Abstract
Genetic manipulation of Staphylococcus aureus is limited by the availability of only a single strain, RN4220, that is capable of easily accepting foreign DNA. Inactivation of the hsdR gene of the SauI type I restriction-modification system was shown previously to be responsible for the high transformation efficiency of RN4220 (D. E. Waldron and J. A. Lindsay, J Bacteriol. 188:5578-5585, 2006). However, deletion of this gene in three different S. aureus strains was not sufficient to make them readily transformable, which would be remarkably useful for genetic studies of this pathogenic organism. These results indicate that another unknown factor(s) is required for the transformable phenotype in S. aureus.
Staphylococcus aureus is a major pathogen that causes both nosocomial and community-acquired infections, which range from superficial skin infections to severe systemic diseases. It is an extremely versatile pathogen which has developed resistance to virtually all known classes of antibiotics and which expresses various virulence factors that allow it to cause infection in different environments.
Molecular genetic studies of S. aureus have resulted in a better understanding of both the virulence and antibiotic resistance mechanisms of this organism, which is crucial for discovery of new approaches to treat staphylococcal infections. One of the limitations in genetically manipulating S. aureus is the fact that there is only a single strain available which can easily accept plasmid DNA isolated from Escherichia coli. This strain, RN4220, is a chemical mutant obtained in the early 1980s by highly mutagenizing NCTC8325-4 (= RN450) with nitrosoguanidine and selecting for a mutant that was able to accept and maintain S. aureus plasmids (10; B. Kreiswirth, personal communication).
One of the most important bacterial defenses against uptake of foreign DNA is restriction-modification (R-M) systems. These systems, comprising restriction endonucleases and methyltransferases, recognize and modify specific DNA sequences, protecting “self” DNA from restriction while eliminating potentially harmful foreign DNA which lacks appropriate modification (12). There are three distinct well-characterized types of classical R-M systems, including type II restriction enzymes, which cut DNA within specific recognition sequences and are therefore widely used as molecular biology tools (3). S. aureus strains may contain different R-M systems, including the Sau3AI and Sau96I type II systems (present in isolates of particular lytic groups) (19, 20) or the Sau42I BcgI-like R-M system expressed by S. aureus φ42 lysogens (5). However, the only chromosomal R-M system widely distributed in the sequenced S. aureus isolates is the SauI type I system (21). Type I R-M systems require the products of three genes, hsdR (restriction), hsdM (modification), and hsdS (sequence specificity), and cut DNA at sites remote from the recognition sequence (12). The staphylococcal SauI system includes a single hsdR gene and two copies of the hsdM and hsdS genes, and there is substantial variation between the hsdS genes from different isolates (21). It was recently shown that transformable RN4220 carries a stop mutation in the sauI hsdR gene and that complementation with a functional copy of this gene restores a nontransformable phenotype (21). An hsdR mutant does not cleave foreign unmodified DNA, but it does modify incoming DNA, which therefore is not cleaved when it is transferred to other S. aureus strains that contain an identical R-M system. For this reason, RN4220 has become an essential intermediate for laboratory manipulation of S. aureus, despite its limited clinical relevance. DNA first introduced into RN4220 by electroporation can then be transferred into other laboratory strains (for example, by phage transduction).
The relevance of the SauI type I R-M system for horizontal transfer of foreign DNA in different S. aureus isolates in nature has been confirmed with bovine isolates that are hypersusceptible to gene transfer from enterococci and have stop mutations in each of the two SauI hsdS gene copies (18). These “hyperrecipient” strains may be ideal backgrounds for the acquisition of new antibiotic resistance markers, such as the vanA gene complex present in vancomycin-resistant S. aureus strains (18). However, the existence of a second pathway in S. aureus that blocks horizontal transfer of foreign DNA or of an unknown but necessary factor essential for SauI activity has also been proposed based on the fact that some strains that are hypersusceptible to gene transfer (such as B111 used by Noble et al. [13]) do not have a mutation in any of the five hsd genes (18).
The availability of laboratory and clinical strains other than RN4220 that are capable of accepting foreign DNA would be remarkably useful for genetic studies of S. aureus, including studies of virulence and antibiotic resistance mechanisms present only in relevant clinical isolates. Therefore, we have tried to reproduce the hsdR mutation in RN4220 in different backgrounds of widely used laboratory strains in order to generate useful, easily transformable strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The following bacterial strains were used in this study: methicillin-susceptible S. aureus strains NCTC8325-4 (14) and SH1000 (8), methicillin-resistant S. aureus strain COL (6, 7), and restriction-deficient S. aureus strain 879R4RF (17). Unless otherwise stated, all S. aureus strains were grown in tryptic soy broth (TSB) (Difco) at 37°C with aeration. When required, the medium was supplemented with erythromycin (10 μg/ml), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (100 μg/ml), or chloramphenicol (10 μg/ml).
Construction of hsdR null mutants.
hsdR null mutants were constructed using the backgrounds of strains NCTC8325-4, SH1000, and COL and the thermosensitive plasmid pMAD (1), which contains an erythromycin resistance marker as well as the lacZ reporter gene under control of a constitutive promoter. Two PCR fragments, corresponding to the upstream and downstream regions of the hsdR gene, were amplified from chromosomal DNA of strain NCTC8325-4 using Phusion high-fidelity DNA polymerase (Finnzymes) and primer pairs HsdRP1/HsdRP2 (5′-TGAGGATCCTTTGTGACGAATCAAGGTAGTCATAGTG-3′ and 5′-GTATTTTTCAGTATTCACTTTGGTATGCCATTCATATCCC-3′) and HsdRP3/HsdRP4 (5′-AAGTGAATACTGAAAAATACGGTGTGTAATGATTCAGCCC-3′ and 5′-GCTGAATTCTATAACAAGAACTTAATTTCAGCCGGCG-3′), respectively (Fig. 1A). In the second step, the two fragments were joined by overlapping PCR using primers HsdRP1 and HsdRP4. The 2-kb PCR final product was restricted with BamHI and EcoRI (at the cleavage sequences underlined above; NEB), cloned into pMAD, resulting in pΔHsdR-1, and sequenced (STABVida). The latter plasmid was electroporated, as previously described (11), into the transformable strain RN4220 at 30°C with erythromycin selection and was subsequently transduced into NCTC8325-4, SH1000, and COL using phage 80α (15). Strains containing pΔHsdR-1 were incubated at a nonpermissive temperature (43°C) in the presence of erythromycin to select for recombinants in which the plasmid had integrated into the chromosome, and then they were incubated at a permissive temperature (30°C) in the absence of antibiotic selection to select for white colonies in which the pΔHsdR-1 plasmid (and consequently lacZ and erm genes) had been excised. To identify mutants in which the hsdR gene had been deleted, chromosomal DNA was extracted from erythromycin-sensitive, white colonies and tested by performing PCR using GoTaq polymerase (Promega) and primers HsdRP5 (5′-CAAGTCCCTCCATTAATCGTAG-3′) and HsdRP6 (5′-TGATGGTTGCCAAACACATG-3′), which hybridize 211 bp upstream and 84 bp downstream of the hsdR gene, respectively (Fig. 1A).
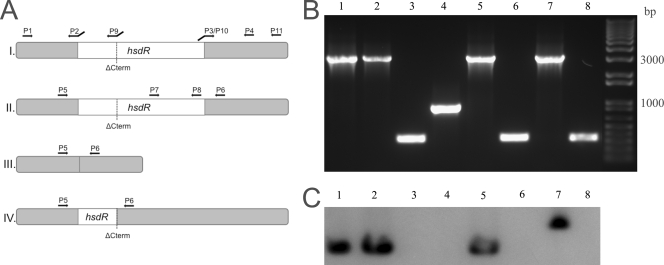
FIG. 1.
Genetic characterization of hsdR mutants. The strains used were RN4220 (lane 1), NCTC8325-4 (lane 2), NCTC8325-4ΔHsdR (lane 3), NCTC8325-4ΔC-term HsdR (lane 4), SH1000 (lane 5), SH1000ΔHsdR (lane 6), COL (lane 7), and COLΔHsdR (lane 8). (A) Localization of primers used to construct hsdR mutants (panel I) (see Materials and Methods for details); of primers used to characterize the hsdR mutants by PCR for wild-type strains RN4220, NCTC8325-4, SH1000, and COL (panel II), for hsdR null mutants NCTC8325-4ΔHsdR, SH1000ΔHsdR, and COLΔHsdR (panel III), and for strain NCTC8325-4ΔC-term HsdR, which contains a truncated version of the hsdR gene (panel IV); and of primers HsdRP7 and HsdRP8 used to amplify the hsdR probe used for Southern blotting (panel II). (B) PCR fragments obtained by amplifying the DNA of the eight strains indicated above using primers HsdRP5 and HsdRP6, which flank the hsdR gene. (C) Results of Southern blot hybridization of SmaI-restricted genomic DNA of the eight strains indicated above with an internal probe for the hsdR gene amplified using primers HsdRP7 and HsdRP8, showing that this gene is absent from the hsdR mutant strains. Note that the probe hybridized with the DNA fragment which was deleted in strain NCTC8325-4ΔC-term HsdR.
The transformable RN4220 strain expresses a truncated HsdR product containing only the first 192 amino acids of the wild-type protein (21). Therefore, we constructed an NCTC8325-4 strain in which the sequence coding for the C terminus of HsdR was deleted. For this purpose, a 2,598-bp DNA fragment encompassing the upstream region, the first 576 bp (192 codons) followed by a stop codon, and the downstream region of the hsdR gene was amplified in the following way. The initial PCRs were performed using primer pairs HsdRP1/HsdRP9 (5′-TGAGGATCCTTTGTGACGAATCAAGGTAGTCATAGTG-3′ and 5′-GTATTTTTCAACTCTTCAATAGTTCGCTATCATTATTAGA-3′) and HsdRP10/HsdRP11 (5′-ATTGAAGAGTTGAAAAATACGGTGTGTAATGATTCAGCCC-3′ and 5′-GCTGAATTCGATGCCTTTGTTTGGAATATTACGC-3′) (Fig. 1A). The two resulting fragments were joined by overlap PCR using primers HsdRP1 and HsdRP11 and cloned into pMAD, resulting in plasmid pΔHsdR-2, which was sequenced. This plasmid was electroporated into RN4220 at 30°C and transduced into NCTC8325-4, where exchange of the wild-type copy of hsdR with the truncated copy was promoted, using the procedure described above for generation of the null mutants. The resulting strain, designated NCTC8325-4ΔC-termHsdR, was verified by performing PCR using primers HsdRP5 and HsdRP6 (Fig. 1A).
All genomic constructs were also verified by Southern blot hybridization using a specific probe for the 3′ region of the hsdR gene. For this purpose, genomic DNA of S. aureus wild-type and hsdR mutant strains was digested with SmaI and separated by pulsed-field gel electrophoresis (PFGE) as previously described (4). The DNA was then transferred to a positively charged nylon membrane (Amersham), hybridized with the hsdR internal probe, a 817-bp DNA fragment amplified by PCR using primers HsdRP7 (5′-GATCGTGATGGTGAAGTGCC-3′) and HsdRP8 (5′-GGTTACGACGTTGTTCCGC-3′) (Fig. 1A), and labeled using the Gene Images AlkPhos direct labeling system (Amersham) according to the manufacturer's instructions.
Electroporation.
Electroporation was performed as previously described (11). Briefly, 45-μl portions of competent cells of RN4220, of wild-type strains NCTC8325-4, SH1000, and COL, and of the corresponding hsdR mutants were mixed with 0.2 μg of the replicative plasmid pGC2 (22) extracted from E. coli DH5α in a 2.0-mm electroporation cuvette (Bio-Rad) and subjected to an electroporation pulse in a Gene Pulser Xcell apparatus (Bio-Rad) set to 2.5 kV, 25 μF, and 100 Ω. Following electroporation, cells were immediately resuspended in 955 μl of SMMP medium (5.5 parts SMM buffer [1 M sucrose, 0.04 M maleic acid, 0.04 M MgCl2; pH 6.5], 4 parts 7% antibiotic medium number 3 [Difco], 0.5 parts 10% bovine serum albumin) and incubated for 1 h at 37°C before they were plated on tryptic soy agar (TSA) supplemented with chloramphenicol. The number of transformants was determined after 16 h of incubation at 37°C. Three independent batches of competent cells were prepared for each strain, and each batch was used in two different electroporation experiments.
The competent cells were also electroporated (in duplicate) with 50 ng of pGC2 extracted from RN4220 using a Wizard Plus SV minipreps kit (Promega) after 15 min of incubation with 100 μg/ml lysostaphin at 37°C to lyse the cells.
To determine the transformation efficiency of the wild-type and mutant strains after heat treatment, the competent cells were incubated for 2 min at 56°C immediately before they were subjected to the electroporation pulse. Two independent experiments were performed, in which heat-treated cells were transformed with 0.2 μg of plasmid pGC2 DNA extracted from E. coli.
Transduction.
Phages 80α and φ75 were used to transduce pGC2 from strain 879R4RF into NCTC8325-4 and SH1000 wild-type and hsdR mutant strains. The transduction experiments were performed essentially as previously described (15). To prepare the phage lysates, recipient 879R4RF cells containing plasmid pGC2 were grown on TSA (Difco) plates for 16 h at 37°C and resuspended in 1 ml of TSB supplemented with 5 mM CaCl2. Dilutions (10−1 to 10−6) of the 80α and φ75 stock lysates were prepared in phage buffer (1 mM MgSO4, 4 mM CaCl2, 50 mM Tris-HCl, 0.1 M NaCl, 0.1% gelatin; pH 7.8), and 10 μl of each dilution was mixed with 10 μl of the recipient cells and 3 ml of top phage agar (3 g/liter Casamino Acids, 3 g/liter yeast extract, 5.9 g/liter NaCl, 5 mM CaCl2, 5 g/liter Bacto agar; pH 7.8). The mixture was plated on bottom phage agar (having the same composition as top phage agar except for 15 g/liter Bacto agar) and incubated for 16 h at 30°C. Phage buffer (3 ml) was added to the plates showing confluent lysis, which were subsequently incubated at 4°C for 1 h. The phage buffer and top phage agar were collected and incubated for 1 h at 4°C. After a centrifugation step the phage lysate was recovered and filtered through a sterile 0.45-μm filter.
For transduction, recipient cells were plated on TSA, grown overnight at 37°C, and resuspended in 1 ml of TSB supplemented with 5 mM CaCl2. Cells (100 μl) were mixed with phage lysate (1 μl and 10 μl), and phage buffer was added to obtain a final volume of 300 μl. The mixture was incubated at 37°C for 20 min, added to 3 ml of 0.3GL top agar (3 g/liter Casamino Acids, 3 g/liter yeast extract, 5.9 g/liter NaCl, 3.3 ml/liter sodium lactate, 2 ml/liter 50% glycerol, 0.5 g/liter trisodium citrate, 7.5 g/liter Bacto agar; pH 7.8) preheated to 50°C, and plated in plates containing 0.3GL bottom agar (having the same composition as 0.3GL top agar except for 15 g/liter Bacto agar) with a layer of 20 ml of 0.3GL bottom agar without antibiotics over a layer of 10 ml of 0.3GL bottom agar with chloramphenicol (30 μg/ml). The plates were used within 1 h after preparation. The number of transductants carrying pGC2 was determined after overnight incubation at 37°C.
Bacteriophage susceptibility assays.
NCTC8325-4 and SH1000 wild-type and hsdR mutant strains were tested for susceptibility to bacteriophages 80α and φ75 propagated in the nonlysogenic S. aureus 879R4RF strain.
Recipient cells were grown on TSA plates for 16 h at 37°C and resuspended in 1 ml of TSB supplemented with 5 mM CaCl2. Serial dilutions of the 80α and φ75 stock lysates (10 μl of 10−1 to 10−6 dilutions) were mixed with 10 μl of the recipient cells, added to 3 ml of top phage agar, plated on bottom phage agar, and incubated overnight at 30°C. The efficiency of plaquing was calculated by dividing the number of phage PFU per milliliter of phage lysate for each strain tested by the number of phage PFU per milliliter of phage lysate for RN4220.
RESULTS AND DISCUSSION
Effect of deletion of the hsdR gene on the transformation efficiency of S. aureus strains.
Developing a method to generate various clinical and laboratory S. aureus strains capable of easily accepting foreign DNA would be an important step forward in our ability to genetically manipulate S. aureus. With this aim, we decided to reproduce the hsdR mutation present in transformable S. aureus strain RN4220 in three different backgrounds of widely used laboratory strains, methicillin-susceptible S. aureus strains NCTC8325-4 (14) and SH1000 (8) and methicillin-resistant S. aureus strain COL (6, 7). The approach used consisted of constructing a complete null mutant in the three backgrounds by removing the hsdR gene, leaving no resistance marker in the chromosome (see Materials and Methods), so that the resulting strains would be more versatile for future genetic studies. The absence of the hsdR gene in strains NCTC8325-4ΔHsdR, SH1000ΔHsdR, and COLΔHsdR was confirmed by PCR (Fig. 1B) using primers HsdRP5 and HsdRP6, which flank the hsdR gene (Fig. 1A). Furthermore, SmaI-digested genomic DNA of wild-type and hsdR mutant strains was separated by PFGE and hybridized with an hsdR gene internal probe. The PFGE restriction patterns of the wild-type strains and corresponding hsdR null mutants were identical, as expected (data not shown), and Southern blotting showed that the hsdR gene is not present in the genomes of the three null mutant strains (Fig. 1C). Overexposure of the Southern blot autoradiography film revealed two weak bands corresponding to other SmaI DNA fragments in all wild-type and knockout strains (data not shown). However, as no hsdR homologue is present in either the NCTC8325-4 or COL genome, these weak bands probably resulted from nonspecific hybridization.
In order to assess the ability of the hsdR null mutants to accept foreign DNA, electroporation was performed using plasmid pGC2 DNA extracted from E. coli DH5α. Contrary to our expectations, complete deletion of the hsdR gene was not sufficient to generate readily transformable NCTC8325-4, SH1000, and COL strains (Table 1). Similar results were obtained when cells were transformed with the temperature-sensitive pMAD plasmid at 30°C (data not shown).
TABLE 1.
Electroporation efficiency of S. aureus wild-type and hsdR mutant strains transformed with plasmid pGC2 DNA extracted from E. coli DH5α or from S. aureus RN4220
| Strain | pGC2 from E. coli DH5 α
|
pGC2 from S. aureus RN4220
|
pGC2 from E. coli DH5α (heat-treated competent cells)c
|
|||
|---|---|---|---|---|---|---|
| Transformation efficiency (CFU/μg DNA)a | SD (CFU/μg DNA) | Transformation efficiency (CFU/μg DNA)b | SD (CFU/μg DNA) | Transformation efficiency (CFU/μg DNA)b | SD (CFU/μg DNA) | |
| RN4220 | 3.7 × 105 | 2.3 × 105 | 6.1 × 105 | 3.8 × 105 | 2.0 × 103 | 4.7 × 102 |
| NCTC8325-4 | 1.7 | 2.6 | 3.0 × 104 | 0.2 × 104 | 0.0 | 0.0 |
| NCTC8325-4ΔHsdR | 3.4 | 6.2 | 3.9 × 104 | 1.9 × 104 | 8.1 × 101 | 3.6 × 101 |
| NCTC8325-4ΔC-term HsdR | 2.5 | 2.8 | 1.9 × 104 | 0.3 × 104 | 1.0 × 103 | 9.7 × 102 |
| SH1000 | 0 | 0 | 7.0 × 104 | 2.6 × 104 | 0 | 0 |
| SH1000ΔHsdR | 0 | 0 | 2.7 × 104 | 0.3 × 104 | 3.0 × 101 | 0 |
| COL | 0 | 0 | 3.0 × 101 | 4.2 × 101 | 0 | 0 |
| COLΔHsdR | 0 | 0 | 5.0 × 101 | 4.2 × 101 | 0 | 0 |
Average values for six electroporation experiments.
Average values for two electroporation experiments.
S. aureus competent cells were incubated at 56°C for 2 min prior to electroporation.
As the transformable RN4220 strain is not an hsdR null mutant but expresses a truncated HsdR product containing the first 192 amino acids (approximately 20%) of the wild-type protein (21), we hypothesized that this peptide could somehow have a role in the transformable phenotype of RN4220. Therefore, we replaced the wild-type copy of the hsdR gene in the chromosome of NCTC8325-4, a strain closely related to RN4220, with an open reading frame containing only the first 192 codons of hsdR. The 3′ hsdR gene deletion genotype was confirmed by PCR and Southern blotting (Fig. 1). Failure to electroporate NCTC8325-4ΔC-termHsdR with replicative plasmid pCG2 extracted from E. coli (Table 1) showed that the ability of NCTC8325-4 to accept foreign DNA was not improved by the presence of the truncated form of the HsdR protein.
Importantly, hsdR mutant strains had growth rates identical to those of the parental strains (data not shown) and were competent if they were transformed with appropriately modified S. aureus DNA. This was tested by electroporating hsdR mutants with the pGC2 plasmid mentioned above but extracted from S. aureus strain RN4220 instead of E. coli. Since RN4220 belongs to the same lineage (CC8) as NCTC8325-4, SH1000, and COL, it was expected that its DNA, modified by its R-M system(s), would be accepted by the other strains. In fact, in contrast to the results for pGC2 extracted from E. coli, all strains tested were able to accept pGC2 DNA from RN4220 (Table 1), suggesting that hsdR mutants are competent but do not accept foreign DNA due to the presence of an additional factor(s) that degrades unmodified DNA.
Effect of deletion of the hsdR gene on S. aureus susceptibility to infection by bacteriophages.
One of the proposed roles of R-M systems is protection of bacterial strains against phage infections (3). Additionally, phage transduction is a method commonly used in the laboratory to move DNA between staphylococcal strains. Therefore, in an experiment complementary to the electroporation experiment, we tested if we could introduce foreign DNA into the hsdR mutants by transduction. For this, we used phages 80α and φ75 to transduce plasmid pGC2 from S. aureus strain 879R4RF into NCTC8325-4 and SH1000 wild-type and hsdR mutant strains. Strain 879R4RF belongs to the CC51 lineage, and DNA propagated in this strain is recognized as “foreign” DNA by members of the CC8 lineage, such as NCTC8325-4 and SH1000 (21). In agreement with the electroporation data, NCTC8325-4ΔHsdR and SH1000ΔHsdR were not capable of accepting pGC2 DNA coming from 879R4RF by transduction (Table 2).
TABLE 2.
Efficiency of transduction and infection of S. aureus wild-type and hsdR mutant strains using phages 80α and φ75
| Strain | Transduction of plasmid pGC2 from strain 879R4RF (no. of transductants/106 phage particles)
|
Efficiency of plaquinga
|
||
|---|---|---|---|---|
| Phage 80α | Phage φ75 | Phage 80α from 879R4RF | Phage φ75 from 879R4RF | |
| RN4220 | 158 | 620 | 1.0 | 1.0 |
| NCTC8325-4 | 0 | 0 | 3.1 × 10−5 | 3.7 × 10−6 |
| NCTC8325-4ΔHsdR | 0 | 0 | 5.8 × 10−5 | 3.9 × 10−3 |
| NCTC8325-4ΔC-term HsdR | 0 | 0 | 1.0 × 10−4 | 6.1 × 10−3 |
| SH1000 | 0 | 0 | 4.5 × 10−5 | 4.7 × 10−5 |
| SH1000ΔHsdR | 0 | 0 | 1.6 × 10−4 | 3.6 × 10−3 |
The efficiency of plaquing was calculated by dividing the number of phage PFU per milliliter of phage lysate in each strain tested by the corresponding number in RN4220.
We also tested the susceptibility of the hsdR mutants to infection by phages propagated in 879R4RF. When NCTC8325-4 and SH1000 and the corresponding hsdR mutants were infected with phage 80α, again there was no significant increase (threefold or less) in the efficiency of infection associated with deletion of hsdR (Table 2). However, when the same strains were infected with phage φ75, there was a 100- to 1,000-fold increase in the efficiency of infection of the hsdR mutants compared with their parental strains (Table 2). It is possible that phage φ75 (but not phage 80α) is able to resist the action of the additional factor(s) that degrades foreign DNA (possibly a second R-M system) which we postulate exists in NCTC8325-4 and SH1000. In fact, many phages have some means of reducing their susceptibility to R-M systems, such as unusual modifications of DNA, a low frequency of target sequences, or the production of a protein that inhibits the activity of an R-M system (3, 12).
Heat treatment of competent cells increases the transformation efficiency of hsdR mutants.
Some restriction enzymes (including type II enzymes) can be inactivated by heat, and brief heat treatment of staphylococcal cells has been used to increase phage sensitivity (2, 17). In order to determine if the restriction activity which remains in the hsdR mutants can be inactivated by heat, we examined the transformation efficiency of wild-type and hsdR mutant strains after exposure of competent cells to 56°C for 2 min. Indeed, after the heat treatment we were finally able to introduce foreign DNA (pGC2 extracted from E. coli) into the NCTC8325-4ΔHsdR and SH1000ΔHsdR strains by electroporation, while the parental strains remained nontransformable (Table 1). For unknown reasons, we were not able to transform COLΔHsdR. Importantly, the heat treatment resulted in a 100-fold decrease in the transformation efficiency of control strain RN4220 (Table 1). The low efficiencies obtained in the NCTC8325-4 and SH1000 backgrounds do not allow us to propose the use of heat-treated hsdR mutants as standard tools for transformation experiments with S. aureus, particularly when transformation of nonreplicative plasmids is required as a step in procedures for genetically manipulating S. aureus.
Conclusion.
It seems to be unambiguous that a mutation in SauI hsdR is required for the transformable phenotype of RN4220; this strain does not have mutations in any of the other hsd genes or their promoters, and complementation with a plasmid containing the intact SauI hsdR gene completely blocks uptake of E. coli DNA by RN4220 (21). However our data indicate that deleting or truncating SauI hsdR is not enough to allow the S. aureus strains tested to easily accept foreign DNA either via electroporation or via phage infection or transduction, and therefore the ability of RN4220 to accept foreign DNA must be dependent on an additional unknown factor(s), possibly a second R-M system. Interestingly, NCTC8325 (the parental strain of the prophage-cured strain NCTC8325-4) has been suggested to have two distinct R-M systems based on analysis of restriction- and modification-deficient mutants (9, 16), but the second R-M system could be associated with phages present in this strain and is not annotated in the genome.
Constructing S. aureus mutants of relevant laboratory and clinical strains that are capable of efficiently accepting foreign DNA by inactivating the SauI type I R-M system does not seem to be a viable option. Full sequencing of the RN4220 genome should indicate which additional, unidentified factors contribute to its transformable phenotype. The information obtained should contribute to development of better genetic tools for manipulating S. aureus, as well as to a better understanding of gene transfer in this pathogenic organism.
Acknowledgments
This work was supported by grant POCI/BIA-BCM/66449/2006 from Fundação para a Ciência e Tecnologia, Portugal, awarded to M.G.P. H.V. was supported by fellowship SFRH/BD/38732/2007.
We thank Jodi Lindsay for helpful discussions.
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asheshov, E. A., and M. P. Jevons. 1963. The effect of heat on the ability of a host strain to support the growth of a Staphylococcus phage. J. Gen. Microbiol. 31:97-107. [DOI] [PubMed] [Google Scholar]
- 3.Bickle, T. A., and D. H. Kruger. 1993. Biology of DNA restriction. Microbiol. Rev. 57:434-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leao, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Dempsey, R. M., D. Carroll, H. Kong, L. Higgins, C. T. Keane, and D. C. Coleman. 2005. Sau42I, a BcgI-like restriction-modification system encoded by the Staphylococcus aureus quadruple-converting phage Phi42. Microbiology 151:1301-1311. [DOI] [PubMed] [Google Scholar]
- 6.Dyke, K. G., M. P. Jevons, and M. T. Parker. 1966. Penicillinase production and intrinsic resistance to penicillins in Staphylococcus aures. Lancet i:835-838. [DOI] [PubMed] [Google Scholar]
- 7.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iordanescu, S., and M. Surdeanu. 1976. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96:277-281. [DOI] [PubMed] [Google Scholar]
- 10.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 11.McNamara, P. J. 2008. Genetic manipulation of Staphylococcus aureus, p. 89-129. In J. A. Lindsay (ed.), Staphylococcus molecular genetics. Caister Academic Press, Norfolk, United Kingdom.
- 12.Murray, N. E. 2000. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 64:412-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noble, W. C., Z. Virani, and R. G. Cree. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72:195-198. [DOI] [PubMed] [Google Scholar]
- 14.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 15.Oshida, T., and A. Tomasz. 1992. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J. Bacteriol. 174:4952-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sjostrom, J. E., S. Lofdahl, and L. Philipson. 1978. Biological characteristics of a type I restriction-modification system in Staphylococcus aureus. J. Bacteriol. 133:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stobberingh, E. E., and K. C. Winkler. 1977. Restriction-deficient mutants of Staphylococcus aureus. J. Gen. Microbiol. 99:359-367. [DOI] [PubMed] [Google Scholar]
- 18.Sung, J. M., and J. A. Lindsay. 2007. Staphylococcus aureus strains that are hypersusceptible to resistance gene transfer from enterococci. Antimicrob. Agents Chemother. 51:2189-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sussenbach, J. S., C. H. Monfoort, R. Schiphof, and E. E. Stobberingh. 1976. A restriction endonuclease from Staphylococcus aureus. Nucleic Acids Res. 3:3193-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sussenbach, J. S., P. H. Steenbergh, J. A. Rost, W. J. van Leeuwen, and J. D. van Embden. 1978. A second site-specific restriction endonuclease from Staphylococcus aureus. Nucleic Acids Res. 5:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waldron, D. E., and J. A. Lindsay. 2006. SauI: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188:5578-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu, S., H. de Lencastre, A. Sali, and A. Tomasz. 1996. A phosphoglucomutase-like gene essential for the optimal expression of methicillin resistance in Staphylococcus aureus: molecular cloning and DNA sequencing. Microb. Drug Resist. 2:277-286. [DOI] [PubMed] [Google Scholar]