Abstract
Synthetic metabolic pathways have been constructed for the production of enantiopure (R)- and (S)-3-hydroxybutyrate (3HB) from glucose in recombinant Escherichia coli strains. To promote maximal activity, we profiled three thiolase homologs (BktB, Thl, and PhaA) and two coenzyme A (CoA) removal mechanisms (Ptb-Buk and TesB). Two enantioselective 3HB-CoA dehydrogenases, PhaB, producing the (R)-enantiomer, and Hbd, producing the (S)-enantiomer, were utilized to control the 3HB chirality across two E. coli backgrounds, BL21Star(DE3) and MG1655(DE3), representing E. coli B- and K-12-derived strains, respectively. MG1655(DE3) was found to be superior for the production of each 3HB stereoisomer, although the recombinant enzymes exhibited lower in vitro specific activities than BL21Star(DE3). Hbd in vitro activity was significantly higher than PhaB activity in both strains. The engineered strains achieved titers of enantiopure (R)-3HB and (S)-3HB as high as 2.92 g liter−1 and 2.08 g liter−1, respectively, in shake flask cultures within 2 days. The NADPH/NADP+ ratio was found to be two- to three-fold higher than the NADH/NAD+ ratio under the culture conditions examined, presumably affecting in vivo activities of PhaB and Hbd and resulting in greater production of (R)-3HB than (S)-3HB. To the best of our knowledge, this study reports the highest (S)-3HB titer achieved in shake flask E. coli cultures to date.
The synthesis of chiral molecules is of significant interest in the pharmaceutical industry because frequently one stereoisomer of a drug has efficacy while the other has either substantially reduced or no activity or may even have adverse effects (20, 23). Additionally, chiral molecules serve as building blocks for many pharmaceuticals and high-value compounds. Thus, the ability to prepare chiral molecules with high optical purity is important. Stereoselective chemical processes generally employ expensive chiral catalysts, require harsh physical conditions and solvents, and suffer from extensive by-product formation. In contrast, enzyme-catalyzed reactions are highly stereoselective and can be performed in aqueous solutions under mild conditions (21). As a result, the use of biological processes for chiral molecule production has been extensively investigated (4, 28, 32, 36). One example of such a process is the biosynthesis of 3-hydroxybutyric acid (3HB), a versatile chiral molecule containing one hydroxyl group and one carboxyl group, used as a building block for the synthesis of optically active fine chemicals, such as vitamins, antibiotics, pheromones, and flavor compounds (5, 6, 18, 27).
The biosynthesis of 3HB has typically been achieved by two different mechanisms: depolymerization (in vitro or in vivo) of microbially synthesized poly-(R)-3-hydroxybutyric acid (PHB) (8, 13) or direct synthesis of 3HB without a PHB intermediate (9, 12, 15). However, due to the stereospecific constraints of PHB synthesis, in which polymers are composed exclusively of (R)-3HB monomer units, the synthesis of (S)-3HB from PHB is effectively impossible. In contrast, direct synthesis of both enantiopure (R)-3HB and (S)-3HB is possible. Pathways facilitating (R)-3HB synthesis have been constructed in Escherichia coli by simultaneous expression of phaA (encoding acetoacetyl coenzyme A [CoA] thiolase) and phaB [encoding (R)-3HB-CoA dehydrogenase] from Ralstonia eutropha H16, and ptb (encoding phosphotransbutyrylase) and buk (encoding butyrate kinase) from Clostridium acetobutylicum ATCC 824 (9). In addition to the use of ptb and buk to catalyze the conversion of (R)-3HB-CoA to (R)-3HB, tesB (encoding thioesterase II from E. coli) has also been used for the direct hydrolysis of (R)-3HB-CoA to yield (R)-3HB (15). The production of (S)-3HB in E. coli has recently been reported using a biosynthetic pathway consisting of phaA from R. eutropha H16, hbd [encoding (S)-3HB-CoA dehydrogenase] from C. acetobutylicum ATCC 824, and bch (encoding 3-hydroxyisobutyryl-CoA hydrolase) from Bacillus cereus ATCC 14579 (12).
In E. coli, the synthesis of both enantiomers of 3HB begins with the condensation of two molecules of acetyl-CoA, catalyzed by a thiolase, to give acetoacetyl-CoA (Fig. 1). The acetoacetyl-CoA is then reduced either to (R)-3HB-CoA via ketone reduction mediated by an NADPH-dependent (R)-3HB-CoA dehydrogenase (PhaB) or to (S)-3HB-CoA via an NADH-dependent (S)-3-HB-CoA dehydrogenase (Hbd). (R)-3HB-CoA and (S)-3HB-CoA can each be further modified via a suitable CoA removal reaction to form (R)-3HB and (S)-3HB, respectively. In an effort to increase chiral 3HB production, it is essential to identify a thiolase capable of efficiently catalyzing the first reaction in the 3HB biosynthetic pathways, to draw acetyl-CoA from competing endogenous pathways. Thus, we examined three different thiolases (BktB and PhaA from R. eutropha H16 and Thl from C. acetobutylicum ATCC 824) to determine which is most proficient for 3HB synthesis. (R)-3HB-CoA and (S)-3HB-CoA synthesized via the reduction reaction catalyzed by PhaB and Hbd, respectively, must be converted to their respective free acid forms before transport or diffusion out of the cell. We have compared two sets of CoA-removing enzyme mechanisms, including the phosphotransbutyrylase (Ptb) and butyrate kinase (Buk) system encoded by the ptb-buk operon from C. acetobutylicum ATCC 824 and acyl-CoA thioesterase II (TesB) from E. coli MG1655. Moreover, it has long been argued whether B strains or K-12 strains of E. coli would serve as better hosts for the biosynthesis of small molecules. Microarrays and Northern blot analyses have suggested that several metabolic pathways, including the tricarboxylic acid (TCA) cycle, glyoxylate shunt, glycolysis, and fatty acid degradation are different between these two strains (22, 25, 34, 35), implying that they may differ significantly in their abilities to supply significant levels of acetyl-CoA as the precursor for 3HB synthesis. Thus, we have also compared 3HB synthesis across two representative E. coli strains: BL21Star(DE3) (B strain) and MG1655(DE3) (K-12 strain). 3HB chirality was examined and verified by high-performance liquid chromatography (HPLC) analysis using a chiral stationary phase to provide separation.
FIG. 1.
Schematic representation of (S)-3HB or (R)-3HB synthesis from glucose in engineered E. coli. BktB, acetoacetyl-CoA thiolase from R. eutropha H16; Thl, acetoacetyl-CoA thiolase from C. acetobutylicum ATCC 824; PhaA, acetoacetyl-CoA thiolase from R. eutropha H16; Hbd, (S)-3HB-CoA dehydrogenase from C. acetobutylicum ATCC 824; PhaB, (R)-3HB-CoA dehydrogenase from R. eutropha H16; Ptb, phosphotransbutyrylase from C. acetobutylicum ATCC 824; Buk, butyrate kinase from C. acetobutylicum ATCC 824; TesB, acyl-CoA thioesterase II from E. coli MG1655.
Altogether, we have explored the production of each stereoisomer of 3HB across different strains of E. coli, different thiolases, and different CoA removal systems to engineer E. coli strains for enhanced chiral 3HB production.
MATERIALS AND METHODS
Microorganisms.
The bacterial strains used are listed in Table 1. C. acetobutylicum ATCC 824 was purchased from the American Type Culture Collection (ATCC, Manassas, VA). R. eutropha H16 was provided by Anthony Sinskey, Department of Biology, Massachusetts Institute of Technology, Cambridge. E. coli DH10B (Invitrogen, Carlsbad, CA) and ElectroTen-Blue (Stratagene, La Jolla, CA) were used for transformation of cloning reactions and propagation of all plasmids. BL21Star(DE3) (Invitrogen, Carlsbad, CA) and MG1655(DE3) were used as host strains for the biosynthesis of chiral 3HB, where MG1655(DE3) was constructed using a λDE3 lysogenization kit (Novagen, Darmstadt, Germany) for site-specific integration of λDE3 prophage into E. coli MG1655 (kindly donated by Gregory Stephanopoulos, Department of Chemical Engineering, Massachusetts Institute of Technology.
TABLE 1.
E. coli strains, plasmids, and oligonucleotide primers used
| Strain, plasmid, or primer | Relevant genotype or primer sequence (5′→3′)a | Reference or source |
|---|---|---|
| Strains | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139Δ(ara leu)7697 galU galK λ−rpsL nupG | Invitrogen |
| ElectroTen-Blue | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Kanr [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| MG1655 | F− λ−ilvG rfb-50 rph-1 | ATCC 700926 |
| BL21Star(DE3) | F−ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) | Invitrogen |
| MG1655(DE3) | F− λ−ilvG rfb-50 rph-1 (DE3) | This study |
| Plasmids | ||
| pGEM-T Easy | PCR cloning vector; F1 ori Ampr | Promega |
| pETDuet-1 | ColE1(pBR322) ori lacI T7lac Ampr | Novagen |
| pCDFDuet-1 | CloDF13 ori lacI T7lac Strr | Novagen |
| pET-H | pETDuet-1 harboring hbd from C. acetobutylicum ATCC 824 | This study |
| pET-H-Bb | pETDuet-1 harboring bktB from R. eutropha H16 and hbd from C. acetobutylicum ATCC 824 | This study |
| pET-H-Tb | pETDuet-1 harboring thl and hbd from C. acetobutylicum ATCC 824 | This study |
| pET-H-Pb | pETDuet-1 harboring phaA from R. eutropha H16 and hbd from C. acetobutylicum ATCC 824 | This study |
| pET-P | pETDuet-1 harboring phaB from R. eutropha H16 | This study |
| pET-P-Bb | pETDuet-1 harboring bktB and phaB from R. eutropha H16 | This study |
| pET-P-Tb | pETDuet-1 harboring thl from C. acetobutylicum ATCC 824 and phaB from R. eutropha H16 | This study |
| pET-P-Pb | pETDuet-1 harboring phaA and phaB from R. eutropha H16 | This study |
| pCDF-T | pCDFDuet-1 harboring tesB from E. coli MG1655 | This study |
| pCDF-PB | pCDFDuet-1 harboring ptb-buk operon from C. acetobutylicum ATCC 824 | This study |
| Primersc | ||
| bktB_US_EcoRI | GAATTCATGACGCGTGAAGTGGTAGTG | Sigma-Genosys |
| bktB_DS_XhoI | CTCGAGCGCAAGGCTAACCTCAGAT | Sigma-Genosys |
| thil_US_EcoRI | ATAGAATTCCATGAGAGATGTAGTAATAGTAAGTG | Sigma-Genosys |
| thil_DS_XhoI | TATTGAACCTCCTCGAGAACTTAGTTATAT | Sigma-Genosys |
| phaA_US_EcoRI | GAATTCGACTACACAATGACTGACGTTGTC | Sigma-Genosys |
| phaA_DS_XhoI | CTCGAGAAAACCCCTTCCTTATTTGC | Sigma-Genosys |
| hbd_US_EcoRI | GAATTCGGGAGGTCTGTTTAATGAAAA | Sigma-Genosys |
| hbd_DS_NotI | GCGGCCGCTGTAAACTTATTTTG | Sigma-Genosys |
| phaB_US_EcoRI | GAATTCAACGAAGCCAATCAAGGAG | Sigma-Genosys |
| phaB_DS_NotI | GCGGCCGCGCAGGTCAGCCCATATG | Sigma-Genosys |
| tesB_US_EcoRI | GAATTCTACTGGAGAGTTATATGAGTCAGG | Sigma-Genosys |
| tesB_DS_tSalI | GTCGACTTAATTGTGATTACGCATC | Sigma-Genosys |
Restriction enzyme sites used in the cloning are underlined.
Each gene is under the control of the T7lac promoter with a ribosome binding site.
Primers were synthesized at Sigma-Genosys, St. Louis, MO.
Plasmid construction.
Genes derived from C. acetobutylicum ATCC 824 (thl, hbd, and ptb-buk operon), R. eutropha H16 (phaA, bktB, and phaB), and E. coli K-12 (tesB) were obtained by PCR using genomic DNA (gDNA) templates. All gDNAs were prepared using the Wizard gDNA purification kit (Promega, Madison, WI). Custom oligonucleotides (primers) were purchased for all PCR amplifications (Sigma-Genosys, St. Louis, MO) as shown in Table 1. In all cases, HotStar HiFidelity polymerase (Qiagen, Valencia, CA) was used for DNA amplification. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). Recombination DNA techniques were performed according to standard procedures (24).
Two coreplicable vectors, pETDuet-1 and pCDFDuet-1 (Novagen, Darmstadt, Germany), were used for construction of chiral 3HB biosynthetic pathways (33). Both Duet vectors contain two multiple cloning sites (MCS), each of which is preceded by a T7lac promoter and a ribosome binding site, affording high-level expression of each gene.
For cloning genes, PCR products incorporated with desired restriction sites within the 5′ and 3′ primers were first cloned into pGEM-T Easy vector (Promega, Madison, WI) by TA cloning. The resulting plasmids were then digested and the genes were subcloned into pETDuet-1 or pCDFDuet-1. The hbd gene was inserted in between the EcoRI and NotI sites (MCS I) of pETDuet-1 to create pET-H. The phaB gene was inserted between the NcoI and NotI sites (MCS I) of pETDuet-1 to create pET-P. Plasmids pET-H-B, pET-H-T, and pET-H-P were constructed by inserting bktB, thl, and phaA, respectively, between the MfeI and XhoI sites (MCS II) of pET-H. In a similar manner, plasmids pET-P-B, pET-P-T, and pET-P-P were constructed by inserting bktB, thl, and phaA, respectively, between the MfeI and XhoI sites (MCS II) of pET-P. Plasmid pCDF-PB was created by inserting the ptb-buk operon between the EcoRI and SalI sites of pCDFDuet-1. Cloning the tesB gene between the EcoRI and SalI sites of pCDFDuet-1 resulted in plasmid pCDF-T. All constructs were confirmed to be correct by restriction enzyme digestion and nucleotide sequencing. Plasmids constructed in the present work are listed in Table 1.
Culture conditions.
Seed cultures of the recombinant strains were grown in LB medium at 30°C overnight on a rotary shaker at 250 rpm. For the biosynthesis of chiral 3HB, the seed cultures were used to inoculate 50 ml LB medium in 250-ml flasks supplemented with 20 g liter−1 glucose at an inoculation volume of 5%. Cultures were incubated at 30°C on a rotary shaker until the optical density at 600 nm (OD600) reached 0.4 to ∼0.8. At this point, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the cultures to induce recombinant protein expression. In all cases, LB medium was supplemented with 50 mg liter−1 ampicillin and 25 mg liter−1 streptomycin and cultures were incubated aerobically. Culture broth was sampled at 4 h postinduction for enzyme assays and at 48 h postinduction for HPLC analysis.
Metabolite analysis and DCW determination.
Samples were centrifuged to pellet cells, while the aqueous supernatant was collected for HPLC analysis. Concentrations of 3HB, glucose, and acetate were analyzed via HPLC using an Agilent 1100 series instrument equipped with a refractive index detector. Analyte separation was achieved using an Aminex HPX-87H anion-exchange column (Bio-Rad Laboratories, Hercules, CA) according to the method of Buday et al. (3) using 5 mM H2SO4 as the mobile phase.
The dry cell weight (DCW) was determined in triplicates by filtering culture broth (25 ml) through a preweighed filter paper (0.45-μm-pore cellulose acetate filters; Whatman, Maidstone, United Kingdom), followed by washing with deionized water (25 ml). The filter paper was then dried for 2 days in a 70°C oven and weighed for DCW determination. One OD600 unit was equivalent to 0.42 g DCW liter−1 for BL21Star(DE3) and 0.39 g DCW liter−1 for MG1655(DE3).
Methyl esterification of 3HB.
Fifty milliliters of 3HB-containing LB microbial culture was centrifuged at 6,000 × g for 10 min, and the supernatant was transferred to an evaporation dish for evaporation overnight in a 50°C oven, resulting in a brown residue. To this residue, 2 ml of acidic methanol (4:1 methanol/concentrated HCl ratio) was added, and the dish was shaken for 2 min to remove 3HB from the residue. The methanol solution was transferred into a test tube, sealed, and heated at 100°C for 3 h. After heating, the solution was allowed to cool to room temperature. After cooling, 200 μl of the methanol solution was mixed with 1 ml of pure isopropanol in a 1.7-ml tube, vortexed, and allowed to stand for 5 min. Precipitate in this sample was removed by centrifugation at 13,200 × g for 10 min. The supernatant was taken for chiral HPLC analysis.
Chiral HPLC analysis of methyl-3HB.
Chiral HPLC analysis was performed on an Agilent 1100 series instrument equipped with a Chiralcel OD-H column (0.46 cm φ by 25 cm) purchased from Daicel Chemical Industries (West Chester, PA). The column temperature was maintained at 40°C. Methyl-3HB was detected on a diode array detector at 210 nm. The mobile phase was 9:1 n-hexane-isopropanol, and the flow rate through the column was 0.7 ml min−1. Methyl-(R)-3HB and methyl-(S)-3HB purchased from Sigma-Aldrich (St. Louis, MO) were used as standards and had retention times of 7.78 and 9.37 min, respectively.
Enzyme assays.
Samples of 5 ml culture broth were centrifuged at 5,000 × g and 4°C for 10 min. The pellets were resuspended in 200 μl of lysing buffer (1 Roche protease inhibitor cocktail tablet in 10.5 ml of 10 mM Tris-HCl supplemented with 1 mg ml−1 lysozyme; pH 8.0). Crude cell lysates were prepared by freeze-thaw lysis, carried out by subjecting cells to five cycles of freezing in liquid nitrogen followed by thawing at 37°C. After centrifugation at 14,000 × g and 4°C for 20 min, the supernatant was taken for enzyme assays. The total protein concentration of this supernatant was determined by the method of Bradford (1) using bovine serum albumin as a standard.
Activities of thiolases, including BktB, Thl, and PhaA, were assayed using acetoacetyl-CoA and CoA as substrates, and the decrease in acetoacetyl-CoA concentration was followed at 303 nm (10). Activities of PhaB and Hbd were assayed using acetoacetyl-CoA as the substrate and NADPH or NADH as the cofactor, respectively. The decrease in NADPH or NADH concentration resulting from 3HB-CoA formation from acetoacetyl-CoA was measured at 340 nm (10, 26).
Quantification of intracellular cofactor levels.
The NAD+, NADH, NADP+, and NADPH were each determined using Enzychrom NAD+/NADH and NADP+/NADPH assay kits (Bioassay Systems, Hayward, CA). Manufacturer's protocols were followed, except that for sample preparation cells were resuspended in 100 μl extraction buffer and sonicated for 10 s on ice before heat extraction. The assays utilized alcohol dehydrogenase and glucose dehydrogenase for NAD(H) and NADP(H) quantifications, respectively, and colorimetric changes in the samples were measured at 565 nm.
RESULTS
Production of chiral 3HB in BL21Star(DE3).
We first constructed the chiral 3HB pathways in BL21Star(DE3), an E. coli B strain, by cotransforming one pETDuet-1 derivative (pET-H-B, pET-H-T, pET-H-P, pET-P-B, pET-P-T, or pET-P-P) and one pCDFDuet-1 derivative (pCDF-T or pCDF-PB) (Table 1). All strains were cultured at 30°C for 48 h in LB medium supplemented with 20 g liter−1 glucose, and accumulation of chiral 3HB in the culture medium was measured. We then compared capabilities of two sets of CoA-removing enzymes, Ptb-Buk and TesB, for (R)-3HB or (S)-3HB production. TesB successfully removed the CoA moiety from both (R)-3HB-CoA and (S)-3HB-CoA to release the free acids (Table 2). In comparison, Ptb-Buk could also convert (R)-3HB-CoA but was essentially inactive on (S)-3HB-CoA. Titers as high as 1.58 g liter−1 and 2.41 g liter−1 of (S)-3HB and (R)-3HB, respectively, could be achieved when TesB was used to remove CoA. In the case of Ptb-Buk, only a trace amount of (S)-3HB (less than 0.10 g liter−1) was produced, and 1.26 g liter−1 of (R)-3HB was observed. It was also noted that less acetate (1.02 g liter−1 on average) was produced with the TesB system than with the Ptb-Buk system, which produced 1.55 g liter−1 on average.
TABLE 2.
Extracellular production of chiral 3HB by E. coli BL21Star(DE3) grown in shake flasksa
| Chiral 3HB production | 3HB synthesis pathway
|
Amt of acetate (g liter−1) | Amt of 3HB (g liter−1) | 3HB specific content (g g DCW−1) | 3HB yield (g g glucose−1) | ||
|---|---|---|---|---|---|---|---|
| Step 1 | Step 2 | Step 3 | |||||
| CoA-removing enzyme set 1 | |||||||
| (S)-3HB | BktB | Hbd | Ptb-Buk | 1.53 ± 0.13 | <0.1 | NAb | NA |
| Thl | Hbd | Ptb-Buk | 1.79 ± 0.03 | <0.1 | NA | NA | |
| PhaA | Hbd | Ptb-Buk | 1.55 ± 0.14 | <0.1 | NA | NA | |
| (R)-3HB | BktB | PhaB | Ptb-Buk | 1.33 ± 0.02 | 1.23 ± 0.06 | 1.19 ± 0.05 | 0.18 ± 0.01 |
| Thl | PhaB | Ptb-Buk | 1.66 ± 0.08 | 1.03 ± 0.02 | 0.69 ± 0.01 | 0.15 ± 0.01 | |
| PhaA | PhaB | Ptb-Buk | 1.41 ± 0.11 | 1.26 ± 0.06 | 1.19 ± 0.15 | 0.20 ± 0.03 | |
| CoA-removing enzyme set 2 | |||||||
| (S)-3HB | BktB | Hbd | TesB | 0.89 ± 0.07 | 1.55 ± 0.05 | 1.50 ± 0.09 | 0.27 ± 0.01 |
| Thl | Hbd | TesB | 1.09 ± 0.12 | 1.58 ± 0.07 | 1.22 ± 0.06 | 0.24 ± 0.01 | |
| PhaA | Hbd | TesB | 1.01 ± 0.03 | 1.44 ± 0.04 | 1.43 ± 0.04 | 0.26 ± 0.02 | |
| (R)-3HB | BktB | PhaB | TesB | 0.88 ± 0.03 | 2.41 ± 0.04 | 2.24 ± 0.04 | 0.30 ± 0.00 |
| Thl | PhaB | TesB | 1.20 ± 0.03 | 1.90 ± 0.06 | 1.52 ± 0.03 | 0.26 ± 0.01 | |
| PhaA | PhaB | TesB | 1.05 ± 0.09 | 2.36 ± 0.08 | 2.06 ± 0.17 | 0.30 ± 0.02 | |
Cells were grown aerobically in LB medium with the addition of 2% glucose at 30°C for 48 h. IPTG (1 mM) was added when the OD600 reached 0.4 to ∼0.8. Data are presented as the average value and standard deviation of measurements from three independent cultures.
NA, not applicable.
The efficacies of three different thiolases on chiral 3HB production were then compared. Both BktB and PhaA were found to yield similar titers of (R)-3HB when coexpressed with PhaB and Ptb-Buk (∼1.25 g liter−1) or PhaB and TesB (∼2.39 g liter−1). However, Thl gave approximately 20% lower titers of (R)-3HB and increased acetate production compared to BktB and PhaA. This phenomenon was not observed in (S)-3HB production, where each of the thiolases resulted in similar titers of (S)-3HB, with an average of 1.52 g liter−1. In general, TesB could outperform Ptb-Buk for CoA removal, resulting in significantly higher titers of both (R)-3HB and (S)-3HB (Table 2).
Production of chiral 3HB in MG1655(DE3).
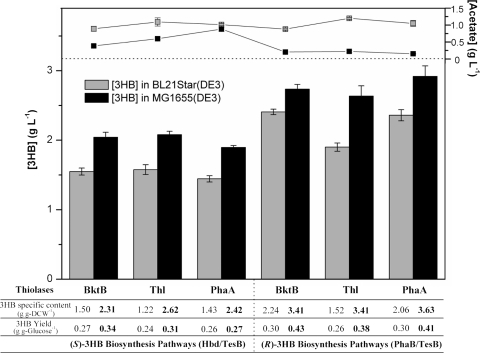
Since TesB was identified as the most effective enzyme among those tested for CoA removal in BL21Star(DE3), further investigation of MG1655(DE3), an E. coli K-12 strain, exclusively employed 3HB pathways using TesB. In all cases, (R)-3HB or (S)-3HB production was substantially higher with MG1655(DE3) than with BL21Star(DE3) under the same culture conditions (Fig. 2). MG1655(DE3) produced up to 2.08 g liter−1 of (S)-3HB and 2.92 g liter−1 of (R)-3HB, ∼30% and ∼20% higher titers, respectively, than those produced by BL21Star(DE3). It is also interesting to note that generally less acetate was produced in MG1655(DE3) than in BL21Star(DE3), suggesting that more acetyl-CoA carbon flux was directed toward 3HB biosynthesis in MG1655(DE3) than toward acetate production. These two production systems were also compared in terms of 3HB-specific content (g g DCW−1) and 3HB yield (g g glucose−1). 3HB specific contents in MG1655(DE3) strains were ∼50 to 120% greater than their respective BL21Star(DE3) counterparts as a result of increased 3HB production and reduced biomass accumulation (Fig. 2). The comparison of 3HB yield on glucose shows that the efficiency of 3HB production from glucose in MG1655(DE3) was higher than that in BL21Star(DE3). Generally, MG1655(DE3) was able to produce more chiral 3HB and less acetate, while also accumulating less biomass than BL21Star(DE3).
FIG. 2.
Extracellular production of chiral 3HB by E. coli BL21Star(DE3) and MG1655(DE3) grown in shake flasks. (S)-3HB was produced when Hbd was employed (left), while (R)-3HB was produced when PhaB was employed (right). In all cases, TesB was used to mediate CoA removal.
Confirmation of 3HB stereochemistry.
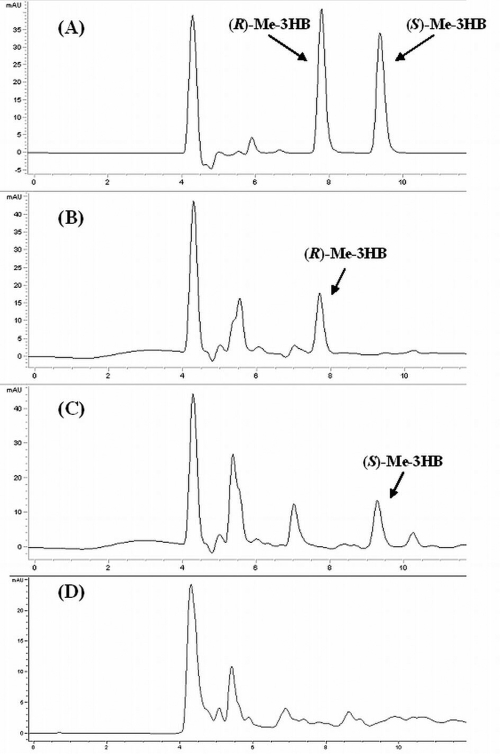
E. coli BL21Star(DE3) harboring bktB, tesB, and either phaB or hbd was grown at 30°C in 50 ml LB supplemented with 20 g liter−1 glucose for 48 h. The stereochemistry of the resulting 3HB in the media from these cultures was determined by methyl esterification of the 3HB present followed by chiral HPLC analysis as described in Materials and Methods. The results confirm previous reports that cells harboring a thiolase and CoA removal enzyme produce (R)-3HB if the cells are concomitantly expressing phaB and synthesize (S)-3HB if they are expressing hbd (Fig. 3) (12, 15).
FIG. 3.
HPLC spectra of methyl-(R)-3HB and methyl-(S)-3HB standards (A); culture medium from E. coli BL21Star(DE3) expressing bktB, phaB, and tesB after boiling in methanol (B); culture medium from E. coli BL21Star(DE3) expressing bktB, hbd, and tesB after boiling in methanol (C); and culture medium from E. coli BL21Star(DE3) after boiling in methanol as a control (D).
Measurement of specific activities of 3HB synthesis enzymes expressed in E. coli.
To better understand the differences in 3HB titers between the different 3HB pathways in E. coli BL21Star(DE3) and MG1655(DE3), we sought to find a correlation between in vivo chiral 3HB production and in vitro enzyme activities. The activities of BktB, Thl, PhaA, Hbd, and PhaB were measured in recombinant E. coli BL21Star(DE3) and MG1655(DE3). The background acetoacetyl-CoA thiolase activities in E. coli BL21Star(DE3) and MG1655(DE3) were very weak (<0.04 U mg protein−1), and there were undetectable 3HB-CoA dehydrogenase activities (Table 3). Plasmid-based expression of BktB, Thl, PhaA, Hbd, and PhaB was able to give functional enzymes in both E. coli strains (Table 3). It is interesting to note that all enzymes analyzed had greater specific activities in BL21Star(DE3) than in MG1655(DE3), showing approximately 130% to 360% higher thiolase activity and 24% to 44% higher 3HB-CoA dehydrogenase activity, although 3HB production was lower. For comparison of alternative acetoacetyl-CoA thiolases, Thl had approximately sixfold higher specific activity than BktB and PhaA in BL21Star(DE3) and sixfold and threefold higher specific activities than BktB and PhaA, respectively, in MG1655(DE3). For 3HB-CoA dehydrogenase activities of PhaB and Hbd, results show that the activities of Hbd (NADH dependent) were approximately 140-fold and 120-fold higher than those of PhaB (NADPH dependent) in BL21Star(DE3) and MG1655(DE3), respectively.
TABLE 3.
Enzyme specific activities of crude extracts of E. coli BL21Star(DE3) and MG1655(DE3)
| Enzyme | Sp act(U mg−1) for E. coli straina:
|
|
|---|---|---|
| BL21Star(DE3) | MG1655(DE3) | |
| Acetoacetyl-CoA thiolase | ||
| Control | 0.04 ± 0.01 | 0.03 ± 0.00 |
| BktB | 5.05 ± 0.69 | 1.10 ± 0.24 |
| Thl | 29.66 ± 3.87 | 6.85 ± 0.97 |
| PhaA | 5.90 ± 0.87 | 2.56 ± 0.36 |
| 3-HB-CoA dehydrogenase | ||
| Control | NDb | ND |
| Hbd | 58.78 ± 7.76 | 40.76 ± 8.08 |
| PhaB | 0.41 ± 0.18 | 0.33 ± 0.05 |
One unit was defined as the conversion of 1 μmol of substrate to product per min at 25°C. Data are presented as the average value and standard deviation of measurements from three independent cultures.
ND, not detected.
Measurement of cofactor levels in engineered MG1655(DE3) strains.
Although Hbd consistently showed higher in vitro activity than PhaB, the accumulated titers of (S)-3HB were lower than those of (R)-3HB. Since these two enzymes have different pyridine nucleotide cofactor (NADPH/NADH) specificities, we measured intracellular levels of the reduced and oxidized cofactors to gain additional insights. At the late exponential phase (4 h postinduction), the specific contents of NADH and NADPH in all tested recombinant strains were found to be significantly lower than those of their respective oxidized forms (NAD+ and NADP+), with the ratios of NADH/NAD+ and NADPH/NADP+ ranging from 0.08 to 0.16 and 0.26 to 0.31, respectively. At the stationary phase (24 h postinduction), reduced cofactor increased concomitantly with the decrease of oxidized cofactor, resulting in higher ratios of NADH/NAD+ and NADPH/NADP+ ratios, ranging from 0.20 to 0.44 and 0.79 to 0.99, respectively. Consistent with previously published reports (2, 11), in general NADH was found to be the predominant reducing equivalent in our E. coli strains while the NADPH/NADP+ ratio was considerably higher than the NADH/NAD+ ratio under the culture conditions examined.
DISCUSSION
The underlying objective of this study was to explore the high-level production of both (R)- and (S)-3HB in recombinant E. coli by investigating different host strains, thiolase homologs, and CoA removal mechanisms. We have achieved shake flask-scale production of enantiomerically pure (R)-3HB and (S)-3HB to concentrations of up to 2.92 g liter−1 and 2.08 g liter−1, respectively. During the preparation of this article, biosynthesis of enantiopure (S)-3HB was reported at titers of 0.61 g liter−1 (12), which we believe is the highest reported concentration prior to this study.
E. coli B versus E. coli K-12 in chiral 3HB production.
As previously described (22), there exist several intrinsic differences in metabolic pathways between E. coli B and K-12 strains, suggesting that the availability of metabolic intermediates as precursors for engineered biosynthetic pathways may also differ between these two strains. In addition, it has been generally concluded that the E. coli B strains are capable of producing greater amounts of proteins than the E. coli K-12 strains, making E. coli B strains better for protein production (31). However, in terms of their role as microbial chemical factories, higher expression levels of recombinant proteins may not necessarily result in higher product titers, especially when substrate availability rather than enzymatic activity is rate limiting in the pathway. The results presented here support the hypothesis that host strain selection can critically influence the activity of recombinant enzymes as well as the productivity of a nonnatural pathway. Although strains constructed in BL21Star(DE3) showed much higher acetoacetyl-CoA thiolase and 3HB-CoA dehydrogenase activities than those constructed in MG1655(DE3), the chiral 3HB titers from recombinant strain BL21Star(DE3) were roughly 20% to 30% lower than those from MG1655(DE3). Such discrepancy between product titers and enzyme activities in those strains suggests that distribution of precursors, i.e., acetyl-CoA, and not enzyme expression levels was the limiting factor for chiral 3HB production. It further implies that metabolic networks play an important role in chiral 3HB synthesis (30). This hypothesis is supported by the observation that significantly more cell mass (data not shown) and acetate were accumulated in BL21Star(DE3), probably due to a large fraction of carbon flux drawn into the TCA cycle and acetate production pathway at the acetyl-CoA node (Fig. 1 and 2). This flux distribution resulted in less acetyl-CoA directed into engineered chiral 3HB pathways in BL21Star(DE3). In contrast, MG1655(DE3), likely with a different distribution within its metabolic network, achieved high-level production of chiral 3HB while accumulating less cell mass and acetate despite its relatively lower enzyme activities. Since excess accumulation of cell mass and by-product formation represent inefficient use of carbon resources that will reduce the yield of desired products, MG1655(DE3) served as the superior production strain by better balancing its growth while maintaining efficient production of chiral 3HB.
Effect of alternative acetoacetyl-CoA thiolases on chiral 3HB production.
It was originally anticipated that the three different acetoacetyl-CoA thiolases (step 1 in Fig. 1) examined in this work would differ in terms of chiral 3HB production by directing different amounts of carbon flux (in the form of acetyl-CoA) into the engineered 3HB pathways. With the exception of Thl for (R)-3HB synthesis in BL21Star(DE3), the choice of thiolase had little effect on chiral 3HB titers, even though the enzymes were found to display different specific activities in vitro in both recombinant strains (Table 3). Thl showed the highest in vitro specific activity in both strains, but recombinant BL21Star(DE3) with either Thl/PhaB/TesB or Thl/PhaB/Ptb-Buk yielded reduced (R)-3HB titers compared to other BL21Star(DE3) counterparts. To explain this contradiction, it should be noted that in vitro thiolase activity was assayed in the thiolytic direction, where two acetyl-CoA molecules were synthesized from one acetoacetyl-CoA molecule and one CoA molecule. Thus, as a result of the combined effect of a 100-fold-lower PhaB activity compared to Hbd and a higher Thl activity compared to BktB and PhaA in BL21Star(DE3), the acetoacetyl-CoA synthesized by Thl could accumulate in the cell. Since the thiolytic reaction is highly exergonic, thiolysis of acetoacetyl-CoA by thiolase is thermodynamically favored (17). The accumulated acetoacetyl-CoA would then be cleaved into two acetyl-CoA molecules in the thiolytic direction, thereby supplying more acetyl-CoA to cell mass and acetate accumulation. In contrast, this did not occur in MG1655(DE3), probably due to the less active competing pathways toward cell mass and acetate accumulation and a negligible thiolytic reaction as a result of lower enzyme activities compared to BL21Star(DE3).
TesB versus Ptb-Buk as a CoA removal system.
It has been suggested that the efficient removal of CoA from (R)-3HB-CoA can lead to enhanced (R)-3HB production (9), which could also be true for (S)-3HB production. To test this concept, two CoA removal systems were assessed. The first is Ptb-Buk, encoded by an operon from C. acetobutylicum, which has been used for direct synthesis of polyhydroxyalkanoate (PHA) together with a PHA synthase utilizing the reverse reaction (i.e., the formation of the CoA thioester) (16). The second is TesB from E. coli, which is reported to possess a broad substrate specificity but unknown physiological function in E. coli (37). While Ptb-Buk uses a two-step CoA-removal scheme through a phosphorylated intermediate, TesB catalyzes one-step CoA-removal by direct hydrolysis (Fig. 1). More chiral 3HB was likely produced in the TesB system than in the Ptb-Buk system due to the essentially irreversible hydrolysis by TesB. In addition, it was noted that pathways incorporating the Ptb-Buk system do not yield (S)-3HB (Table 2), which is consistent with a previous report by Lee et al. (12). The low-level production (<0.10 g liter−1) of (S)-3HB produced by strains containing the Ptb-Buk system may have been due to the endogenous TesB activity in E. coli. In fact, in recombinant strains of E. coli BL21Star(DE3) in which phaA and hbd were solely expressed (i.e., no overexpression of tesB or ptb-buk), similarly low levels of (S)-3HB were also produced (data not shown).
Discrepancies between enzyme activities of Hbd and PhaB and production titers of (S)-3HB and (R)-3HB.
Although Hbd demonstrated much higher in vitro specific activities than PhaB, significantly lower titers of (S)-3HB than (R)-3HB were achieved in all strains expressing Hbd (Table 2 and 3). This contradictory behavior may have been influenced by the following three factors in vivo: (i) the cofactor balance between NADH and NADPH and their respective oxidized counterparts, (ii) the substrate preference of TesB for (R)-3HB-CoA over (S)-3HB-CoA, and (iii) the competing pathway of fatty acid β-oxidation where (S)-3HB-CoA is an intermediate.
Given that Hbd and PhaB are NADH- and NADPH-dependent dehydrogenases, respectively, the physiological levels of NADH and NADPH and their redox ratios in E. coli most likely influence the in vivo catalytic activities of Hbd and PhaB, affecting chiral 3HB titers accordingly. Since NADH is the predominant reducing equivalent found in E. coli under normal conditions (2) as well as the conditions examined here (Table 4), these results indicate that Hbd should theoretically show higher in vivo activity than PhaB, resulting in greater production of (S)-3HB than (R)-3HB. The opposite, however, was observed in this study, suggesting that the physiological NADH/NADPH ratio alone can not resolve the contradiction. It may instead be explained by differences in the physiological ratios of NADH to NAD+ and NADPH to NADP+. We have shown that the ratio of NADPH to NADP+ is substantially higher than that of NADH to NAD+ in E. coli MG1655(DE3) under our culture conditions. These results, together with previous findings that the PHB synthesis is likely affected by the intracellular NADPH/NADP+ ratio (11), suggest that in the case of chiral 3HB synthesis, a higher NADPH/NADP+ ratio may result in more favorable reduction by PhaB while reduction by Hbd may be limited by the lower NADH/NAD+ ratio. Overall, these observations are consistent with correspondingly higher yields of (R)-3HB than (S)-3HB in the present study.
TABLE 4.
Levels and ratios of NAD+, NADH, NADP+, and NADPH cofactors in engineered MG1655(DE3) strainsa
| Construct and postinduction time | Cofactor level (nmol/mg DCW)
|
Cofactor ratio
|
||||
|---|---|---|---|---|---|---|
| NAD+ | NADH | NADP+ | NADPH | NADH/NAD+ | NADPH/NADP+ | |
| pET-H-P + pCDF-T | ||||||
| 4 h | 6.15 ± 0.75 | 0.97 ± 0.11 | 1.58 ± 0.16 | 0.50 ± 0.08 | 0.16 ± 0.00 | 0.31 ± 0.02 |
| 24 h | 3.95 ± 0.29 | 1.75 ± 0.02 | 0.96 ± 0.08 | 0.94 ± 0.09 | 0.44 ± 0.03 | 0.99 ± 0.18 |
| pET-P-P + pCDF-T | ||||||
| 4 h | 7.11 ± 0.08 | 0.59 ± 0.07 | 1.89 ± 0.18 | 0.49 ± 0.03 | 0.08 ± 0.01 | 0.26 ± 0.01 |
| 24 h | 4.94 ± 0.13 | 1.49 ± 0.21 | 1.22 ± 0.05 | 0.97 ± 0.23 | 0.30 ± 0.03 | 0.79 ± 0.16 |
| pETDuet + pCDFDuet | ||||||
| 4 h | 8.05 ± 1.09 | 0.85 ± 0.08 | 2.12 ± 0.04 | 0.59 ± 0.02 | 0.11 ± 0.00 | 0.28 ± 0.02 |
| 24 h | 4.79 ± 0.44 | 0.96 ± 0.06 | 1.12 ± 0.12 | 0.97 ± 0.09 | 0.20 ± 0.01 | 0.87 ± 0.01 |
Data are presented as the average and standard deviation of measurements from two independent cultures.
In a similar manner to the substrate preference of Ptb-Buk for (R)-3HB-CoA, TesB might cleave (R)-3HB-CoA more efficiently than (S)-3HB-CoA. To verify this possibility, it will be informative to perform in vitro enzyme assays using (S)-3HB-CoA and (R)-3HB-CoA as substrates. Unfortunately, the unavailability of these chemicals prevented us from pursuing this experiment.
The third hypothesis was tested using an E. coli mutant with an impaired fatty acid β-oxidation pathway. The E. coli MG1655(DE3) mutant was created by the method of Datsenko and Wanner (7) by insertional inactivation of fadB, encoding 3-hydroxyacyl-CoA dehydrogenase. This fadB mutation should presumably attenuate or block the fatty acid β-oxidation cycle, thereby reducing the degradation of (S)-3HB-CoA. However, this mutant failed to achieve higher titers of (S)-3HB compared to its native counterpart (data not shown). It is possible that other FadB homologs were involved in the degradation of (S)-3HB-CoA. For example, fadJ (previously called yfcX), encodes a subunit of enoyl-CoA hydratase that has been shown to possess the same catalytic function as FadB (19, 29).
Conclusions.
The findings of this study suggest that the distribution of acetyl-CoA is likely the key factor affecting the production of chiral 3HB between E. coli BL21Star(DE3) and MG1655(DE3). Thus, in order to alter the distribution, further research should focus on blocking competing pathways for acetyl-CoA, for example, by deletion of acetate synthesis pathways comprised of acetate kinase and phosphotransacetylase (encoded by ackA-pta) or pyruvate oxidase (encoded by poxB) (14). Also, we cannot entirely rule out the possibility that the overall higher titers of chiral 3HB in recombinant MG1655(DE3) might have been due to their higher TesB specific activities compared to BL21Star(DE3) counterparts in the case where CoA removal is rate limiting. Therefore, in vitro enzyme assays of both TesB and Ptb-Buk should be able to further elucidate the cause of superior titers of chiral 3HB in MG1655(DE3) compared with in BL21Star(DE3). Overall, production of ∼3 g liter−1 (R)-3HB and ∼2 g liter−1 (S)-3HB was achieved in shake flask cultures within 2 days. Further strain engineering should lead to more economical production of chiral 3HB.
Acknowledgments
This work was supported by the Synthetic Biology Engineering Research Center (SynBERC) funded by the National Science Foundation (grant no. 0540879), as well as by a seed grant from the MIT Energy Initiative (grant no. 6917278). D.R.N. is supported by a fellowship from the Natural Sciences and Engineering Research Council (NSERC) of Canada.
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Brumaghim, J. L., Y. Li, E. Henle, and S. Linn. 2003. Effects of hydrogen peroxide upon nicotinamide nucleotide metabolism in Escherichia coli: changes in enzyme levels and nicotinamide nucleotide pools and studies of the oxidation of NAD(P)H by Fe(III). J. Biol. Chem. 278:42495-42504. [DOI] [PubMed] [Google Scholar]
- 3.Buday, Z., J. Linden, and M. Karim. 1990. Improved acetone-butanol fermentation analysis using subambient HPLC column temperature. Enzyme Microb. Technol. 12:24-27. [Google Scholar]
- 4.Chen, G. Q., and Q. Wu. 2005. Microbial production and applications of chiral hydroxyalkanoates. Appl. Microbiol. Biotechnol. 67:592-599. [DOI] [PubMed] [Google Scholar]
- 5.Chiba, T., and T. Nakai. 1987. A new synthetic approach to the carbapenem antibiotic PS-5 from ethyl(S)-3-hydroxybutanoate. Chem. Lett. 11:2187-2188. [Google Scholar]
- 6.Chiba, T., and T. A. Nakai. 1985. Synthetic approach to (1)-thienamycin from methyl (R)-(2)-3-hydroxybutanoate. A new entry to (3R,4R)-3-[(R)-1-hydroxyethyl]-4-acetoxy-2-azetidinone. Chem. Lett. 161:651-654. [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Roo, G., M. B. Kellerhals, Q. Ren, B. Witholt, and B. Kessler. 2002. Production of chiral R-3-hydroxyalkanoic acids and R-3-hydroxyalkanoic acid methylesters via hydrolytic degradation of polyhydroxyalkanoate synthesized by pseudomonads. Biotechnol. Bioeng. 77:717-722. [DOI] [PubMed] [Google Scholar]
- 9.Gao, H. J., Q. Wu, and G. Q. Chen. 2002. Enhanced production of D-(−)-3-hydroxybutyric acid by recombinant Escherichia coli. FEMS Microbiol. Lett. 213:59-65. [DOI] [PubMed] [Google Scholar]
- 10.Inui, M., M. Suda, S. Kimura, K. Yasuda, H. Suzuki, H. Toda, S. Yamamoto, S. Okino, N. Suzuki, and H. Yukawa. 2008. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl. Microbiol. Biotechnol. 77:1305-1316. [DOI] [PubMed] [Google Scholar]
- 11.Lee, I. Y., M. K. Kim, Y. H. Park, and S. Y. Lee. 1996. Regulatory effects of cellular nicotinamide nucleotides and enzyme activities on poly(3-hydroxybutyrate) synthesis in recombinant Escherichia coli. Biotechnol. Bioeng. 52:707-712. [DOI] [PubMed] [Google Scholar]
- 12.Lee, S. H., S. J. Park, S. Y. Lee, and S. H. Hong. 2008. Biosynthesis of enantiopure (S)-3-hydroxybutyric acid in metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 79:633-641. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. Y., and Y. Lee. 2003. Metabolic engineering of Escherichia coli for production of enantiomerically pure (R)-(−)-hydroxycarboxylic acids. Appl. Environ. Microbiol. 69:3421-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, H., G. N. Bennett, and K. Y. San. 2005. Metabolic engineering of aerobic succinate production systems in Escherichia coli to improve process productivity and achieve the maximum theoretical succinate yield. Metab. Eng. 7:116-127. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Q., S. P. Ouyang, A. Chung, Q. Wu, and G. Q. Chen. 2007. Microbial production of R-3-hydroxybutyric acid by recombinant E. coli harboring genes of phbA, phbB, and tesB. Appl. Microbiol. Biotechnol. 76:811-818. [DOI] [PubMed] [Google Scholar]
- 16.Liu, S. J., and A. Steinbuchel. 2000. Exploitation of butyrate kinase and phosphotransbutyrylase from Clostridium acetobutylicum for the in vitro biosynthesis of poly(hydroxyalkanoic acid). Appl. Microbiol. Biotechnol. 53:545-552. [DOI] [PubMed] [Google Scholar]
- 17.Masamune, S., C. T. Walsh, A. J. Sinskey, and O. P. Peoples. 1989. Poly-(R)-3-hydroxybutyrate (PHB) biosynthesis: mechanistic studies on the biological Claisen condensation catalyzed by β-ketoacyl thiolase. Pure Appl. Chem. 61:303-312. [Google Scholar]
- 18.Mori, K. 1981. A simple synthesis of (S)-(+)-sulcatol, the pheromone of Gnathotrichus tetusus employing baker's yeast for asymmetric reduction. Tetrahedron 37:1341-1342. [Google Scholar]
- 19.Park, S. J., and S. Y. Lee. 2004. New fadB homologous enzymes and their use in enhanced biosynthesis of medium-chain-length polyhydroxyalkanoates in fadB mutant Escherichia coli. Biotechnol. Bioeng. 86:681-686. [DOI] [PubMed] [Google Scholar]
- 20.Patel, R. N. 2006. Biocatalysis: synthesis of chiral intermediates for drugs. Curr. Opin. Drug Discov. Dev. 9:741-764. [PubMed] [Google Scholar]
- 21.Patel, R. N. 2000. Stereoselective biocatalysis, CRC Press, Boca Raton, FL.
- 22.Phue, J. N., S. B. Noronha, R. Hattacharyya, A. J. Wolfe, and J. Shiloach. 2005. Glucose metabolism at high density growth of E. coli B and E. coli K: differences in metabolic pathways are responsible for efficient glucose utilization in E. coli B as determined by microarrays and Northern blot analyses. Biotechnol. Bioeng. 90:805-820. [DOI] [PubMed] [Google Scholar]
- 23.Pollard, D. J., and J. M. Woodley. 2007. Biocatalysis for pharmaceutical intermediates: the future is now. Trends Biotechnol. 25:66-73. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Schneider, D., E. Duperchy, J. Depeyrot, E. Coursange, R. Lenski, and M. Blot. 2002. Genomic comparisons among Escherichia coli strains B, K-12, and O157:H7 using IS elements as molecular markers. BMC Microbiol. 2:18-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schubert, P., A. Steinbuchel, and H. G. Schlegel. 1988. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-β-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J. Bacteriol. 170:5837-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seebach, D., H. F. Chow, R. F. W. Jackson, M. A. Sutter, S. Thaisrivongs, and J. Zimmermann. 1986. (+)-11,11′-Di-O-methylelaiophylidene-preparation from Elaiophylin and total synthesis from (R)-3-hydroxybutyrate and (S)-malate. Liebigs Ann. Chem. 1986:1281-1308. [Google Scholar]
- 28.Shiraki, M., T. Endo, and T. Saito. 2006. Fermentative production of (R)-(−)-3-hydroxybutyrate using 3-hydroxybutyrate dehydrogenase null mutant of Ralstonia eutropha and recombinant Escherichia coli. J. Biosci. Bioeng. 102:529-534. [DOI] [PubMed] [Google Scholar]
- 29.Snell, K. D., F. Feng, L. Zhong, D. Martin, and L. L. Madison. 2002. YfcX enables medium-chain-length poly(3-hydroxyalkanoate) formation from fatty acids in recombinant Escherichia coli fadB strains. J. Bacteriol. 184:5696-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stelling, J., S. Klamt, K. Bettenbrock, S. Schuster, and E. D. Gilles. 2002. Metabolic network structure determines key aspects of functionality and regulation. Nature 420:190-193. [DOI] [PubMed] [Google Scholar]
- 31.Terpe, K. 2006. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 72:211-222. [DOI] [PubMed] [Google Scholar]
- 32.Tokiwa, Y., and B. P. Calabia. 2008. Biological production of functional chemicals from renewable resources. Can. J. Chem. 86:548-555. [Google Scholar]
- 33.Tolia, N. H., and L. Joshua-Tor. 2006. Strategies for protein coexpression in Escherichia coli. Nat. Methods 3:55-64. [DOI] [PubMed] [Google Scholar]
- 34.van de Walle, M., and J. Shiloach. 1998. Proposed mechanism of acetate accumulation in two recombinant Escherichia coli strains during high density fermentation. Biotechnol. Bioeng. 57:71-78. [DOI] [PubMed] [Google Scholar]
- 35.Xia, X. X., M. J. Han, S. Y. Lee, and J. S. Yoo. 2008. Comparison of the extracellular proteomes of Escherichia coli B and K-12 strains during high cell density cultivation. Proteomics 8:2089-2103. [DOI] [PubMed] [Google Scholar]
- 36.Zhao, K., G. Tian, Z. Zheng, J. C. Chen, and G. Q. Chen. 2003. Production of D-(−)-3-hydroxyalkanoic acid by recombinant Escherichia coli. FEMS Microbiol. Lett. 218:59-64. [DOI] [PubMed] [Google Scholar]
- 37.Zheng, Z., Q. Gong, T. Liu, Y. Deng, J.-C. Chen, and G.-Q. Chen. 2004. Thioesterase II of Escherichia coli plays an important role in 3-hydroxydecanoic acid production. Appl. Environ. Microbiol. 70:3807-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]