATCTATTATTCCT-3′ | 5′-TTTTTATTATGTTGGCTGAGTTAC-3′ | 50.3 | |
| A13 | 5′-ATGCCAGCCACCACCAACTCCAC-3′ | 5′-CTTACCCGCGCCGAACTTATCCTT-3′ | 58.2 | |
| B01 | 5′-AAGCCCCCACGTAATTCCCTGACA-3′ | 5′-CATTTCCCGCGCCTGACTGAGA-3′ | 60.7 | |
| B02 | 5′-AAGCGGCATAAATGGCAGACAGAC-3′ | 5′-AACGCCACGCTTTACCTTTACCC-3′ | 57.8 | |
| B03 | 5′-AATCCGCCCGTTTTTACTGA-3′ | 5′-TGTGCCTGATAGCTCCTTCTTTTT-3′ | 54.9 | |
| B06 | 5′-CCCCCTGTTGCGTCTGCTGAAAA-3′ | 5′-CGGCCATCCATGCGGTGACTT-3′ | 60.8 | |
| B07 | 5′-TAAAGCGCTGGTGGGGAAAGGTAG-3′ | 5′-CCAGAACGCGGGCACAAAAA-3′ | 54.5 | |
| B08 | 5′-TGCCGACATCGCCAGTTG-3′ | 5′-GCCATGCTTCAGGGAGATAAAT-3′ | 55.7 | |
| B10 | 5′-TGTGCGTCGTTTCAGTTCGTCA-3′ | 5′-TAGGTGTTCCGCGTCGTGTAAAAG-3′ | 57.8 | |
| B16 | 5′-CATGCTTCTGCCCTGAGTT-3′ | 5′-GCGCCGTGCTTATGAAA-3′ | 54.8 | |
| C | A14 | 5′-TGCCGACAACCTCCCACAGATACT-3′ | 5′-TCACTTCCGCTACAACACGCACAT-3′ | 57.8 |
| A15 | 5′-CGCCAAAGCAGCACAGCAGGATA-3′ | 5′-AGAAAAACAGGCGAAGGCGATGAT-3′ | 53.4 | |
| D | A06c | 5′-CCGCCTGCGATGGTGGTTGC-3′ | 5′-GGGCGCGGGTGATTTTGCTCTC-3′ | 60.2 |
| A07 | 5′-ATGCGATCGCCTTCTTCAA-3′ | 5′-TACCATACACGCCACAGTTTTTA-3′ | 52.3 | |
| B15 | 5′-CCCGCTGGCAGGCATTGAAG-3′ | 5′-GGCGGCAGCGGACACGAG-3′ | 60.4 | |
| E | A01 | 5′-ACCAAGGCATCCCCCGTGTGAA-3′ | 5′-ATAATCCGCTGGGGCTGGCTGAC-3′ | 60.8 |
| B05 | 5′-GGTTTTCCGGCACTTTCCACTCCA-3′ | 5′-ATCCTGCCGGGCGAACATCCTTAT-3′ | 58.1 | |
| B12 | 5′-TGAACACCCGCAGCAACA-3′ | 5′-CGCCGCATCTACTCCTATCG-3′ | 54.2 | |
| B18 | 5′-AATAACTCGGCTTTTGCTTTTT-3′ | 5′-AATACTCCGGTTCTGTCTAATCC-3′ | 52.5 | |
| F | A12 | 5′-GGGCGGACTTTGTTTGGTTGAA-3′ | 5′-GCCTGGCGGAAATGGACTGTAT-3′ | 55.9 |
| B13 | 5′-CTGGATGCGGCAAAACCTGT-3′ | 5′-GCCCCTTCTCTACGCAAATCAT-3′ | 55.0 |
Analysis of distribution of lineage II conserved regions in E. coli O157:H7 strains.
Results of PCR screening assays targeting lineage II conserved markers within the 44 strains tested were converted into presence/absence binary data. A neighbor-joining dendrogram was generated based on a matrix of pairwise distances calculated from the binary data, using the proportion of shared markers as a distance metric. Based on similar BLASTN homologies and strain distributions, some PCR primer sets appeared to recognize the same locus. To avoid biasing the dendrogram results, a single representative PCR assay from each of these 18 loci was included in the final binary data. The data were subsequently bootstrapped, and a neighbor-joining consensus dendrogram was created using the neighbor and consense programs from Phylip v3.16 (11). E. coli K-12 (5) was used as an out-group strain in the dendrogram since in silico analysis indicated that none of the primer sets would have generated a PCR product of the correct size in this strain.
RESULTS AND DISCUSSION
SSH DNA library sequences.
The three lineage II-minus-lineage I SSH libraries yielded 811 contigs, which were obtained from 1,085 clones. Each contig represented between 1 and 27 of the clones obtained, with an average of 1.58 ± 1.90 clones per contig (Table 3). Sequence identities of the contigs were obtained from BLASTN searches, and sequences were binned into six different classes (A to F), based on sequence identity to E. coli O157:H7 strain Sakai (A), non-Sakai bacteriophage (B), transitional sequences between lineage-common and lineage II strain-specific DNA (C), sequence deletions in the E. coli O157:H7 lineage II strain with respect to strain Sakai (D), nonbacteriophage DNA found in bacteria other than O157 strain Sakai (E), and those bearing little or no sequence identity to sequences in the GenBank NR database (F). Because many contigs contained sequences that could be ascribed to more than one class, the relative proportions of the different classes exceed 100%.
TABLE 3.
Frequency of sequence classes within SSH DNA libraries
| Sequence classa | No. (%) of clones present in:
|
Range | Avg no. of clones/contig | |||
|---|---|---|---|---|---|---|
| All contigs (n = 811) | 3 SSH (n = 16) | 2 SSH (n = 73) | 1 SSH (n = 722) | |||
| A | 503 (62.0) | 5 (31.3) | 30 (41.1) | 468 (64.7) | 1-7 | 1.30 ± 0.82 |
| B | 92 (11.3) | 10 (66.7) | 34 (46.6) | 48 (6.6) | 1-16 | 2.90 ± 2.64 |
| C | 72 (8.9) | 3 (18.8) | 12 (16.4) | 57 (7.9) | 1-7 | 1.60 ± 1.22 |
| D | 19 (2.3) | 0 (0) | 6 (8.2) | 13 (1.8) | 1-5 | 1.79 ± 1.13 |
| E | 80 (9.9) | 1 (13.3) | 8 (12.7) | 72 (9.7) | 1-27 | 3.01 ± 4.67 |
| F | 188 (23.3) | 5 (31.3) | 14 (19.2) | 177 (24.5) | 1-11 | 1.67 ± 1.65 |
| All | 1-27 | 1.58 ± 1.90 | ||||
Class A, sequences with homology to E. coli Sakai (14) genes; class B, sequences with homology to bacteriophage sequences not found in E. coli Sakai; class C, putative mosaic sequences with homology to both E. coli Sakai and non-Sakai DNA sequences within the same contig; class D, sequences with homology to E. coli Sakai genes but lacking synteny with the E. coli Sakai chromosome; class E, sequences with homology to previously published nonbacteriophage and non-Sakai DNA sequences; class F, sequences with regions containing no homology to any annotated DNA sequences in GenBank.
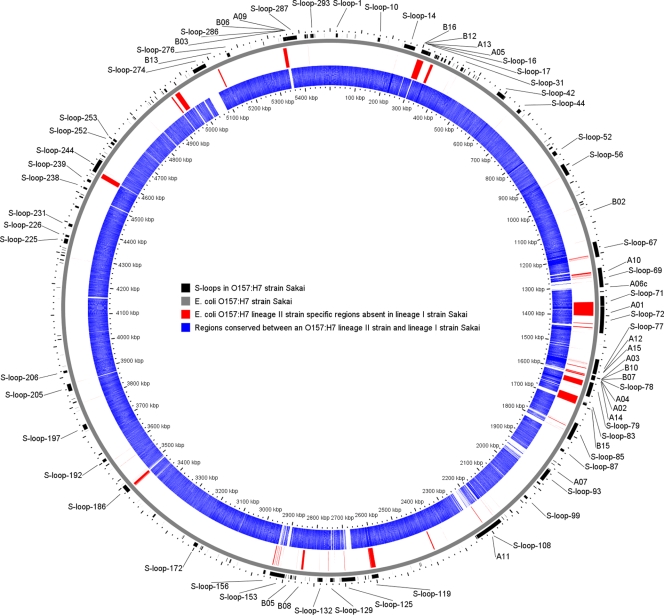
Class A contigs possessed at least 90% identity over at least 90% of their length to sequences from E. coli Sakai (14) and likely represent driver DNA contamination or lineage II sequences with only minor deviations from that in the lineage I E. coli Sakai genome. These sequences accounted for 503 (62.0%) of the contigs and were not analyzed further. The remaining 308 contigs were reasoned to be enriched for sequences that are unique to lineage II strains. In order to determine the distribution of these DNA sequences in our E. coli O157:H7 collection of 44 strains of different LSPA-6 genotypes (Table 1), PCR primers were designed to detect representative target sequences from these contigs (Table 2). The 27 PCR targets tested included 11 of the 16 sequences detected in all three SSH experiments, 11 of the 73 sequences detected in two SSH experiments, and five sequences detected in single SSH experiments (Tables 3 and 4). Class B, C, E, and F sequences represent insertions of DNA segments in lineage II strains with respect to the lineage I E. coli Sakai genome, and class D sequences represent deletions in lineage II strains with respect to the position of the element in the E. coli Sakai genome. The approximate location of these contigs and their corresponding PCR targets in the E. coli O157:H7 genome are shown in Fig. 1.
TABLE 4.
Distribution of putative lineage II conserved markers among E. coli O157:H7 strains from various lineages
| Class | Primer set | SSH(s)a | Predicted amplicon size (bp) | No. of indicated markers/total no. (%)
|
Sequence homology | |||
|---|---|---|---|---|---|---|---|---|
| Lineage I (LSPA-6 111111) | Lineage II (LSPA-6 222222) | Lineage II (LSPA-6 211111) | Other lineage II | |||||
| B | A10 | 2, 3 | 435 | 0/17 (0) | 17/17 (100) | 3/5 (60) | 4/5 (80) | E. coli UTI89 (7) (96% homology over 1,388 nt) genes with homology to bacteriophage HK97 gp56 and bacteriophage Nil2p gene dnaB homologue |
| A11 | 1, 2, 3 | 365 | 0/17 (0) | 17/17 (100) | 3/5 (60) | 4/5 (80) | Bacteriophage BP-4795 (8) (95% homology over 762 nt), encoding a hypothetical protein | |
| A13 | 1, 2, 3 | 204 | 0/17 (0) | 17/17 (100) | 0/5 (0) | 5/5 (100) | E. coli K-12 (5) ORFs b1155 to b1157 (81% homology over 1,260 nt), encoding hypothetical proteins and a putative tail fiber assembly protein within the cryptic prophage e14 | |
| B02 | 1, 2, 3 | 136 | 0/17 (0) | 13/17 (76.5) | 5/5 (100) | 3/5 (60) | Bacteriophage P2 (accession no. AF063097) genes J and I (94% homology over 1,019 nt), encoding baseplate assembly proteins | |
| A05 | 1, 2, 3 | 246 | E. coli Sakai (14) ORF ECs0284 (88% homology over 338 nt), encoding a DNA invertase, adjacent to 213 nt of DNA sequence, without significant homology to known DNA sequences adjacent to bacteriophage P2 (accession no. AF063097) FI tail sheath protein gene (95% homology over 546 nt) | |||||
| B03 | 1, 2, 3 | 746 | 0/17 (0) | 17/17 (100) | 0/5 (0) | 5/5 (100) | E. coli O109I P4-like bacteriophage ORFs ecO109Im and psu (23) (98% homology over 1,248 nt), encoding methyltransferase and Psu-like proteins | |
| B06 | 1, 2, 3 | 327 | 0/17 (0) | 17/17 (100) | 0/5 (0) | 5/5 (100) | S. boydii strain Sb227 (45) ORFs SBO4168 to SBO4170 (97% homologous over 1,352 nt) and SBO3220 (98% homology over 170 nt), encoding a putative bacteriophage protein and conserved hypothetical proteins | |
| A09 | 2, 3 | 572 | S. boydii strain Sb227 (45) ORFs SBO4164 and SBO4165 (95% homology over 1,366 nt), encoding a prophage P4 integrase and a putative DNA primase | |||||
| B08 | 2, 3 | 616 | 0/17 (0) | 17/17 (100) | 3/5 (60) | 5/5 (100) | Bacteriophage HK022 (19) (93% homology over 632 nt, with gaps), intergenic region between genes encoding an excisionase and an Ea22 protein | |
| B10 | 1, 2, 3 | 272 | 0/17 (0) | 17/17 (100) | 5/5 (100) | 5/5 (100) | S. boydii strain Sb227 (45) ORF SBO2131 (99% homology over 892 nt), encoding conserved hypothetical protein | |
| A02 | 1, 2 | 582 | S. boydii strain Sb227 (45) ORFs SBO2122 to SBO2125 (94% homology over 1,013 nt), encoding conserved hypothetical protein and prophage capsid proteins | |||||
| A03 | 2, 3 | 483 | S. boydii strain Sb227 (45) ORFs SBO2131 to SBO2132 (98% homology over 995 nt), encoding conserved hypothetical proteins | |||||
| A04 | 2, 3 | 438 | S. boydii strain Sb227 (45) ORFs SBO2126 to SBO2127 (97% homology over 684 nt), encoding a conserved hypothetical protein and a hypothetical bacteriophage protein | |||||
| B16 | 1, 2, 3 | 830 | 0/17 (0) | 17/17 (100) | 0/5 (0) | 5/5 (100) | S. flexneri bacteriophage V (16) ORFs int, xis, orf28, orf29, orf30, and orf31 (97% homologous over 2,453 nt), encoding an integrase, an excisionase, and hypothetical proteins | |
| B01 | 1, 2, 3 | 316 | S. flexneri bacteriophage V (16) ORFs orf39, orf40, and dam (96% homology over 823 nt), encoding a replication protein, a hypothetical protein, and a DNA adenine methylase | |||||
| B07 | 1,3 | 97 | S. flexneri bacteriophage V (16) ORF5 and ORF6 (98% homology over 803 nt), encoding a capsid protein and a hypothetical protein | |||||
| C | A14 | 2, 3 | 474 | 0/17 (0) | 17/17 (100) | 4/5 (80) | 5/5 (100) | 599 nt of DNA sequence without significant homology to known DNA sequences adjacent to E. coli Sakai (14) ORFs ECs1599 and ECs1600 (90 to 91% homologous over 367 nt), encoding hypothetical proteins |
| A15 | 1, 2, 3 | 1,068 | 0/17 (0) | 17/17 (100) | 4/5 (80) | 4/5 (80) | 247 nt of DNA sequence without significant homology to known DNA sequences adjacent to E. coli Sakai (14) ORFs ECs1557 and ECs1558 (96% homology over 1,968 nt), encoding a putative antirepressor protein and a putative tail assembly protein | |
| D | A06c | 2 | 1,212 | 1/17 (5.9) | 17/17 (100) | 5/5 (100) | 5/5 (100) | E. coli Sakai (14) intergenic region between ORFs ECs1159 and ECs1160 (98% homology over 365 nt), encoding a hypothetical protein and a putative integrase, adjacent to ECs1252 (95% homologous over 929 nt with a single 82-nt gap), encoding a putative transport protein |
| A07 | 1, 3 | 226 | 0/17 (0) | 15/17 (88.2) | 5/5 (100) | 4/5 (80) | E. coli Sakai (14) ORF ECs1928 (97% homology over 148 nt), encoding a hypothetical protein, adjacent to ECs1997 (99% homology over 269 nt), encoding a putative filament protein | |
| B15 | 3 | 558 | 0/17 (0) | 17/17 (100) | 1/5 (20) | 5/5 (100) | E. coli Sakai (14) ORF ECs1688 (100% homology over 378 nt), encoding a murein transglycosylase, adjacent to ORFs ECs1705 and ECs1706 (99% homologous over 672 nt), encoding a putative dihydroxyacetone kinase and a hypothetical protein | |
| E | A01 | 1 | 432 | 0/17 (0) | 12/17 (70.6) | 0/5 (0) | 2/5 (40) | E. coli strain EC93 contact-dependent inhibition (cdiB) gene (3) (98% homologous over 523 nt) |
| B05 | 1 | 512 | 0/17 (0) | 1/17 (5.9) | 0/5 (0) | 0/5 (0) | ColE6-CT14 colicin plasmid (1) colicin E6 gene (99% homologous over 1,605 nt) | |
| B12 | 2, 3 | 258 | 0/17 (0) | 17/17 (100) | 0/5 (0) | 5/5 (100) | E. coli UTI89 (7) ORFs (91% homologous over 710 nt), encoding a regulatory protein and a hypothetical protein | |
| B18 | 1 | 290 | 5/17 (29.4) | 8/17 (47.1) | 0/5 (0) | 2/5 (40) | pColD-157 plasmid (15) colicin D-157 activity protein (99% homology over 921 nt) | |
| F | A12 | 1, 2, 3 | 446 | 0/17 (0) | 16/17 (94.1) | 4/5 (80) | 4/5 (80) | 643 nt of DNA sequence without significant homology to known DNA sequences |
| B13 | 1, 2 | 507 | 0/17 (0) | 14/17 (82.4) | 4/5 (80) | 5/5 (100) | DNA sequence without significant homology to known DNA sequences | |
Sequence found in the following SSH experiments: 1, E. coli FRIK 920-E. coli Sakai; 2, E. coli FRIK 1999-E. coli Sakai; 3, E. coli FRIK 2001-E. coli 93-001.
FIG. 1.
Approximate location of lineage II-specific regions with respect to the E. coli O157:H7 strain Sakai genome. Contigs containing lineage II-specific regions together with regions with identity to the Sakai genome were used to place the lineage II-specific regions on the circular map, using the CGView program (43). Red regions represent lineage II-specific loci which are centered at their approximate insertion points with respect to the Sakai genome. Twenty-five of the 27 primers were placed on the Sakai genome in an analogous fashion. Two primer sets (B01 and B18) could not be placed on the map because they targeted lineage II-specific contigs that were missing sequences with identity to the Sakai genome to serve as reference points.
Distribution of lineage II SSH sequences among E. coli O157:H7 strains.
Class B loci possessed at least 80% sequence identity over at least 50 bp to bacteriophage sequences not found in E. coli O157:H7 lineage I strain Sakai (Table 4). These sequences were present in 92 (11.3%) of the 811 contigs, ranged in size from 66 to 3,180 bp, and represented 73,574 of the 565,341 nucleotides (13.0%) from all of the contigs. The 16 primer sets from class B loci amplified DNA from the majority of lineage II strains but not from any of the lineage I strains. Some of these primer sets generated identical strain distribution patterns and were derived from contigs sharing homology with different segments of a single prophage that may be unique to lineage II strains. The class B target sequences share identity with several genes of other lambdoid prophage, including prophage from E. coli UTI89 (7), stx1-converting bacteriophage BP-4795 from E. coli O84:H4 4795/97 (8), cryptic prophage e14 from E. coli K-12 (5), E. coli bacteriophage P2 (accession number AF063097), P4-like bacteriophage from E. coli H709c (23), two different Shigella boydii Sb227 bacteriophages (45), and E. coli bacteriophage HK022 (19). It is well known that bacteriophages are major contributors of genetic diversity in E. coli O157 and other enteric bacteria (21, 22, 34, 39, 40). Further characterization of these lineage II conserved bacteriophages will be required to determine their effect on the phenotype of this group of E. coli O157:H7 strains.
Class C loci likely represent transitional sequences between lineage-common and strain- or lineage-specific DNA. They consist of stretches of DNA sequence with at least 80% sequence identity over at least 50 bp to E. coli Sakai genes adjacent to non-Sakai DNA sequences, and they were found in 8.9% (72/811) of the contigs, ranged in size from 277 to 2,232 bp, and represented 54,989 (9.7%) of all of the contig nucleotides. The two class C primer sets, A14 and A15, have one primer in each set within sequences conserved among lineage I and lineage II strains and the other member of the pair in adjacent non-Sakai DNA. A14 targets a DNA segment within the non-LEE effector (NLE)-encoding Sakai prophage (Sp) 6, and A15 targets a DNA segment in Sp7. PCR products from these primer sets amplified the same-size segments in nearly all lineage II strains but did not produce a PCR product with DNA from any of the lineage I strains. These novel segments in lineage II strains are likely the result of insertion/deletion events resulting in the loss of DNA (in lineage I) or acquisition of DNA (in lineage II).
In contrast to class E loci, class D loci were missing chromosomal DNA segments found in lineage I strain Sakai. These sequences were found in 2.3% (19/811) of the contigs, ranged in size from 438 to 1,427 bp, and represented 17,213 (3.0%) of all contig nucleotides. Two of the class D primer sets, A06c and A07, amplify sequences flanking the insertion sites of the Sakai Stx2-encoding Sp5 and the NLE-encoding Sp10, respectively. In contrast to all E. coli O157:H7 lineage I strains, most lineage II strains produced amplicons with these primers, showing that Sp5 and Sp10 are not present in the respective lineage I chromosomal prophage insertion sites. This finding is supported by other studies that have shown that the chromosomal location of the stx2-transducing prophage Sp5 varies among E. coli O157:H7 strains (35, 40). The third primer set was based on a contig with identity to S-loop 83. In lineage II strains, the E. coli Sakai open reading frame (ORF) ECs1688 is juxtaposed next to ECs1705 due to an apparent insertion/deletion in the intervening ORFs. This region encodes a putative iron transport gene cluster with identity to the prrA-modD-yc73-fepC region of the pyonephritis and cystitis pathogenicity island of E. coli CFT073 (13). All three of the class D loci were also identified in a previous study of lineage I-specific genomic segments (42). In the latter study, a segment of DNA in the tellurite resistance and adherence-conferring genomic island carrying the perC-like regulatory protein gene, pchD, was also found to be missing in lineage II strains. The fact that the genes contained within these regions include stx2 in Sp5, a putative iron transport gene cluster in S-loop 83, and a gene with similarity to the perC-like LEE1 regulator in the tellurite resistance and adherence-conferring genomic island provides strong evidence of key differences in virulence-associated traits between these two lineages.
Class E loci possessed at least 80% sequence identity over at least 50 bp to nonbacteriophage DNA sequences found in bacteria other than E. coli O157:H7 strain Sakai, ranged in size from 51 to 3,110 bp, and represented 9.9% (80 of 811) of the contigs or 54,989 (9.7%) of all contig nucleotides. Primer set A01 amplified a target with identity to the E. coli strain EC93 cdiB gene (3). The cdiB gene is part of a cell contact-dependent growth inhibition system in E. coli EC93 and uropathogenic E. coli strains, which has been shown to inhibit in vitro growth of E. coli K-12 (3). While the cdiB gene was highly prevalent among lineage II strains of LSPA-6 222222 (Table 4), lineage I strains were missing this operon. The lineage II conserved region amplified by primer set B12 has identity to a putative regulatory gene and two hypothetical genes found in the uropathogenic E. coli strain UTI89 (7) and the avian pathogenic E. coli serotype O1 strain (18). This sequence was absent from lineage I strains but present in all lineage II strains except for genotype LSPA-6 211111 strains.
The other two class E PCR primer sets, B05 and B18, represent colicin genes identified at high frequency within the E. coli FRIK 920-E. coli Sakai SSH library but not in either of the two remaining SSH experiments. These targets were not lineage specific. The colicin E gene was amplified by the B05 primers and was found only in the strain FRIK 920, while the colicin D gene targeted by primer set B18 was found among both lineages but more commonly among genotype LSPA-6 222222 strains (8/17 or 47.1%) than lineage I genotype LSPA-6 111111 strains (5/17 or 29.4%) (Table 4).
Class F contigs had very little or no identity to sequences in the GenBank NR database. These were >50 bp in length and contained <80% sequence identity over any 50-bp stretch within the contig or contained no region of >50 bp in length with >80% sequence identity (criteria used for inclusion in classes B, C, and E). Class F represented 23.2% (188/811) of the contigs, ranged in size from 51 bp to 3,485 bp, and included 150,034 (26.5%) of all contig nucleotides. PCR analysis with the A12 and B13 primer sets confirmed that these segments are present in a large number of lineage II strains and are therefore not due to artifacts of the SSH methodology. Although the function of these genes is unknown, the fact that they are conserved in lineage II strains, coupled with their prevalence within the SSH libraries (23.3%), would argue that they play an important functional role in the survival of the organism.
PCR amplicons generated from tester strains were sequenced and compared with the original contigs to confirm correct assembly of the SSH sequences. The sequences obtained were 89 to 100% identical to the original contigs, suggesting that the original assemblies were correct and representative of the actual genomic sequences. The high proportion of primer sets (25/27) that amplified these sequences in the E. coli O157:H7 lineage II strains tested illustrates that SSH is a robust method for isolating strain-specific sequences.
Genotyping of E. coli O157:H7 strains based on lineage II conserved loci.
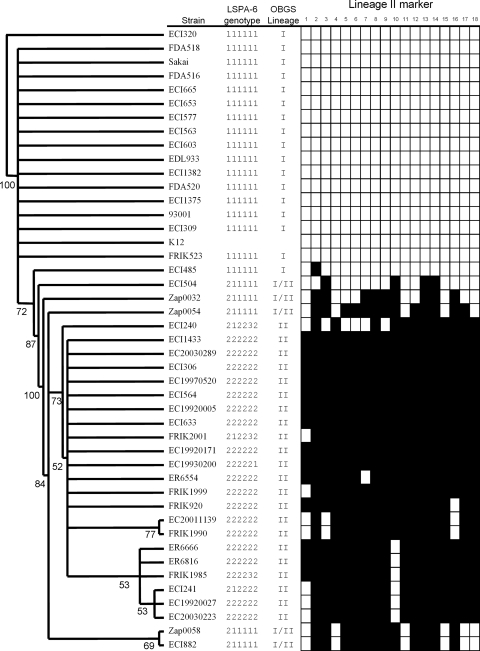
Sequences identified in this study suggest that genomic content and organization vary within lineage I and lineage II populations. Even within a lineage, diversity can be observed, as few loci are conserved in every isolate tested within a lineage. Nonetheless, when heritability of the SSH markers is examined as a whole, the extent of genomic diversity distinguishing the two lineages is impressive. As illustrated in Fig. 2, neighbor-joining analysis of the PCR data (converted to binary strings) shows that the major LSPA-6 genotypes behave as a group with respect to genomic diversity. With the exception of a single LSPA-6 111111 strain, human phage type 14 strain ECI-485, E. coli O157:H7 strains from lineage I (LSPA-6 111111) and from lineage II (other LSPA-6 genotypes) clustered separately (ECI-485 clustered with the lineage II strains, not with the other lineage I strains). While the LSPA-6 markers are within backbone regions of the genome, many of the lineage-specific loci found in this study are located within mobile genetic elements such as phage; this congruence implies that selection is uniquely shaping the genomes of these two lineages.
FIG. 2.
Phylogenetic relatedness of E. coli O157:H7 strains based on the presence of 18 lineage II conserved molecular markers. A neighbor-joining dendrogram was generated based on a matrix of pairwise distances calculated from the binary data by using the proportion of shared lineage II markers as a distance metric. Marker distribution is shown on the right, with positive PCR results indicated by black boxes and negative results indicated by white boxes. The level of support as a proportion of 500 bootstrap replicates is indicated on each branch on the dendrogram.
Within lineage II, the LSPA-6 genotype 211111 strains showed significant strain-strain diversity (Fig. 2). We have previously shown that LSPA-6 genotype 211111 strains group separately from other E. coli O157:H7 strains and share genomic characteristics of lineage I or lineage II populations at independent loci (42), leading us to categorize them as lineage I/II strains. This finding, coupled with the fact that the lineage II SSH markers are not uniformly distributed within LSPA-6 genotype 211111, suggests that there is significant diversity among lineage I/II strains and that they are distinct from lineage I and lineage II strains (47).
Statistical analysis of the distribution of lineage I and lineage II strains has previously shown biases in the frequency with which some subtypes are found in bovine or human clinical samples (46). In this report and our previous studies, we have identified genomic features that differentiate populations of these lineages. As observed in other studies, bacteriophages are important contributors to genomic diversity in this organism. Whether these phages carry unique combinations of virulence genes and regulatory genes or otherwise influence physiological traits of the two lineages remains to be determined.
A large number of studies now have confirmed that genomic alterations associated with bacteriophage differentiate subpopulations of E. coli O157:H7 (34, 39, 40). Unfortunately, many of these studies have not been done within a phylogenetic context, so it is difficult to determine how those results relate to lineage I and II groupings. Nonetheless, collectively, the studies show that the phage-mediated events are common and that they are also the common events discriminating the two lineages. Since both lineages coexist temporally and spatially, we would expect that they would be exposed to similar phage pools in nature. However, it would appear that these lineages have different host distributions and that they may have different selective pressures or differ in the strategies used to deal with these selective forces. It is also possible that they are uniquely susceptible to lysogeny by certain bacteriophages. Recently we have shown phage type to be lineage-related in E. coli O157:H7 strains (48). The mode of acquisition of these phage-related elements and their effect on phenotype are at present unknown but will become clearer as these lineage-specific loci begin to be studied.
A number of the differentially distributed elements have the potential to affect the ecology and pathogenicity of E. coli O157:H7. Within lineage I strains, these include genomic regions containing the stx2-transducing Sakai prophage Sp5 and a putative iron transport gene cluster with homology to the prrA-modD-yc73-fepC gene cluster of E. coli CFT073 (13). Within lineage II strains, these include genes for a putative contact-dependent inhibition system (3) and a putative regulatory gene found within the uropathogenic E. coli strain UTI89. In addition, this study identified the existence of other bacteriophage-related elements that appear to be conserved among lineage II E. coli O157:H7 strains. These may possess factors which influence the virulence and host distribution of E. coli O157:H7 lineages.
Acknowledgments
We thank Shelley Frost for technical assistance and the Canadian Food Inspection Agency (CFIA) for allowing part of this research to be conducted at the Animal Diseases Research Institute, Lethbridge, Alberta, Canada.
This research was supported by grants from Health Canada's Office of Biotechnology and Science.
Footnotes
Published ahead of print on 27 March 2009.
REFERENCES
- 1.Akutsu, A., H. Masaki, and T. Ohta. 1989. Molecular structure and immunity specificity of colicin E6, an evolutionary intermediate between E-group colicins and cloacin DF13. J. Bacteriol. 171:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, S. K., R. Pamma, A. D. Hernday, J. E. Bickham, B. A. Braaten, and D. A. Low. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245-1248. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, M., N. L. Padola, A. Kruger, M. E. Sanz, J. E. Blanco, E. A. Gonzalez, G. Dahbi, A. Mora, M. I. Bernardez, A. I. Etcheverria, G. H. Arroyo, P. M. Lucchesi, A. E. Parma, and J. Blanco. 2004. Virulence genes and intimin types of Shiga-toxin-producing Escherichia coli isolated from cattle and beef products in Argentina. Int. Microbiol. 7:269-276. [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. L., C. S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooton, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 103:5977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creuzburg, K., J. Recktenwald, V. Kuhle, S. Herold, M. Hensel, and H. Schmidt. 2005. The Shiga toxin 1-converting bacteriophage BP-4795 encodes an NleA-like type III effector protein. J. Bacteriol. 187:8494-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowd, S. E., and H. Ishizaki. 2006. Microarray based comparison of two Escherichia coli O157:H7 lineages. BMC Microbiol. 6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.Griffin, P. M. 1998. Epidemiology of Shiga toxin-producing Escherichia coli infections in humans in the United States, p. 15-22. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, DC.
- 13.Guyer, D. M., J. S. Kao, and H. L. Mobley. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect. Immun. 66:4411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 15.Hofinger, C., H. Karch, and H. Schmidt. 1998. Structure and function of plasmid pColD157 of enterohemorrhagic Escherichia coli O157 and its distribution among strains from patients with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 36:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huan, P. T., B. L. Whittle, D. A. Bastin, A. A. Lindberg, and N. K. Verma. 1997. Shigella flexneri type-specific antigen V: cloning, sequencing and characterization of the glucosyl transferase gene of temperate bacteriophage SfV. Gene 195:207-216. [DOI] [PubMed] [Google Scholar]
- 17.Jarvis, K. G., and J. B. Kaper. 1996. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect. Immun. 64:4826-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, T. J., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. J. Johnson, C. Doetkott, J. A. Skyberg, A. M. Lynne, J. R. Johnson, and L. K. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189:3228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophage HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophage. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 20.Karmali, M. A., B. T. Steele, M. Petric, and C. Lim. 1983. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet i:619-620. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J., J. Nietfeldt, and A. K. Benson. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. USA 96:13288-13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J., J. Nietfeldt, J. Ju, J. Wise, N. Fegan, P. Desmarchelier, and A. K. Benson. 2001. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, β-glucuronidase-negative enterohemorrhagic Escherichia coli O157. J. Bacteriol. 183:6885-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kita, K., J. Tsuda, T. Kato, K. Okamoto, H. Yanase, and M. Tanaka. 1999. Evidence of horizontal transfer of the EcoO109I restriction-modification gene to Escherichia coli chromosomal DNA. J. Bacteriol. 181:6822-6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laegreid, W. W., R. O. Elder, and J. E. Keen. 1999. Prevalence of Escherichia coli O157:H7 in range beef calves at weaning. Epidemiol. Infect. 123:291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laing, C., C. Pegg, D. Yawney, K. Ziebell, M. Steele, R. Johnson, J. E. Thomas, E. N. Taboada, Y. Zhang, and V. P. J. Gannon. 2008. Rapid determination of Escherichia coli O157:H7 lineage types and molecular subtypes by using comparative genomic fingerprinting. Appl. Environ. Microbiol. 74:6606-6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lejeune, J. T., S. T. Abedon, K. Takemura, N. P. Christie, and S. Sreevatsan. 2004. Human Escherichia coli O157:H7 genetic marker in isolates of bovine origin. Emerg. Infect. Dis. 10:1482-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Locking, M. E., S. J. O'Brien, W. J. Reilly, E. M. Wright, D. M. Campbell, J. E. Coia, L. M. Browning, and C. N. Ramsay. 2001. Risk factors for sporadic cases of Escherichia coli O157 infection: the importance of contact with animal excreta. Epidemiol. Infect. 127:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone, A. S., A. E. Yousef, and J. T. LeJeune. 2007. Association of prophage antiterminator Q. alleles and susceptibility to food-processing treatments applied to Escherichia coli O157 in laboratory media. J. Food Prot. 70:2617-2619. [DOI] [PubMed] [Google Scholar]
- 29.Manning, S. D., A. S. Motiwala, A. C. Springman, W. Qi, D. W. Lacher, L. M. Ouellette, J. M. Mladonicky, P. Somsel, J. T. Rudrik, S. E. Dietrich, W. Zhang, B. Swaminathan, D. Alland, and T. S. Whittam. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. USA 105:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNally, A., A. J. Roe, S. Simpson, F. M. Thomson-Carter, D. E. Hoey, C. Currie, T. Chakraborty, D. G. Smith, and D. L. Gally. 2001. Differences in levels of secreted locus of enterocyte effacement proteins between human disease-associated and bovine Escherichia coli O157. Infect. Immun. 69:5107-5114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Brien, A. D., V. L. Tesh, A. Donohue-Rolfe, M. P. Jackson, S. Olsnes, K. Sandvig, A. A. Lindberg, and G. T. Keusch. 1992. Shiga toxin: biochemistry, genetics, mode of action, and role in pathogenesis. Curr. Top. Microbiol. Immunol. 180:65-94. [DOI] [PubMed] [Google Scholar]
- 33.O'Brien, S. J., G. K. Adak, and C. Gilham. 2001. Contact with farming environment as a major risk factor for Shiga toxin (Vero cytotoxin)-producing Escherichia coli O157 infection in humans. Emerg. Infect. Dis. 7:1049-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophage the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 35.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 37.Sargeant, J. M., M. W. Sanderson, R. A. Smith, and D. D. Griffin. 2003. Escherichia coli O157 in feedlot cattle feces and water in four major feeder-cattle states in the USA. Prev. Vet. Med. 61:127-135. [DOI] [PubMed] [Google Scholar]
- 38.Saridakis, C. E., R. P. Johnson, A. Benson, K. Ziebell, and C. L. Gyles. 2004. Influence of animal origin and lineage on survival of Escherichia coli O157:H7 strains in strong and weak acid challenges. J. Food Prot. 67:1591-1596. [DOI] [PubMed] [Google Scholar]
- 39.Sato, T., T. Shimizu, M. Watarai, M. Kobayashi, S. Kano, T. Hamabata, Y. Takeda, and S. Yamasaki. 2003. Distinctiveness of the genomic sequence of Shiga toxin 2-converting phage isolated from Escherichia coli O157:H7 Okayama strain as compared to other Shiga toxin 2-converting phages. Gene 309:35-48. [DOI] [PubMed] [Google Scholar]
- 40.Shaikh, N., and P. I. Tarr. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophage: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, D., M. Blackford, S. Younts, R. Moxley, J. Gray, L. Hungerford, T. Milton, and T. Klopfenstein. 2001. Ecological relationships between the prevalence of cattle shedding Escherichia coli O157:H7 and characteristics of the cattle or conditions of the feedlot pen. J. Food Prot. 64:1899-1903. [DOI] [PubMed] [Google Scholar]
- 42.Steele, M., K. Ziebell, Y. Zhang, A. Benson, P. Konczy, R. Johnson, and V. Gannon. 2007. Identification of Escherichia coli O157:H7 genomic regions conserved in strains with a genotype associated with human infection. Appl. Environ. Microbiol. 73:22-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stothard, P., and D. S. Wishart. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537-539. [DOI] [PubMed] [Google Scholar]
- 44.Tarr, P. I., S. S. Bilge, J. C. J. Vary, S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, F., J. Yang, X. Zhang, L. Chen, Y. Jiang, Y. Yan, X. Tang, J. Wang, Z. Xiong, J. Dong, Y. Xue, Y. Zhu, X. Xu, L. Sun, S. Chen, H. Nie, J. Peng, J. Xu, Y. Wang, Z. Yuan, Y. Wen, Z. Yao, Y. Shen, B. Qiang, Y. Hou, J. Yu, and Q. Jin. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 33:6445-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, Z., J. Kovar, J. Kim, J. Nietfeldt, D. R. Smith, R. A. Moxley, M. E. Olson, P. D. Fey, and A. K. Benson. 2004. Identification of common subpopulations of non-sorbitol-fermenting, β-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70:6846-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., C. Laing, M. Steele, K. Ziebell, R. Johnson, A. K. Benson, E. Taboada, and V. P. Gannon. 2007. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziebell, K., M. Steele, Y. Zhang, A. Benson, E. N. Taboada, C. Laing, S. McEwen, B. Ciebin, R. Johnson, and V. Gannon. 2008. Genotypic characterization and prevalence of virulence factors among Canadian Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 74:4314-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]