Abstract
In silico genome analysis of Lactobacillus acidophilus NCFM coupled with gene expression studies have identified putative genes and regulatory networks that are potentially important to this organism's survival, persistence, and activities in the gastrointestinal tract. Correlation of key genotypes to phenotypes requires an efficient gene replacement system. In this study, use of the upp-encoded uracil phosphoribosyltransferase (UPRTase) of L. acidophilus NCFM was explored as a counterselection marker to positively select for recombinants that have resolved from chromosomal integration of pORI-based plasmids. An isogenic mutant carrying a upp gene deletion was constructed and was resistant to 5-fluorouracil (5-FU), a toxic uracil analog that is also a substrate for UPRTase. A 3.0-kb pORI-based counterselectable integration vector bearing a upp expression cassette, pTRK935, was constructed and introduced into the Δupp host harboring the pTRK669 helper plasmid. Extrachromosomal replication of pTRK935 complemented the mutated chromosomal upp allele and restored sensitivity to 5-FU. This host background provides a platform for a two-step plasmid integration and excision strategy that can select for plasmid-free recombinants with either the wild-type or mutated allele of the targeted gene in the presence of 5-FU. The efficacy of the system was demonstrated by in-frame deletion of the slpX gene (LBA0512) encoding a novel 51-kDa secreted protein associated with the S-layer complex of L. acidophilus. The resulting ΔslpX mutant exhibited lower growth rates, increased sensitivity to sodium dodecyl sulfate, and greater resistance to bile. Overall, this improved gene replacement system represents a valuable tool for investigating the mechanisms underlying the probiotic functionality of L. acidophilus.
Lactobacillus acidophilus NCFM is a commercially established probiotic bacterium that is widely used in dietary supplements and in milk and fermented dairy products (47). Originally a human intestinal isolate from the 1970s (8), this strain has since been examined extensively for various desirable traits. Due to the importance of understanding the molecular mechanisms involved in probiotic functions, the complete genome sequence of L. acidophilus NCFM was determined (2). The NCFM genome sequence serves as a blueprint for in silico identification of candidate gene loci and gene regulatory networks that may play essential roles in the survival and host interactions of this microorganism in the gastrointestinal (GI) tract, including genes involved in acid tolerance (5), bile tolerance (38, 42), adherence factors (20), environmental sensing and response (7), prebiotic sugar utilization (9), polysaccharide biosynthesis, oxalate degradation (6), and bacteriocin production (22). In addition, ongoing microarray gene expression studies have revealed specific gene sets of interest that are being investigated further. The fundamental approach for establishing functional roles for important gene features involves inactivation of the genes and subsequent phenotypic analyses of the associated isogenic mutants. Hence, an efficient gene knockout system is a valuable genetic tool for correlating key genotypes to phenotypes and for functional characterization of genes associated with probiotic attributes of L. acidophilus.
The development of a site-directed lactococcal chromosomal integration system by Law and coworkers (34) involving the simultaneous use of a broad-host-range nonreplicative pWV01-derived vector (Ori+ RepA−) or so-called pORI-based vector and a temperature-sensitive helper plasmid, pVE6007 (35), that provides repA in trans for conditional replication of the pORI-based plasmids has greatly facilitated genetic studies of various gram-positive bacteria. This gene knockout strategy was adapted for use in L. acidophilus and L. gasseri with an alternate helper plasmid, pTRK669, that provides a higher permissive temperature range for thermophilic lactobacilli (46). This system has since been used successfully for generating numerous chromosomal insertion derivatives (1, 5, 9, 20, 23, 26, 38, 53). Nevertheless, the stability of the insertional mutations after single-crossover homologous recombination requires maintenance of antibiotic selection, and the same selection marker cannot be used to introduce multiple mutations into a strain. Furthermore, insertional inactivation of a specific target within an operon may have polar effects on downstream regions. These limitations were overcome by construction of markerless gene deletions via a double-crossover homologous recombination process involving plasmid integration and excision, where the wild-type allele is replaced with the mutant allele carrying an internal deletion in the target gene (6, 42). However, due to low efficiency of plasmid excision and allelic replacement and the lack of a selectable marker to detect these events, extensive screening is often necessary to isolate the desired recombinants.
One practical approach to address this issue is to incorporate counterselectable genetic markers in allelic replacement systems to facilitate the recovery of plasmid-free derivatives following the second recombination event. Among the established counterselectable markers are the Bacillus subtilis sacB gene (encoding a levansucrase), which results in sucrose sensitivity (44), and the galK gene (encoding galactokinase), which mediates galactose or galactose analog toxicity (39, 49) in merodiploids that have not undergone the second homologous crossover event. Genes involved in purine and pyrimidine salvage pathways, specifically the genes coding for phosphoribosyltransferases (PRTases), such as upp (encoding uracil PRTase [UPRTase]), hprT (encoding hypoxanthine PRTase), pyrE/ura5 (encoding orotate PRTase), and pryF/ura3 (encoding orotidine-5′-phosphate decarboxylase), are also counterselectable markers that are widely used in bacterial, archaeal, and eukaryotic gene knockout systems (14, 15, 24, 25, 33, 41, 43, 54). These PRTases convert preformed purine and pyrimidine bases into corresponding nucleotide monophosphates for de novo nucleotide biosynthesis. The presence of a base analog that is also recognized as a substrate by the targeted PRTase can be lethal to cells when the base is incorporated into the nucleotide precursors, whereas a mutant with a nonfunctional PRTase is affected less by the toxicity of the base analog. The counterselection approach involved the use of an integration vector that provides ectopic expression of the PRTase-encoding gene in a PRTase-defective background host for gene-targeting deletions. Single-crossover integration of the recombinant plasmid is initially selected, and the recombination event renders the host sensitive to the PRTase-specific base analog due to expression of the plasmid-borne PRTase gene. Excision of the integrated plasmid following a second recombination event restores the base analog resistance phenotype of the host, which then serves as a counterselection strategy for rapid identification of plasmid-free recombinants that bear either the wild-type allele or the mutated allele of the targeted gene. The upp-based counterselection approach has previously been used for allelic replacement in B. subtilis and Enterococcus faecalis (24, 33).
In the present study, we report the development of an improved markerless gene replacement system for L. acidophilus NCFM that involves the use of upp as a counterselectable marker for the existing pORI-based knockout system for positive selection of double recombinants. We demonstrated the efficiency of the upp counterselection scheme by deleting the slpX gene (LBA0512) encoding a novel secreted protein that was found to be associated with the S-layer complex in L. acidophilus. This study demonstrated the first functional counterselective gene replacement system for the genus Lactobacillus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. L. acidophilus strains were propagated in MRS broth (Difco Laboratories, Inc., Detroit, MI) statically under ambient atmospheric conditions or on MRS solid medium supplemented with 1.5% (wt/vol) agar (Difco) under anaerobic conditions at 37°C or 42°C, as indicated below. Recombinant strains were selected in the presence of 2 μg/ml of erythromycin (Em) (Sigma-Aldrich, St. Louis, MO) and/or 5 μg/ml of chloramphenicol (Cm) (Sigma) when appropriate. For counterselection of plasmid-free double recombinants, 5-fluorouracil (5-FU) (Sigma), a uracil analog, was added to presterilized semidefined agar medium (SDM) (30) at a final concentration of 100 μg/ml from a 60-mg/ml stock solution prepared with dimethyl sulfoxide (DMSO) (Sigma) as described by Kristich et al. (33). For growth experiments, overnight cultures were inoculated (2% inoculum) into fresh MRS medium (optical density at 600 nm [OD600], ∼0.05), and growth was monitored at OD600. Cells were also plated onto MRS agar at various time points using a Whitley automatic spiral plater (Don Whitley Scientific Ltd., West Yorkshire, England). CFU were enumerated using a ProtoCOL colony counter (Synoptics Ltd., Cambridge, United Kingdom).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristics | Reference or source |
|---|---|---|
| L. acidophilus strains | ||
| NCFM | Human intestinal isolate | 8 |
| NCK1392 | NCFM harboring pTRK669 | 46 |
| NCK1909 | NCFM carrying a 315-bp in-frame deletion in the upp gene | This study |
| NCK1910 | NCK1909 harboring pTRK669; host for pORI-based counterselective integration vector | This study |
| NCK1962 | NCK1909 carrying a 1,356-bp in-frame deletion in the slpX gene | This study |
| E. coli strains | ||
| EC101 | RepA+ JM101; Kmr; repA from pWV01 integrated in chromosome; host for pORI-based plasmids | 34 |
| NCK1391 | E. coli DH10B harboring pTRK669; host for pORI-based plasmids | W. M. Russell and T. R. Klaenhammer, unpublished data |
| Plasmids | ||
| pORI28 | Emr Ori+ (pWV01), replicates only with repA provided in trans | 34 |
| pORI19 | lacZ′ derivative of pORI28 | 34 |
| pTRK669 | Ori (pWV01), Cmr RepA+ | 46 |
| pTRK934 | 3.4 kb; pORI19 with a mutated copy of upp cloned into XbaI/SacI sites | This study |
| pTRK935 | 3.0 kb; pORI28 with a upp expression cassette and lacZ′ from pUC19 cloned into BglII/XbaI sites; serves as counterselective integration vector | This study |
| pTRK956 | 4.5 kb; pTRK935 with a mutated copy of slpX cloned into BamHI/SacI sites | This study |
Escherichia coli strains were grown in brain heart infusion (BHI) (Difco) medium at 37°C with aeration. E. coli EC101 was propagated in the presence of 40 μg/ml of kanamycin. When necessary, Em and Cm were added at final concentrations of 200 μg/ml and 15 μg/ml, respectively.
DNA isolation and manipulations.
Genomic DNA of L. acidophilus NCFM was isolated by the method described by Walker and Klaenhammer (52) or by using a Mo Bio UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA). Plasmid DNA from E. coli was isolated using a QIAprep Spin miniprep kit (Qiagen Inc., Valencia, CA). Restriction enzymes (Roche Molecular Biochemicals, Indianapolis, IN) were used according to the instructions supplied by the manufacturer. DNA ligation was performed using T4 DNA ligase (New England Biolabs, Beverly, MA) or a Fast-Link DNA ligation kit (Epicentre Biotechnologies, Madison, WI) based on the manufacturers' recommendations. PCR primers (see Table S1 in the supplemental material) were synthesized by Integrated DNA Technologies (Coralville, IA). For cloning purposes, PCR amplicons were generated using PfuUltra II Fusion HS DNA polymerase (Stratagene Corp., La Jolla, CA) according to the supplier's instructions. Routine PCR amplifications for screening of recombinants were performed by using standard protocols and Choice-Taq Blue DNA polymerase (Denville Scientific Inc., Metuchen, NJ). PCR products were analyzed on 0.8% agarose gels and purified using a Zymoclean gel DNA recovery kit (Zymo Research Corp., Orange, CA) or a QIAquick gel extraction kit (Qiagen). DNA sequencing was performed by Davis Sequencing, Inc. (Davis, CA).
E. coli chemically competent cells were prepared and transformed based on procedures previously described by Hanahan (27). L. acidophilus cells were prepared for electrotransformation essentially as described by Walker et al. (51), with the following modifications adapted from the procedure of Wei et al. (55). Briefly, stationary-phase cells were inoculated into MRS broth (2% inoculum) and grown for 3 h (OD600, ∼0.1 to 0.2). A filter-sterilized penicillin G stock solution was then added to obtain a final concentration of 10 μg/ml, and the culture was incubated for 1.5 to 2.0 h before it was harvested.
Sequence analysis.
Deduced protein sequences were compared with the nonredundant protein database using BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Theoretical physical and chemical properties of the deduced proteins were assessed using the ProtParam tool (http://au.expasy.org/tools/protparam.html). Signal peptides were predicted using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (13).
Construction of upp deletion mutant.
To construct a Δupp isogenic mutant with an internal 315-bp deletion in the upp gene (LBA0770), first, an upstream chromosomal DNA segment (596 bp) and a downstream chromosomal DNA segment (610 bp) flanking the deletion region were PCR amplified with the upp1U-F/upp2U-R and upp3D-F/upp4D-R primer pairs (see Table S1 in the supplemental material), respectively. The two PCR fragments were gel purified and joined by using the splicing by overlap extension PCR (SOE-PCR) technique (28), where equimolar concentrations of each PCR product (in 10 to 15 ng) were combined and used as amplification templates in a 50-μl PCR mixture with primers upp1U-F and upp4D-R. All PCRs were performed with 25 to 30 amplification cycles to minimize accumulation of nucleotide misincorporation errors in the PCR products. The resulting SOE-PCR products were gel purified, restricted with XbaI and SacI, ligated into similarly digested vector pORI19, and transformed into E. coli DH10B carrying the pTRK669 helper plasmid (NCK1391) with selection on BHI medium containing both Em and Cm. The resulting recombinant plasmid, pTRK934, was electroporated into L. acidophilus NCK1392. The procedures used for isolation of plasmid-free recombinants following chromosomal integration of pTRK934 and double-crossover recombination were performed essentially as described previously (19, 46). In-frame deletions in the Δupp mutants were confirmed by DNA sequencing of PCR products generated with the upp-up/upp-dw primer pair (see Table S1 in the supplemental material) specific for the region flanking the deletion target.
PM analysis.
Phenotypic microarray (PM) experiments were performed by Biolog Inc. (Hayward, CA). PM technology is based on a chemical reaction between the bacteria and a tetrazolium dye. If bacterial growth is supported by the medium in a well, the metabolizing cells reduce tetrazolium and produce a color, which can be measured with the OmniLog instrument over a 24-h period. Both the inoculating medium and the concentrations of substrates are proprietary information (www.biolog.com). Twelve 96-well PMs (PM09 to PM20) were used in this study. These arrays were used for the following analyses: PM09, osmolytes; PM10, pH; and PM11 to PM20, chemical sensitivity (see Table S2 and S3 in the supplemental material). Overnight cultures of L. acidophilus strains were resuspended in MRS broth according to Biolog procedures (www.biolog.com) and added to each well of the 12 96-well plates. Little or no color is formed in negative reactions, whereas color is formed in positive reactions. The OmniLog instrument captured digital images of the PMs, the files were subsequently displayed as kinetic graphs for respiration and growth, and the software was used to calculate the area under the curve for each strain.
Microarray gene expression study. (i) Generation of L. acidophilus NCFM DNA oligoarray.
OligoArray 2.1 (45) was used to design oligonucleotides for construction of an NCFM DNA oligonucleotide microarray (oligoarray). The range of oligonucleotide lengths used was 60 to 70 nucleotides, the melting temperature range used was 80 to 96°C, and the G+C content range used was 25 to 50% based on the G+C content of the NCFM genome (34.71%). The thresholds for rejecting oligonucleotides that can form stable secondary structures and possible cross-hybridization were both 65°C. Oligonucleotides containing AAAAA, TTTTT, GGGGG, CCCCC, or longer homopolymers were rejected. Oligonucleotides were synthesized by Integrated DNA Technologies and spotted on glass slides. Each oligonucleotide (representing each gene) was spotted six times in a random pattern.
(ii) Experimental conditions and RNA isolation.
The NCFM wild-type strain and the Δupp isogenic mutant were grown in fresh MRS medium using 2% inocula from overnight cultures. Aliquots of cells were collected at OD600 of ∼0.3 (early log phase) and 0.8 (mid-log phase) by quick centrifugation at room temperature. Cell pellets were flash-frozen in an ethanol-dry ice bath and stored at −80°C. Samples were collected from two independent experimental replicates (representing two biological replicates). Isolation of total RNA, DNase treatment, and RNA purification were performed essentially as described previously (26). The absence of genomic DNA in purified RNA samples was confirmed by performing PCR with NCFM gene-specific primers.
(iii) Synthesis of cDNA probe and oligoarray hybridization.
Reverse transcription and fluorescent labeling of identical amounts (20 μg) of purified RNA were performed by using the SuperScript indirect cDNA labeling system for DNA microarrays (Invitrogen, Carlsbad, CA) according to the manufacturer's directions, except that 6 μg of random primers (Invitrogen) was used for each 30 μl of first-strand cDNA synthesis reaction mixture. Labeled cDNA samples representing the NCFM and Δupp mutant strains were coupled with monoreactive Cy3 and Cy5 dyes (GE Healthcare Bio-Sciences Corp., Piscataway, NJ), respectively. Comparative hybridizations were performed with samples representing both NCFM and the Δupp derivative at the same growth phases. Prior to hybridization, oligoarray slides were prehybridized in a buffer containing 0.1% (wt/vol) sodium dodecyl sulfate (SDS) (Invitrogen), 0.1% (wt/vol) bovine serum albumin (Invitrogen), and 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; Invitrogen) at 42°C for 1 h and then rinsed by submerging them five times in sterile distilled water and twice in isopropanol and spin drying. Pairwise Cy3- and Cy5-labeled probes were dried with a Vacufuge speed vacuum system (Epperdorf, Westbury, NY) and resuspended in 48 to 50 μl (total volume) of hybridization buffer containing 5× SSC, 5× Denhardt's solution (Invitrogen), 30% (vol/vol) formamide, 0.1% SDS, and 200 μg/ml of salmon sperm DNA (Invitrogen). The resulting probe mixtures were incubated at 95°C for 2 min and hybridized to the oligoarrays for 18 to 20 h at 42°C. Posthybridization arrays were washed first in prewarmed (42°C) washing buffer containing 1× SSC and 0.03% SDS and then in washing buffer containing 0.5× SSC and 0.03% SDS at room temperature. Subsequently, the arrays were washed in 0.2× SSC, which was followed by a final wash in 0.05× SSC. Each washing step was 5 min long.
(iv) Data acquisition and analysis.
Fluorescence intensities were acquired at a resolution of 10 μm per pixel by using a ScanArray 4000 microarray scanner (Packard Biochip BioScience; Biochip Technologies LLC) and stored as TIFF image files. Signal intensities were quantified, the background value was subtracted, and data were normalized by using the QuantArray 3.0 software package (Packard Bioscience). Spots were analyzed by adaptive quantitation. The data were median normalized (7). The average Cy5/Cy3 ratio for 12 spots for each gene (6 replicate spots per slide per biological replicate) was determined and represented the change in gene expression levels in the Δupp mutant (Cy5) compared to the NCFM strain (Cy3). Confidence intervals and P values for the change were also calculated by using a two-sample t test. P values of ≤0.05 were considered significant.
Construction of a counterselectable integration vector.
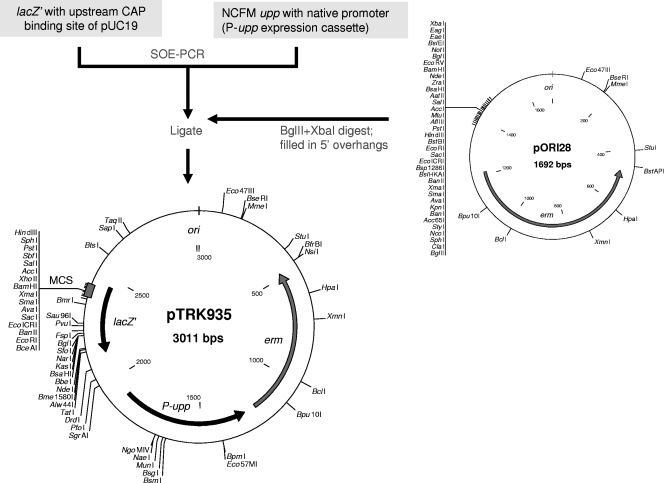
For construction of a pORI-based counterselectable integration vector, the multiple cloning site (MCS) of the pORI28 vector was replaced with a upp expression cassette (P-upp) and a lacZ′ (lacZ-alpha) gene; the lacZ′ gene was used for blue-white screening of E. coli transformants via alpha complementation of the ΔlacZ′ E. coli cloning hosts (Fig. 1). First, plasmid pORI28 DNA was digested with BglII and XbaI to remove the MCS region. The 5′ overhangs of the restricted pORI28 plasmid were subsequently filled in using PfuUltra II Fusion HS DNA polymerase, based on a protocol of the Stratagene PCR polishing kit (Stratagene). Next, the lacZ′ gene with its promoter and catabolite activator protein (CAP) binding site was amplified from the pUC19 vector with the pUC19lacZ-F/pUC19lacZ-R primer pair (see Table S1 in the supplemental material) using 10 ng of pUC19 vector DNA as the amplification template in a 50-μl reaction mixture. The L. acidophilus NCFM upp gene with its putative promoter region was PCR amplified with the upp-F/upp-R primer pair (see Table S1 in the supplemental material). Both purified PCR amplicons were joined by SOE-PCR as described above using the pUC19lacZ-F/upp-R primer pair. The resulting lacZ′-P-upp fusion products were ligated into the blunt-ended pORI28 backbone and transformed into E. coli NCK1391, and transformants were selected on BHI medium plates containing Em and Cm in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and isopropyl-β-d-thiogalactopyranoside (IPTG). Positive clones appeared as blue colonies due to expression of the lacZ′ gene in the recombinant plasmid. The resulting counterselectable integration vector, pTRK935, was subjected to DNA sequencing to confirm the integrity of the P-upp and the MCS in the lacZ′ gene.
FIG. 1.
Construction of the pTRK935 counterselectable integration vector. ori, origin of replication of pWV01; erm, gene encoding Em resistance; lacZ′, lacZ-alpha gene from pUC19; MCS, MCS from pUC19. Only unique restriction sites are shown.
Construction of ΔslpX mutant using the counterselectable gene replacement system.
A 1,356-bp in-frame deletion in the slpX gene (LBA0512) was constructed by first amplifying 767-bp and 747-bp DNA segments flanking the upstream and downstream regions of the deletion target, respectively, using the slpX1U-F/slpX2U-R and slpX3D-F/slpX4D-R primer pairs (see Table S1 in the supplemental material). The two purified PCR products were fused and amplified to generate copies of the ΔslpX allele by the SOE-PCR method as described above, digested with BamHI and SacI, and ligated into the pTRK935 counterselectable integration vector with compatible ends. The ligation mixture was transformed into E. coli EC101 with selection on BHI medium containing kanamycin, Em, X-Gal, and IPTG. The resulting plasmid carrying the ΔslpX allele, pTRK936, was electroporated into the L. acidophilus Δupp host strain harboring pTRK669, NCK1910 (Table 1). One Emr Cmr transformant carrying both plasmids was grown overnight in MRS medium containing 2 μg/ml Em and 2 μg/ml Cm and transferred three times (1% inoculum) in MRS medium with Em (ca. 30 generations) in a 42°C water bath. Chromosomal integrants were selected by replica plating samples onto MRS agar supplemented with Em or Cm. One Emr Cms integrant was selected and propagated in MRS medium with Em at 37°C overnight, which was followed by three to five transfers (1% inoculum) in MRS medium without Em (ca. 30 to 50 generations). Cultures were serially diluted, and 10−3 to 10−4 dilutions were plated onto SDM plates supplemented with 5-FU and incubated at 37°C anaerobically for 48 to 72 h until colonies were visible. Eight 5-FUr colonies were selected for PCR analysis of the slpX region using primers slpX-up and slpX-dw (see Table S1 in the supplemental material) that specifically anneal to the flanking region of slpX. In-frame deletions were confirmed by DNA sequencing as described above.
Extraction and purification of the S-layer proteins.
Cultures were propagated overnight in 100 ml of MRS medium, and the cells were harvested by centrifugation at 8,000 × g for 20 min at 4°C. The dry weights of the cell pellets from 100-ml cultures were between 600 and 700 mg, which correspond to wet weights of 5.4 to 6.3 g. After one wash with cold distilled water, the pellets were suspended in 5 M LiCl at a ratio of 1 ml of LiCl to 10 mg of biomass and incubated for 15 min at 4°C with slight stirring. Following centrifugation, the supernatant (∼60 ml) was removed and dialyzed for 24 h against distilled water at 4°C using a Spectrapor 6-8000 membrane. The white precipitate that formed during dialysis was harvested by centrifugation at 40,000 × g for 20 min at 4°C. To remove proteins that coprecipitated with the S-layer protein, the pellet was resuspended in 1 M LiCl and stirred for 15 min at 4°C. Following centrifugation at 40,000 × g for 20 min at 4°C, the pellet was washed with 15 ml of cold distilled water. Each pellet was then separated into two parts, one of which was used for direct mass spectrometry (MS) analysis and one of which was used for trypsinolysis and matrix-assisted laser desorption ionization analysis.
MS analysis of pure S-layer proteins before and after trypsinolysis.
For direct MS analysis, pure S-layer proteins were resuspended in a solution containing 50% (vol/vol) acetonitrile and 4% (vol/vol) formic acid in distilled water (total volume, 500 μl) overnight at 4°C. Then 200 μl of the same acetonitrile-formic acid solution was added, and the mixture was vigorously stirred. Complete resolubilization of the S-layer protein pellet was never observed; therefore, the suspension was centrifuged (12,000 × g, 10 min, 4°C), and the supernatant was removed and used for MS analysis.
For peptide fingerprinting, pure S-layer proteins (stored at −20°C) were subjected to trypsinolysis. S-layer trypsin digestion was performed overnight at 37°C and was stopped with trifluoroacetic acid (Sigma-Aldrich, St. Quentin-Fallavier, France). The supernatants containing peptides were then dried using a Speed vacuum system and stored at −20°C until an MS analysis was performed.
MS.
All mass spectra were obtained using a hybrid quadrupole time of flight mass spectrometer (QStar XL; MDS Sciex, Toronto, Canada). For the matrix-assisted laser desorption ionization experiments, the samples were ionized with a laser beam (λ = 337 nm, 20 Hz), and the peptide β-casein fragment from residue 193 to residue 209 was used as a calibration standard. The more representative monocharged ions were subjected to fragmentation with an energy of collision between 50 and 90 keV.
For electrospray ionization experiments, a liquid Nano liquid chromatography system (LC-packings; Dionex Voisins, Le Bretonneux, France) was coupled online with an QStar XL mass spectrometer. The mass spectrometer was operated in positive ion mode. A voltage of around 5 kV was applied to the micro-ion source. MS and tandem MS (MS/MS) data were acquired in continuum mode. The instrument was calibrated by multipoint calibration using fragment ions that resulted from the collision-induced decomposition of the peptide β-casein fragment from residue 193 to residue 209. Data-directed analysis was employed to perform MS/MS analysis with doubly and triply charged precursor ions. MS/MS spectra were collected from m/z 50 to m/z 2000. The mass spectrometer was operated in data-dependent mode automatically, with switching between MS and MS/MS acquisition using the Analyst QS 1.1 software. All data (MS and MS/MS) were then analyzed using MASCOT (v.2.1) for searches in several databases (MSDB and an internet database named “UMR”) to identify the proteins present in the samples.
Stress challenge assays.
Strains were grown in MRS broth to an OD600 of 0.25 to 0.3 (early log phase) before they were subjected to the stress challenges used (7). The cultures were divided into equal-volume portions and harvested at room temperature. Cells were resuspended in equal volumes of (i) MRS broth with 0.3% (wt/vol) porcine bile (Sigma), (ii) MRS broth with 2.5% (wt/vol) oxgall (Difco), (iii) MRS broth with 10% (wt/vol) NaCl, (iv) MRS broth with 15% (vol/vol) ethanol, and (v) acidified MRS broth at pH 3.5 (adjusted with lactic acid) and pH 3.0 (adjusted with HCl). Viable cell counts were determined after 2 h of incubation at 37°C by plating on MRS medium. For SDS and Triton X-100 treatment assays, early-log-phase cultures were inoculated (1%, vol/vol) into MRS broth containing 0.01% or 0.02% (wt/vol) SDS or 0.25% or 0.50% (vol/vol) Triton X-100. Growth was assessed by determining the OD600 after 24 h of incubation at 37°C. For the freeze-thaw challenge, 1-ml portions of early-log-phase cultures were centrifuged and resuspended in an equal volume of 0.1× MRS broth. Samples were subjected to repeated freezing (ethanol-dry ice bath, 3 min) and thawing (37°C water bath, 3 min) cycles. Survivors were assessed by plating on MRS agar after 5, 10, 15, and 20 freeze-thaw cycles.
Caco-2 epithelial cell adherence assay.
For Caco-2 cell adherence assays, the Caco-2 cell line (ATCC HTB-37) was obtained from the American Type Culture Collection (Rockville, MD). All cell culture media and reagents were purchased from Gibco (Gibco-Invitrogen Corp., Carlsbad, CA). Caco-2 cells were prepared and maintained for adhesion studies as previously described by Buck et al. (20), with the following modifications. Briefly, cells were routinely grown at 37°C in a 95% air-5% CO2 atmosphere in minimal essential medium supplemented with 1 mM sodium pyruvate, 20% (vol/vol) heat-inactivated (56°C, 30 min) fetal bovine serum, 0.1 mM nonessential amino acids, and antibiotics (100 mg/ml penicillin G, 100 mg/ml streptomycin sulfate, and 0.25 mg/ml amphotericin). For the adhesion assay, monolayers were prepared in 12-well tissue culture plates by seeding approximately 6.5 × 104 cells per well with 2 ml of cell culture medium. The culture medium was replaced every 2 days, and the monolayers were used for adherence assays at 14 to 15 days postconfluence. Cells were used between passages 23 and 40. Prior to adherence assays, monolayers were washed twice with 1 ml of phosphate-buffered saline (PBS) (pH 7.4; Invitrogen), and 1 ml of fresh culture medium without antibiotics was subsequently added to each well.
For preparation of bacterial cells, 10 ml of mid-log-phase cells (OD600, 0.6 to 0.8) grown in MRS broth was pelleted by centrifugation at 3,166 × g for 10 min at room temperature, washed once with 10 ml of PBS, and resuspended in PBS to a final concentration of ∼4 × 108 CFU/ml. One milliliter of the bacterial suspension was added to each well containing a Caco-2 monolayer and incubated at 37°C for 1 h. After this incubation, the monolayers were washed five times with 1 ml PBS to remove unbound bacteria and then incubated with 1 ml of 0.05% (vol/vol) Triton X-100 for 10 min. The monolayer cell-bacterium suspensions were serially diluted in 0.1× MRS broth and plated on MRS agar to determine the number of adherent cells (expressed in CFU). All experiments were performed with at least three independent cultures, each with triplicate wells containing monolayers.
Mucin adherence assay.
For immobilization of porcine stomach mucin (type III; Sigma) on a Costar 96-well round-bottom polystyrene plate (Corning Inc., New York), 10 mg/ml of a mucin solution in distilled water was prepared, dispensed into each well (100 μl per well), and incubated at 4°C overnight. Excess mucin was removed by two washes with 200 μl PBS prior to addition of bacterial cells. Mid-log-phase cultures grown in MRS broth were washed once with PBS, and the final OD600 was adjusted to 0.6 with PBS. A cell suspension (100 μl) was added to each well and incubated at 37°C for 1 h. Subsequently, each well was washed five times with 200 μl PBS to remove unbound cells, which was followed by treatment with 200 μl of 0.05% Triton X-100 to detach the mucin layer and bacterial cells. A cell suspension was diluted in 0.1× MRS broth and plated onto MRS agar to determine the number of adherent cells (expressed in CFU). All experiments were performed with two independent cultures, each with triplicate wells containing a mucin layer.
Assay for microbial adhesion to solvents (MATS).
The assay for microbial adhesion to solvents (MATS) was conducted based on the protocol described by Bellon-Fontaine et al. (12), with the following two pairs of solvents: (i) chloroform (Lewis acid, electron acceptor, polar) and hexadecane (apolar), and (ii) ethyl acetate (Lewis base, electron donor, polar) and decane (nonpolar). Briefly, 5-ml portions of mid-log-phase cultures grown in MRS broth were harvested by centrifugation at 3,166 × g at room temperature, washed once in an equal volume of PBS, and resuspended in PBS to a final OD400 of 0.8. Then 1.2 ml of each cell suspension was mixed with 0.2 ml of each solvent by vortexing for 60 s. The mixtures were incubated at room temperature for 15 min to allow complete separation of the two phases. Then 600 μl of the aqueous phase was carefully removed for determination of the OD400. The level of affinity to solvents of the cells was calculated with the following equation: % affinity = 100 × [1 − (A/A0)], where A0 and A are the OD400 of the cell suspension before and after mixing with the solvents, respectively. All assays were done in duplicate with two independent cultures.
Scanning electron microscopy (SEM).
Cells were grown in MRS medium (14 ml) to an OD600 of 0.6 to 0.8 (at which the culture supernatant pH was ∼5.1 to 5.3) and pelleted by centrifugation at 3,166 × g at room temperature. Cell pellets were resuspended in a fresh 1:1 (vol/vol) fixative mixture containing 6% glutaraldehyde and 0.2 M sodium cacodylate (pH 5.5) and stored at 4°C. Sample processing for scanning electron microscopy (SEM) was performed by the Center for Electron Microscopy at North Carolina State University. Samples were viewed with a JOEL JSM 5900LV scanning electron microscope at 15 kV.
Microarray data accession numbers.
Microarray platform and series data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GPL7426 (platform) and GSE14622 (series).
RESULTS
Construction of an NCFM Δupp mutant as a host for counterselective allelic replacement.
In order to investigate the potential of the upp-based counterselective strategy for L. acidophilus NCFM, this strain was first tested to determine its sensitivity to 5-FU. Overnight cultures in MRS broth were serially diluted in SDM and plated at a density of ca. 105 CFU per plate onto SDM containing different concentrations of 5-FU or the equivalent amounts of DMSO (the agent used to dissolve 5-FU) as a control. Growth was inhibited on medium containing 100 μg/ml of 5-FU. The presence of DMSO in the medium did not affect the growth of L. acidophilus, indicating that growth inhibition was due solely to the toxicity of 5-FU. Therefore, 100 μg/ml 5-FU was an acceptable concentration for selection against integrants that have not resolved the merodiploid state.
The sensitivity of L. acidophilus to 5-FU confirmed the presence of a functional UPRTase. A putative UPRTase-encoding gene, LBA0770, was found in the NCFM genome sequence. The LBA0770 coding region was annotated as upp based on sequence similarity to corresponding enzymes in the GenBank database. The deduced amino acid sequence showed more than 80% sequence identity to the UPRTases from L. delbrueckii, L. gasseri, and L. johnsonii. To associate the upp genotype with the observed 5-FUs phenotype, an in-frame 315-bp deletion in the 630-bp putative upp gene was constructed by using the pORI-based two-step allelic replacement scheme (see Materials and Methods). After propagation of an L. acidophilus pTRK934 chromosomal integrant in nonselective medium to facilitate the second homologous recombination, 1,200 colonies were screened for Ems recombinants that had lost the plasmid backbone. Of 13 Ems colonies isolated, 4 carried the deleted Δupp allele, as confirmed by PCR assay and DNA sequencing of the target locus. All four Δupp mutants were able to grow on SDM in the presence of 100 μg/ml 5-FU, a concentration that inhibits growth of the wild type. This result confirmed that LBA0770 encodes a functional UPRTase in NCFM. All of the Δupp mutants showed no apparent difference in the growth rate compared to the NCFM parent strain when they were cultured in MRS broth (data not shown), indicating that a deletion in the upp locus did not affect the growth behavior of the Δupp mutants. One of the Δupp mutants, designated NCK1909, was chosen as the background host for counterselective gene replacement.
PM analyses of Δupp mutant NCK1909.
Since the NCK1909 mutant serves both as the host for genetic exchange and as the reference strain for subsequent phenotypic studies of deletion mutants that will be generated with the counterselective gene knockout system, we utilized the Biolog PM to detect any phenotypic difference between the Δupp mutant and its parent strain, NCFM. This analysis allowed us to compare the phenotypes of the two strains in response to osmolytes, pH, and 240 different chemical substrates. In this analysis only 1 of the 1,152 wells showed a difference in growth (PM19, D07) (see Fig. S1 and Table S3 in the supplemental material). The difference observed for growth in the presence of iodonitrotetrazolium violet, however, was not considered significant as the growth difference was observed at only one of the four concentrations tested. No difference was observed for pH, osmolytes, or other chemical sensitivities. With the exception of 5-FU resistance, the results demonstrated that the upp mutation did not cause any major detectable differences in the cellular phenotype compared to the parent strain.
Microarray gene expression study of the NCK1909 background host.
Previously, we successfully developed a whole-genome microarray for L. acidophilus NCFM covering approximately 97.4% of the open reading frames predicted in the genome (7). In the present study, we designed a new NCFM whole-genome oligoarray platform using OligoArray 2.1 (45). A total of 1,823 oligonucleotides (representing 97.8% of the genes in the chromosome), with each oligonucleotide representing a gene, were spotted six times in a random pattern onto glass slides (total number of spots, 11,520). To assess the reliability of the platform, we conducted a microarray hybridization experiment by labeling cDNA from the same RNA sample with Cy3 and Cy5. The hybridization data revealed a linear correlation (R value) in the relative expression levels of 0.98 for 11,520 spots, with median, maximum, and minimum Cy5/Cy3 expression ratios of 1.0, 1.48, and 0.46, respectively (data not shown).
To further establish the genotype of NCK1909 as a representative reference strain, microarray hybridizations using the new oligoarray platform were performed to compare the transcriptional profiles of NCK1909 with those of NCFM at the early log (OD600, 0.3) and mid-log (OD600, 0.8) phases when the organisms were cultured in MRS medium at 37°C under ambient atmospheric conditions. These cell densities represent the growth stages when cells were collected for stress challenge assays (OD600, 0.25 to 0.3) and cell adherence assays (OD600, 0.6 to 0.8) (see Materials and Methods). Analysis of gene expression data detected only a few genes that were differentially expressed by a onefold or greater change in NCK1909 and NCFM at both early and mid-log phases when organisms were grown in MRS medium (Table 2). In addition, no gene was detected as being expressed in NCK1909 at a level more than onefold higher than the level in NCFM during the mid-log growth phase. Overall, these results further confirm that NCK1909 is a representative reference strain that can be used for both phenotypic and genotypic studies of deletion mutants generated using the upp-based counterselective gene replacement system.
TABLE 2.
Comparative gene expression profiles of NCFM and the Δupp mutant NCK1909 grown in MRS medium
| Open reading frame | Annotation | Δupp/NCFM expression ratio | Log2 ratio | P value |
|---|---|---|---|---|
| Gene expression in early log phase (OD600, ∼0.3) | ||||
| Open reading frames expressed at higher levels in the Δupp mutant than in strain NCFM | ||||
| 1564 | Membrane protein | 2.05 | 1.04 | 0.005 |
| 1970 | Hypothetical protein (49 amino acids) | 2.03 | 1.02 | 0.02 |
| Open reading frames expressed at lower levels in the Δupp mutant than in strain NCFM | ||||
| 1873 | Acetate kinase | 0.49 | −1.02 | 0.0002 |
| 1568 | Putative surface protein | 0.48 | −1.05 | 0.0006 |
| 0586 | Galactose-1-phosphate uridylyltransferase | 0.47 | −1.09 | 1.06E-06 |
| 1066 | Putative phosphorylase | 0.47 | −1.10 | 2.73E-05 |
| 1871 | Neopullulanase | 0.36 | −1.49 | 9.41E-05 |
| 1362 | Hypothetical protein | 0.31 | −1.67 | 1.41E-05 |
| Gene expression at mid-log phase (OD600, ∼0.8) | ||||
| ORFs expressed at lower levels in the Δupp mutant than in strain NCFM | ||||
| 1357 | ABC transporter ATPase and permease protein | 0.499 | −1.00 | 0.004 |
Construction of a pORI-based counterselectable integration vector.
The pORI28 vector was chosen as the backbone for construction of a counterselectable integration vector due to its established efficacy for generating chromosomal insertions and deletions in NCFM. Previous transcriptome analyses of NCFM grown on various carbohydrate sources showed that the upp gene was highly expressed under all experimental conditions (10). Therefore, the native upp promoter was used for constitutive expression of the ectopic upp gene. The lacZ′ gene from pUC19 was also incorporated into the integration vector, which contains common MCS and provides blue-white screening to facilitate selection for E. coli recombinants.
To construct a counterselective integration vector, the entire multiple cloning region of pORI28 was removed by restriction with BglII and XbaI, blunt ended, and ligated with the SOE-PCR products of P-upp and the lacZ′ gene with its upstream promoter and CAP binding site amplified from the pUC19 vector (Fig. 1). Sequencing of the resulting integration vector, pTRK935, unexpectedly showed the presence of an intact XbaI site and five additional random bases at the ligation junction between lacZ′ and the pORI28 backbone, presumably resulting from the blunt-end treatment of the digested pORI28. Thus, the XbaI restriction site in the multiple cloning region of pTRK935 does not function as a unique site for gene cloning. When pTRK935 was introduced into NCK1909 harboring the helper plasmid pTRK669 (NCK1910), no growth was observed in the presence of 100 μg/ml of 5-FU. This result confirmed that the P-upp in the plasmid was able to complement the Δupp genotype in the NCK1910 integration host and restored sensitivity to 5-FU and that it could serve as a functional marker for maintaining negative selection against merodiploids with 5-FU.
Sequence analysis of the slpX gene.
To assess the efficacy of the upp-based counterselection strategy, the slpX gene was selected for targeted deletion. L. acidophilus NCFM has two major S-layer protein genes, slpA (LBA0169) and the silent slpB gene (LBA0175) (Table 3) (32). Our recent S-layer profiling of NCFM based on MS analysis of peptides derived from purified S-layers revealed coexpression of the slpX-encoded protein SlpX with SlpA (expressed by the wild-type strain) and SlpB (expressed by an slpA insertion mutant, NCK1377) (see Table S4 in the supplemental material). Thus, SlpX is suspected to contribute to the S-layer complex in NCFM. The slpA and slpB genes are located in a gene cluster, whereas slpX is located at a distant chromosomal locus. The deduced SlpX sequence shared moderate sequence identity (53%) with the sequence of a surface layer protein from L. acidophilus JCM1038, the only SlpX ortholog found in the GenBank database. Meanwhile, SlpX shared less than 30% sequence identity with SlpA (26%) and SlpB (24%), and the similarity was confined to the N-terminal and C-terminal regions. Nonetheless, SlpX is similar in size to SlpA and SlpB (46 to 51 kDa), and all three S-layer proteins shared features, such as a predicted basic isoelectric point of ∼9.5 and a high proportion of hydrophobic residues (44 to 48%) (Table 3), which are typical characteristics of lactobacillus S-layer proteins.
TABLE 3.
General characteristics of the S-layer proteins in L. acidophilus NCFM
| Protein | Gene locus | Gene length (bp) | Predicted mature protein size (amino acids) | Molecular mass of predicted mature protein (kDa) | Predicted pI | % Hydrophobic residuesa | Top BLASTP hit
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Protein | GenBank accession no. of ortholog | E-value | Amino acid identityb | |||||||
| SlpA | LBA0169 | 1,335 | 413 | 46 | 9.49 | 47.4 | Surface layer protein (L. helveticus) | CAB46984 | 3e-156 | 329/446 (73) |
| SlpB | LBA0175 | 1,374 | 426 | 47 | 9.45 | 48 | SB protein (L. acidophilus) | CAA61561 | 0.00e + 00 | 449/457 (98) |
| SlpX | LBA0512 | 1,500 | 469 | 51 | 9.43 | 44.1 | Surface layer protein HAP50 (L. acidophilus) | AAF65561 | 4e-127 | 273/506 (53) |
Alanine, glycine, isoleucine, leucine, methionine, phenylalanine, proline, tryptophan, tyrosine, and valine.
The numbers in parentheses are percentages.
In-frame deletion of slpX gene using markerless gene replacement with upp-based counterselection.
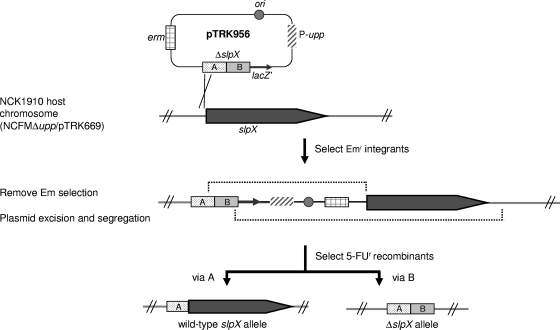
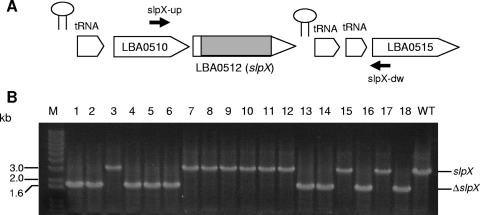
To construct an in-frame 1,356-bp deletion (∼90% of the gene) in the slpX gene, approximately 750-bp chromosomal segments flanking both sides of the region of the deletion target were PCR amplified, joined by SOE-PCR, and cloned into the BamHI/SacI sites of pTRK935. The resulting construct, pTRK956, was electroporated into L. acidophilus NCK1910, and transformants were selected based on resistance to Em and Cm (Fig. 2). One pTRK956 transformant was selected and propagated in MRS broth at 42°C in the presence of Em for 30 generations to facilitate integration of pTRK956 into the chromosome via homologous recombination. The integrated plasmid was subsequently resolved from the chromosome via secondary recombination by passing the integrant in MRS broth without Em at 37°C for 50 generations. Plasmid-free recombinants were recovered on SDM with 5-FU at a frequency of approximately 1.4 × 10−3. Nine of the 18 5-FUr isolates chosen carried the deleted ΔslpX allele, while the remaining isolates retained the wild-type allele (Fig. 3). All ΔslpX recombinants were Ems, confirming that there was excision of the integrated pTRK956 backbone from the chromosome. The genotype was verified by DNA sequencing. One of the ΔslpX mutants, designated NCK1962, was selected for further study.
FIG. 2.
Markerless gene replacement strategy with upp as a counterselectable marker for in-frame deletion of 1,356 bp in the slpX gene. Single-crossover recombination of pTRK956 occurred in the chromosomal slpX region homologous to either fragment A or B (only recombination via fragment A is shown). Removal of Em selection for the integrated pTRK956 plasmid facilitates a second recombination and plasmid excision, generating recombinants with either a wild-type allele or a ΔslpX allele. The resulting recombinants can be selected on SDM containing 100 μg/ml of 5-FU. DNA fragments A (dotted box) and B (shaded box) represent the cloned copy of a ΔslpX allele, where fragment A consists of the 5′ region of slpX (105 bp) and the upstream flanking region and fragment B consists of the 3′ region of slpX (36 bp) and the downstream flanking region.
FIG. 3.
PCR screening for the ΔslpX genotype in random colonies recovered after 5-FU selection. (A) Gene organization of the region surrounding the slpX locus. Arrows indicate the locations of primers slpX-up and slpX-dw used for PCR analysis. The shaded region in the slpX gene represents the deletion target. Hairpin structures indicate putative rho-independent terminators. The chromosomal map is not drawn to scale. (B) Colony PCR analysis of 18 selected 5-FUr recombinants. The expected amplicon sizes generated from wild-type and ΔslpX genotypes are approximately 3.14 kb and 1.78 kb, respectively. Lane M, DNA size marker; lanes 1 to 18, 5-FUr isolates; lane WT, parent strain NCFM (control).
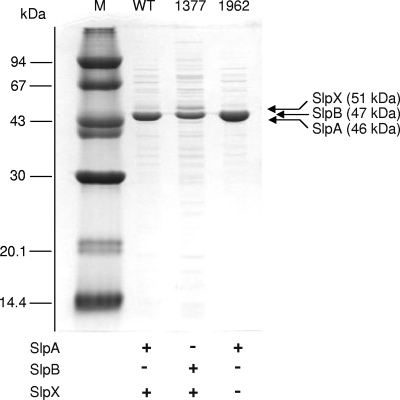
S-layer profile of the NCK1962 mutant.
Resolution of the purified S-layer proteins by SDS-polyacrylamide gel electrophoresis (PAGE) confirmed the absence of a 51-kDa band corresponding to the SlpX protein in NCK1962 (Fig. 4). As expected, the mutant expressed only the dominant SlpA proteins (46 kDa), albeit at a slightly higher level than the wild-type strain based on relative band intensities. In addition, no SlpX-specific peptide was detected for NCK1962 based on MS studies of peptides obtained from trypsinolysis of purified S-layers (see Table S4 in the supplemental material). These results further verified that the lack of the SlpX protein in the S-layers of NCK1962 was due to deletion of the slpX gene. Meanwhile, for both the NCFM strain and the NCK1377 integrant, the overall band intensities of the S-layer proteins expressed suggested that SlpX is present at a significantly lower level in the S-layer complex than the SlpA or SlpB proteins.
FIG. 4.
SDS-PAGE of purified S-layer proteins from L. acidophilus wild-type strain NCFM (lane WT), the NCFM slpA integrant NCK1377 (lane 1377), and the slpX deletion mutant NCK1962 (lane 1962) used for MS analysis. The S-layer profile of each strain is summarized at the bottom based on results of the MS analysis of the purified S-layer proteins. +, present; −, absent. Molecular masses of standard proteins (lane M) are indicated on the left.
Phenotype of the NCK1962 mutant.
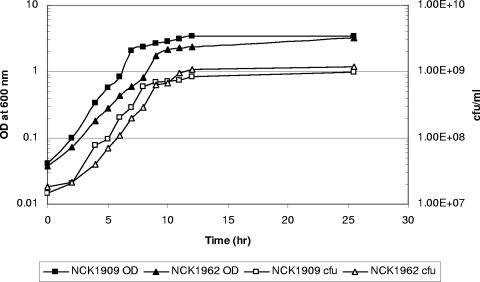
To investigate the role of SlpX in association with the S-layer in L. acidophilus, we examined the impact of slpX deletion on the cell morphotype, the response to environmental stresses, and cell surface properties. Monitoring the growth in MRS medium based on measurement of cell densities and cell counts revealed slightly slower growth of the NCK1962 mutant than of reference strain NCK1909, and the maximum specific growth rates were 0.43 h−1 (after 9 h of growth) and 0.56 h−1 (after 7 h of growth), respectively (Fig. 5). Nonetheless, NCK1962 cultures eventually reached final cell densities similar to those of cultures of the reference strain. Microscopic and SEM examinations of NCK1962 showed no significant difference in the morphology and physical appearance of the cells compared to NCK1909 (data not shown). Based on the MATS assay, both strains displayed high affinity (80 to 95%) for the apolar solvents hexadecane and decane (data not shown), indicating that the NCK1962 mutant retained a hydrophobic cell surface. Also, NCK1962 exhibited basic cell surface properties similar to those of NCK1909, as demonstrated by its higher affinity for chloroform (>99%) than for ethyl acetate (30%).
FIG. 5.
Growth of L. acidophilus NCK1962 and NCK1909 in MRS medium at 37°C under ambient atmospheric conditions. Growth was assessed by determining the optical density and cell counts (CFU/ml).
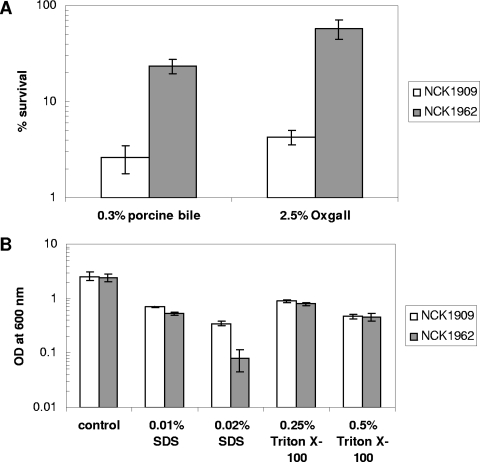
When early-log-phase cultures of the control strain NCK1909 and the mutant strain NCK1962 were exposed to 0.3% porcine bile for 2 h, the mutant had a higher survival rate (23.5% ± 3.9%) than NCK1909 (2.6% ± 0.8%) (Fig. 6A). Likewise, NCK1962 was more resistant to 2.5% oxgall bile treatment; 57.1% ± 12.9% of the survivors recovered, compared to only 4.3% ± 0.7% of the viable NCK1909 cells. On the other hand, NCK1962 was more susceptible to SDS, and severe growth inhibition was observed with 0.02% SDS (Fig. 6B). Meanwhile, NCK1962 showed growth comparable to that of NCK1909 in the presence of Triton X-100. Furthermore, no significant difference in the survival rates of NCK1962 and NCK1909 was observed when the organisms were challenged with 10% NaCl, 15% ethanol, a low pH (pH 3.0 or 3.5) in the presence of an organic acid (lactic acid) or an inorganic acid (HCl), or repeated freeze-thaw cycles (data not shown). The two strains also exhibited comparable abilities to adhere to Caco-2 epithelial cells and porcine gastric mucin in vitro (data not shown).
FIG. 6.
Survival of NCK1962 (ΔslpX mutant) and NCK1909 (reference strain) after stress challenge assays. (A) Survival of early-log-phase cultures of NCK1909 and NCK1962 after exposure to MRS medium supplemented with 0.3% porcine bile or 2.5% oxgall bile for 2 h at 37°C under ambient atmospheric conditions. (B) Early-log-phase cultures of NCK1909 and NCK1962 were inoculated (1% inoculum) into MRS broth supplemented with SDS or Triton X-100 surfactant, and cell density was measured after 24 h of incubation at 37°C under ambient atmospheric conditions. The data are the means ± standard errors of the means for three independent replicates.
DISCUSSION
L. acidophilus belongs to one of the most dynamic groups of commensals among the vast consortium of microflora that inhabits the human upper GI tract. Comparative genomic analysis with other Lactobacillus species and microarray transcriptional studies have revealed critical gene features that may contribute to the survival, adaptation, and colonization of L. acidophilus in the complex gut environment (2, 6, 7, 10, 21, 31, 42). Due to the industrial importance of L. acidophilus as a probiotic and a dietary adjunct, extensive studies have also been devoted to predicting key genes that play major roles in the survival and activities of this organism in both bioproccessing and host environments. Postgenomic analysis of L. acidophilus with the aim of establishing the functional roles of these genes and the associated regulatory networks requires efficient genetic exchange tools since most often, the construction of gene knockouts following the identification of gene targets of interest represents the major bottleneck in functional genomic studies.
In this report, the development of a upp-based counterselection strategy is described for L. acidophilus NCFM to improve the efficiency of the conventional pORI gene knockout system by providing direct selection for plasmid-free recombinants using 5-FU. The upp gene encoding UPRTase (EC 2.4.2.9) is involved in the pyrimidine salvage pathway since UPRTase converts uracil into UMP for nucleotide biosynthesis. Deletion of upp in NCFM confers resistance to 5-FU, confirming the presence of a functional UPRTase. The cytotoxicity of 5-FU is initiated by conversion of 5-FU into 5-fluoro-UMP by UPRTase, which leads to subsequent formation of the very toxic metabolite 5-fluorodeoxyuridine monophosphate (5-FdUMP) (40). The latter compound acts as a thymidylate synthase inhibitor and thereby prevents formation of the dTMP required for synthesis of the dTTP precursor. In some microorganisms, 5-FU can also be directly converted to 5-FdUMP by thymidine phosphorylase and thymidine kinase (3, 36, 37). Previous genome analysis indicated that L. acidophilus NCFM is auxotrophic for UTP and dTTP (2). The absence of genes encoding a nucleoside diphosphate kinase and a nucleoside triphosphate adenylate kinase required for the conversion of UDP (from UMP) to UTP, as well as a thymidine phosphorylase, would prevent formation of 5-FdUMP. Therefore, we speculated that the 5-FUs phenotype of NCFM is likely due to the cumulative toxic effect of the 5-fluoro-UMP or 5-fluoro-UDP intermediates. This observation also implies that, since NCFM does not have a complete pyrimidine de novo biosynthetic pathway, mutation of upp would not result in a significant difference in the physiological behavior and uracil metabolism between two strains. This hypothesis is supported by the phenotypic microarray analysis comparing the NCFM parent strain and the Δupp background host NCK1909, where no phenotypic difference was observed in response to pH, osmolytes, and the chemical substrates examined. Furthermore, microarray transcriptional analysis of NCFM and NCK1909 grown in MRS medium showed little difference in the global gene expression profiles between these two strains. Collectively, these results supported use of the NCK1909 host as a representative of wild-type strain NCFM for both in vitro and in vivo phenotypic studies of mutants constructed with this gene replacement system.
To evaluate the efficacy of the upp counterselective approach in L. acidophilus, the knockout strategy was used to generate an in-frame deletion in the slpX gene. SlpX is a secreted protein that was recently identified as a protein associated with the S-layer complex in L. acidophilus NCFM. In general, S-layers are paracrystalline layers of (glyco)protein subunits that are present in abundance on the cell surface of most eubacteria and archaea (for a review, see reference 48). S-layers have been identified and characterized in numerous Lactobacillus species (for a review, see reference 4). Notably, L. brevis and several species closely related to L. acidophilus, including L. crispatus, L. amylovorus, and L. gallinarum, are among the S-layer-producing lactobacilli that have multiple S-layer protein-encoding genes (16-18, 29). Nonetheless, only a surface layer protein from L. acidophilus JCM1038 that shared moderate sequence similarity with SlpX was found in the current GenBank database (see above). It is tempting to speculate that the SlpX protein is unique to NCFM, although it remains to be determined whether an SlpX homolog is present in unsequenced species that have S-layers. Diverse roles have been proposed for S-layers, including roles as cell shape determinants, as molecular sieves, as layers protecting against phage adsorption or predatory bacteria, and as layers providing anchoring sites for surface-associated enzymes, cell adhesion, and surface recognition. Most of these functions are presumed to be important for the survival of L. acidophilus in the human GI tract and for cellular communication with the host immune and mucosal epithelial cells. Due to the functional potential of S-layers and the close association of SlpX with SlpA or SlpB, it was of great interest to investigate the functional role of SlpX in the formation of the S-layer complex.
Mutation of the slpX gene did not affect the morphology of the NCK1962 mutant, although the mutant strain exhibited a slightly lower growth rate than the NCK1909 background host. A previous study by van der Mei et al. (50) demonstrated that S-layers confer cell surface hydrophobicity to lactobacilli. In the present study, the absence of the SlpX protein in the S-layer structure of NCK1962 did not cause significant change in the hydrophobicity or basic properties of the cell surface based on its affinity for solvents. One possible explanation is that the SlpX proteins are naturally present at much lower levels than SlpA or SlpB, as revealed by analysis of the purified S-layers of the parent strain and the SlpB-expressing slpA integrant NCK1377 (Fig. 4). Thus, the absence of SlpX in NCK1962 might have a negligible effect on the overall hydrophobic and basic properties of the cell surface. Alternatively, the change in surface properties due to the absence of SlpX may be compensated for by the overexpression of SlpA in the NCK1962 mutant.
No major differences were observed between NCK1962 and NCK1909 when they were subjected to a series of environmental stress challenges, such as the presence of 10% NaCl or 15% ethanol or a low pH. NCK1962 also showed a survival rate similar to that of NCK1909 after repeated free-thaw treatment. These results suggest that the lack of SlpX in the S-layer complex did not compromise the ability of the mutant to cope with osmotic and ethanol stresses or to maintain pH homeostasis or physical integrity after freeze-thaw injury. Buck et al. (20) recently showed that the slpA mutant had a significantly reduced ability to adhere to Caco-2 epithelial cells in vitro. These authors, however, indicated that the observed reduced-adherence phenotype of the slpA mutant might also be due to the absence of S-layer-associated surface proteins that contribute to adherence. In the present study, the NCK1962 mutant did not show a reduced ability to adhere to Caco-2 epithelial cells or mucin in vitro. This implies that SlpX is not directly involved in surface adherence or that it may play a minor role as an anchoring site for potential adherence factors.
Unexpectedly, the absence of SlpX proteins in the S-layer complex of the NCK1962 mutant rendered the cells more susceptible to the anionic surfactant SDS but not the nonionic surfactant Triton X-100 than the reference strain. Typically, both surfactants exert their bactericidal effect by causing cell lysis. Due to the anionic properties of SDS that disrupt noncovalent bonds in proteins, the observed SDS-sensitive phenotype of NCK1962 may have been due in part to SDS-induced denaturation of essential surface proteins or transporters in NCK1962 which were otherwise protected by an intact S-layer with SlpX in NCK1909. On the other hand, NCK1962 displayed significantly enhanced tolerance to both biological surfactants, oxgall (bovine bile) and porcine bile, whereas a nearly 2-log reduction was observed in NCK1909 when it was exposed to the same treatments. The major organic constituents of bile are bile salts, cholesterol, and phospholipids (for a review, see reference 11). Bile salts are synthesized de novo by the liver from cholesterol, where the steroid portion of the molecule is conjugated to either glycine (glycoconjugated) or taurine (tauroconjugated) via peptide linkages. We speculated that the lack of SlpX may affect the permeability of the S-layers. This would potentially allow increased intracellular access of conjugated bile salts. Perhaps the anticipated change in the permeability of the SlpX-deficient S-layers in NCK1962 may potentially allow more efficient efflux of intracellular deconjugated bile acids.
At this point, the precise role of slpX remains unclear. The lower abundance of SlpX in the S-layers as revealed by SDS-PAGE suggested that this protein is an auxiliary component of the S-layer complex. Nonetheless, we concluded that SlpX is essential for the maintenance of cellular integrity and other unknown physiological functions. This is due to the observations that inactivation of this protein affected the growth rate and the tolerance to bile salts in the NCK1962 mutant, as well as its relative overexpression in NCK1377, an SlpB-dominant strain that lacks SlpA protein. This implies that SlpX contributes to the survival and probiotic functionality in the host GI environment. Further phenotypic and gene expression studies are currently under way to establish the function of SlpX. Collectively, the results of the present study shed light on the complexity of the biological roles of the S-layers in L. acidophilus.
Overall, we have demonstrated the functionality of the upp counterselection strategy for markerless allelic exchange in L. acidophilus by construction of an in-frame deletion in the slpX gene. To date, this system has been used in our laboratory to generate multiple gene deletions targeted to single or multiple gene loci in L. acidophilus NCFM. The counterselection strategy allows direct selection for double recombinants, thereby facilitating construction of mutants and assignment of roles to candidate genes that are important for the probiotic attributes of L. acidophilus. Since the upp counterselectable marker is recyclable, mutations in countless gene loci can be serially introduced into the same strain for study of complex gene regulatory networks. In addition, mutations can be targeted to genes in an operon without polar effects on neighboring genes. This improved counterselective allelic replacement system should also prove to be useful for introducing desirable probiotic traits into L. acidophilus or to engineer novel strains for delivery of biotherapeutics.
Supplementary Material
Acknowledgments
This work was supported in part by Danisco USA, Inc. (Madison, WI), by the North Carolina Dairy Foundation, and by Dairy Management, Inc.
We are very grateful to Christophe Brandily for his technical assistance, to Sabrina Tachdjian and R. Kelly for sharing their oligoarray hybridization protocol, and to Rosemary Sanozky-Dawes and Richard Tallon for their assistance with and discussions concerning in vitro Caco-2 epithelial cells, mucin adherence studies, and the MATS assay.
Footnotes
Published ahead of print on 20 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altermann, E., B. L. Buck, R. Cano, and T. R. Klaenhammer. 2004. Identification and phenotypic characterization of the cell-division protein CdpA. Gene 342:189-197. [DOI] [PubMed] [Google Scholar]
- 2.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arsene-Ploetze, F., H. Nicoloff, B. Kammerer, J. Martinussen, and F. Bringel. 2006. Uracil salvage pathway in Lactobacillus plantarum: transcription and genetic studies. J. Bacteriol. 188:4777-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avall-Jaaskelainen, S., and A. Palva. 2005. Lactobacillus surface layers and their applications. FEMS Microbiol. Rev. 29:511-529. [DOI] [PubMed] [Google Scholar]
- 5.Azcarate-Peril, M. A., E. Altermann, R. L. Hoover-Fitzula, R. Cano, and T. R. Klaenhammer. 2004. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl. Environ. Microbiol. 70:5315-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azcarate-Peril, M. A., J. M. Bruno-Barcena, H. M. Hassan, and T. R. Klaenhammer. 2006. Transcriptional and functional analysis of oxalyl-coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes from Lactobacillus acidophilus. Appl. Environ. Microbiol. 72:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azcarate-Peril, M. A., O. McAuliffe, E. Altermann, S. Lick, W. M. Russell, and T. R. Klaenhammer. 2005. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl. Environ. Microbiol. 71:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barefoot, S. F., and T. R. Klaenhammer. 1983. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl. Environ. Microbiol. 45:1808-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrangou, R., E. Altermann, R. Hutkins, R. Cano, and T. R. Klaenhammer. 2003. Functional and comparative analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 100:8957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrangou, R., M. A. Azcarate-Peril, T. Duong, S. B. Conners, R. M. Kelly, and T. R. Klaenhammer. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. USA 103:3816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Begley, M., C. G. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29:625-651. [DOI] [PubMed] [Google Scholar]
- 12.Bellon-Fontaine, M.-N., J. Rault, and C. J. van Oss. 1996. Microbial adhesion to solvents: a novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surf. B Biointerfaces 7:47-53. [Google Scholar]
- 13.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 14.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 16.Boot, H. J., C. P. Kolen, B. Pot, K. Kersters, and P. H. Pouwels. 1996. The presence of two S-layer-protein-encoding genes is conserved among species related to Lactobacillus acidophilus. Microbiology 142:2375-2384. [DOI] [PubMed] [Google Scholar]
- 17.Boot, H. J., C. P. Kolen, and P. H. Pouwels. 1995. Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J. Bacteriol. 177:7222-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boot, H. J., C. P. Kolen, J. M. van Noort, and P. H. Pouwels. 1993. S-layer protein of Lactobacillus acidophilus ATCC 4356: purification, expression in Escherichia coli, and nucleotide sequence of the corresponding gene. J. Bacteriol. 175:6089-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruno-Barcena, J. M., M. A. Azcarate-Peril, T. R. Klaenhammer, and H. M. Hassan. 2005. Marker-free chromosomal integration of the manganese superoxide dismutase gene (sodA) from Streptococcus thermophilus into Lactobacillus gasseri. FEMS Microbiol. Lett. 246:91-101. [DOI] [PubMed] [Google Scholar]
- 20.Buck, B. L., E. Altermann, T. Svingerud, and T. R. Klaenhammer. 2005. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71:8344-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callanan, M., P. Kaleta, J. O'Callaghan, O. O'Sullivan, K. Jordan, O. McAuliffe, A. Sangrador-Vegas, L. Slattery, G. F. Fitzgerald, T. Beresford, and R. P. Ross. 2008. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J. Bacteriol. 190:727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobson, A. E., R. B. Sanozky-Dawes, and T. R. Klaenhammer. 2007. Identification of an operon and inducing peptide involved in the production of lactacin B by Lactobacillus acidophilus. J. Appl. Microbiol. 103:1766-1778. [DOI] [PubMed] [Google Scholar]
- 23.Duong, T., R. Barrangou, W. M. Russell, and T. R. Klaenhammer. 2006. Characterization of the tre locus and analysis of trehalose cryoprotection in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 72:1218-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabret, C., S. D. Ehrlich, and P. Noirot. 2002. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol. Microbiol. 46:25-36. [DOI] [PubMed] [Google Scholar]
- 25.Fukagawa, T., N. Hayward, J. Yang, C. Azzalin, D. Griffin, A. F. Stewart, and W. Brown. 1999. The chicken HPRT gene: a counter selectable marker for the DT40 cell line. Nucleic Acids Res. 27:1966-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goh, Y. J., C. Zhang, A. K. Benson, V. Schlegel, J. H. Lee, and R. W. Hutkins. 2006. Identification of a putative operon involved in fructooligosaccharide utilization by Lactobacillus paracasei. Appl. Environ. Microbiol. 72:7518-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning: a practical approach, vol. 1. IRL Press Ltd., Oxford, England. [Google Scholar]
- 28.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 29.Jakava-Viljanen, M., S. Avall-Jaaskelainen, P. Messner, U. B. Sleytr, and A. Palva. 2002. Isolation of three new surface layer protein genes (slp) from Lactobacillus brevis ATCC 14869 and characterization of the change in their expression under aerated and anaerobic conditions. J. Bacteriol. 184:6786-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimmel, S. A., and R. F. Roberts. 1998. Development of a growth medium suitable for exopolysaccharide production by Lactobacillus delbrueckii spp. bulgaricus RR. Int. J. Food Microbiol. 40:87-92. [DOI] [PubMed] [Google Scholar]
- 31.Klaenhammer, T. R., R. Barrangou, B. L. Buck, M. A. Azcarate-Peril, and E. Altermann. 2005. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 29:393-409. [DOI] [PubMed] [Google Scholar]
- 32.Konstantinov, S. R., H. Smidt, W. M. de Vos, S. C. Bruijns, S. K. Singh, F. Valence, D. Molle, S. Lortal, E. Altermann, T. R. Klaenhammer, and Y. van Kooyk. 2008. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. USA 105:19474-19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristich, C. J., D. A. Manias, and G. A. Dunny. 2005. Development of a method for markerless genetic exchange in Enterococcus faecalis and its use in construction of a srtA mutant. Appl. Environ. Microbiol. 71:5837-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinussen, J., P. Glaser, P. S. Andersen, and H. H. Saxild. 1995. Two genes encoding uracil phosphoribosyltransferase are present in Bacillus subtilis. J. Bacteriol. 177:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinussen, J., and K. Hammer. 1994. Cloning and characterization of upp, a gene encoding uracil phosphoribosyltransferase from Lactococcus lactis. J. Bacteriol. 176:6457-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McAuliffe, O., R. J. Cano, and T. R. Klaenhammer. 2005. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71:4925-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merritt, J., P. Tsang, L. Zheng, W. Shi, and F. Qi. 2007. Construction of a counterselection-based in-frame deletion system for genetic studies of Streptococcus mutans. Oral Microbiol. Immunol. 22:95-102. [DOI] [PubMed] [Google Scholar]
- 40.Neuhard, J. 1982. Utilization of preformed pyrimidine bases and nucleosides, p. 95-148. In A. Munch-Petersen (ed.), Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. Academic Press, London, United Kingdom.
- 41.Peck, R. F., S. DasSarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarium with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 42.Pfeiler, E. A., M. A. Azcarate-Peril, and T. R. Klaenhammer. 2007. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 189:4624-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pritchett, M. A., J. K. Zhang, and W. W. Metcalf. 2004. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl. Environ. Microbiol. 70:1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ried, J. L., and A. Collmer. 1987. An nptl-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239-246. [DOI] [PubMed] [Google Scholar]
- 45.Rouillard, J. M., M. Zuker, and E. Gulari. 2003. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31:3057-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell, W. M., and T. R. Klaenhammer. 2001. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl. Environ. Microbiol. 67:4361-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanders, M. E., and T. R. Klaenhammer. 2001. The scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J. Dairy Sci. 84:319-331. [DOI] [PubMed] [Google Scholar]
- 48.Sara, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ueki, T., S. Inouye, and M. Inouye. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153-157. [DOI] [PubMed] [Google Scholar]
- 50.van der Mei, H. C., B. van de Belt-Gritter, P. H. Pouwels, B. Martinez, and H. J. Busscher. 2003. Cell surface hydrophobicity is conveyed by S-layer proteins—a study in recombinant lactobacilli. Colloids Surf. B Biointerfaces 28:127-134. [Google Scholar]
- 51.Walker, D. C., K. Aoyama, and T. R. Klaenhammer. 1996. Electrotransformation of Lactobacillus acidophilus group A1. FEMS Microbiol. Lett. 138:233-237. [DOI] [PubMed] [Google Scholar]
- 52.Walker, D. C., and T. R. Klaenhammer. 1994. Isolation of a novel IS3 group insertion element and construction of an integration vector for Lactobacillus spp. J. Bacteriol. 176:5330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walter, J., P. Chagnaud, G. W. Tannock, D. M. Loach, F. Dal Bello, H. F. Jenkinson, W. P. Hammes, and C. Hertel. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71:979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, G., S. P. Kennedy, S. Fasiludeen, C. Rensing, and S. DasSarma. 2004. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J. Bacteriol. 186:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei, M.-Q., C. M. Rush, J. M. Norman, L. M. Hafner, R. J. Epping, and P. Timms. 1995. An improved method for the transformation of Lactobacillus strains using electroporation. J. Microbiol. Methods 21:97-109. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.