Abstract
A bacteriocin-producing strain of the bacterial spot of tomato plant pathogen, Xanthomonas perforans, with attenuated pathogenicity was deployed for biocontrol of a bacteriocin-sensitive strain of the genetically closely related bacterial spot of tomato plant pathogen, X. euvesicatoria. The attenuated mutant (91-118ΔopgHΔbcnB) of X. perforans was tested in leaf tissue and shown to significantly inhibit internal populations of the wild-type X. euvesicatoria strain although significantly less than the wild-type 91-118 strain, whereas in a phyllosphere inhibition assay, the mutant strain reduced epiphytic populations comparably to 91-118. Thus, the attenuated mutant limited the sensitive bacterium more efficiently on the leaf surface than inside the leaf. In field experiments, weekly application of 91-118ΔopgHΔbcnB significantly reduced X. euvesicatoria populations compared to the growers’ standard control (copper hydroxide and mancozeb applied weekly and acibenzolar-S-methyl applied every 2 weeks). The biological control agent, 91-118ΔopgHΔbcnB, applied every 2 weeks also significantly reduced X. euvesicatoria populations in one season but was not significantly different from the growers’ standard control. Potentially, attenuated pathogenic strains could be deployed as biological control agents in order to improve disease control of foliar plant pathogens.
Bacterial spot of tomato is incited by several Xanthomonas spp., including Xanthomonas euvesicatoria, X. perforans, and X. vesicatoria (13). On tomato plants, three races, designated tomato race 1 (T1), T2, and T3, were originally identified based on their reactions on three tomato genotypes (22, 23, 33, 37). These races, T1, T2, and T3, are principally members of X. euvesicatoria, X. vesicatoria, and X. perforans, respectively. Control of bacterial spot of tomato is extremely difficult when moderate-to-high temperatures and high moisture conditions exist. The disease causes significant damage to the crop, resulting in major losses (27). Management is primarily limited to bactericides, such as fixed coppers (3, 20, 32); however, copper-tolerant strains have become prevalent (31, 32) and chemical control alone is insufficient to control the disease under optimal weather conditions. Additionally, the use of copper compounds has led to soil contamination in some instances (16).
Recently, there has been increased interest in integrated biological control strategies for bacterial diseases (5, 7, 11, 19, 23). However, optimization of biocontrol agents for consistent disease suppression for many bacterial diseases has been difficult. Studies are increasing our understanding of the ecology of nonpathogenic saprophytes as biocontrol agents, but their selection is limited to labor-intensive protocols. New biological control strategies are currently being sought, including the use of bacteriocins, bacteriophages, and attenuated plant pathogens (4, 5, 7, 9, 11, 23, 26, 35, 38).
For many years, only X. euvesicatoria (T1) was present on tomato plants in Florida. In 1991, X. perforans (T3) appeared in Florida tomato fields (15) and eventually became the prevalent race (14). Following that observation, Jones et al. (12) demonstrated that when X. perforans and X. euvesicatoria were coinoculated onto tomato plants in the field, X. perforans displaced X. euvesicatoria and became the predominant strain. Further studies revealed that the competitive nature of X. perforans was due in large part to its antagonism toward X. euvesicatoria strains (9, 12, 36). Tudor-Nelson et al. (36) identified three different bacteriocins, designated BCN-A, BCN-B, and BCN-C, which were found to confer the ability to inhibit X. euvesicatoria strains in plate assays. Hert et al. (9) determined that these bacteriocins provided X. perforans strains with a competitive advantage in the greenhouse and field and that a mutant X. perforans strain expressing only BCN-A and BCN-C was most effective in displacing X. euvesicatoria and outcompeted wild-type (WT) X. perforans. Field experiments conducted with a nonpathogenic Hrp− strain of X. perforans as a potential biocontrol agent for controlling X. euvesicatoria resulted in marginal control (18). Although the WT X. perforans strain has a competitive advantage over X. euvesicatoria populations, Hrp− mutants such as that used in the study by Liu (18) do not appear to have the necessary competitiveness to suppress X. euvesicatoria populations.
Previous research has focused on colonization of the plant by biocontrol agents to determine the relationship between invasion efficiency and biological control efficacy. For example, Frey et al. (8) achieved only low-to-moderate levels of biological control of a WT bacteriocin-sensitive strain of Ralstonia solanacearum when using a bacteriocin-producing nonpathogenic Hrp− mutant strain of R. solanacearum. However, control using a moderately pathogenic hrp mutant (hrcV) capable of higher levels of colonization of the root and stem achieved greater disease suppression (6). Etchebar et al. (6) suggested that there was a positive correlation between colonization of the xylem by the hrp mutant and the level of control of WT R. solanacearum. As a result of previous studies showing that nonpathogenic strains of X. perforans only provide low levels of biological control (18), we hypothesized that using an attenuated pathogenic bacteriocin-producing strain of X. perforans rather than a nonpathogenic strain may increase the efficiency of control under field conditions.
In this study, our strategy was to use an attenuated mutant of X. perforans that colonizes leaf tissue more effectively than nonpathogenic strains do and potentially provides more effective colonization and increases the likelihood for interaction between X. perforans and X. euvesicatoria. We selected strain 91-118ΔopgHΔbcnB as the biocontrol agent since it was previously shown that deletion of the osmoregulated periplasmic glucan gene opgH resulted in a pathogenic phenotype with a significantly reduced ability to cause disease and internal colonization in susceptible tomato tissue (22). The selected mutant also lacked BcnB activity based on a previous study in which BcnB appeared to negatively affect competitive ability in that a ΔbcnB mutant was more effective at colonizing tomato leaves in field experiments than WT X. perforans was (9).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The origins and relevant characteristics of the bacterial strains and plasmids used in this study are listed in Table 1. Strains of X. perforans and X. euvesicatoria were grown on nutrient agar (NA) medium (Difco Laboratories, Detroit, MI) at 28°C. Strains of Escherichia coli were grown on Luria-Bertani medium (Difco Laboratories, Detroit, MI) at 37°C (21). All strains were stored in 20% glycerol in sterile tap water at −80°C. Bacterial cultures for plant inoculations were grown in nutrient broth (Difco Laboratories, Detroit, MI) for 18 h at 28°C on a rotary shaker at 100 rpm. Cells were pelleted by centrifugation (4,000 × g, 15 min) and resuspended in sterile tap water. Bacterial suspensions were standardized to an optical density at 600 nm of 0.3 (5 × 108 CFU/ml) with a Spectronic 20 spectrophotometer (Spectronic UNICAM, Rochester, NY) and subsequently diluted in sterile tap water to appropriate cell densities for individual experiments. Antibiotics were used to maintain selection for resistance markers at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 12.5 μg/ml; rifampin, 100 μg/ml; spectinomycin, 50 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 34 μg/ml; streptomycin, 200 μg/ml; nalidixic acid, 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or referencea |
|---|---|---|
| X. euvesicatoria | ||
| E3-1 | Nalr Smr | 9 |
| 91-106 | 36 | |
| X. perforans | ||
| 91-118 | Rifr | 36 |
| 91-118ΔbcnB | BcnB− Rifr Kmr | 9 |
| 91-118ΔopgH | OpgH− Rifr | 23 |
| 91-118ΔopgHΔbcnB | OpgH− BcnB− Rifr Kmr | This study |
| E. coli | ||
| DH5α | F−recA | BRL |
| C2110 | Nalr | BRL |
| λPIR | Host for pOK1; Spr oriR6K RK2 replicon UB | |
| Plasmids | ||
| pOK1 | Suicide vector; SacB | 10 |
| pBluescript-KS+ | Phagemid, pUC derivative; Ampr | Stratagene |
| pLAFR3 | Tcrrlx+ RK2 replicon | BJS |
| pRK2013 | Helper plasmid; Kmr Tra+ | 4 |
| pXV442-255 | pLAFR3; bcnB with Kmr insertion | STN |
BRL, Bethesda Research Laboratories, Gaithersburg, MD; Stratagene, Stratagene Inc., La Jolla, CA; BJS, B. J. Staskawicz, University of California, Berkeley; UB, U. Bonas, Martin-Luther-Universität, Halle, Germany; STN, Simone Tudor-Nelson.
Generation of the 91-118ΔopgHΔbcnB attenuation mutant.
Triparental matings were performed with E. coli DH5α containing pRK2013Km as the helper plasmid (Table 1), E. coli DH5α containing pXV442-255 (pXV442Tc with insertion of a Kmr cassette for inactivation of BcnB) as the donor, and strain 91-118ΔopgH as the recipient (22). Marker exchange was achieved by standard methods (29). Candidate colonies were screened for loss of BCN activity and confirmed for insertion by Southern hybridization (with subclone BcnB as the probe) and PCR (with primers BCN-1 and BCN-2) as previously described (9).
Plant materials.
Tomato (Lycopersicon esculentum) cv. Bonny Best or FL47 seeds were planted in Plugmix (W. R. Grace & Co., Cambridge, MA). After 2 weeks, the seedlings that emerged were transferred to Metromix 300 (W. R. Grace & Co., Cambridge, MA) in 10-cm plastic pots. Seedlings were grown in a glass house at temperatures ranging from 25 to 35°C.
In planta growth curve assays.
In planta assays were conducted in controlled environmental growth chambers to compare the growth curves of the 91-118ΔopgH and 91-118ΔopgHΔbcnB mutants with that of the WT parent strain, 91-118. Inoculum was prepared as described above and diluted to a final concentration of 3 × 105 CFU/ml. Leaflets of 6-week-old Bonny Best tomato seedlings were infiltrated (15 leaflets per strain) with a hypodermic syringe and needle as described previously (12). Following inoculation, plants were kept in a growth room on a 12-h photoperiod of cool, white fluorescent light at 24 to 28°C. Three samples were taken from each treatment every 24 h for 5 days. Bacterial populations were quantified by macerating 1-cm2 leaf disks in 1 ml sterile tap water, running 10-fold serial dilutions in sterile tap water, and plating them onto NA medium amended with the appropriate antibiotic. After incubation at 28°C for 4 to 5 days, colonies typical of Xanthomonas were counted. Population data were log10 transformed, and standard errors were determined. The overall growth curve was determined by calculating the area under the population progress curve (AUPPC). The AUPPC is a modification of the area under the disease progress curve, which has been used to analyze disease progress (30): standardized AUPPC = Σ [(xi + xi − 1)/2](ti − ti − 1), where x is population density in log10 CFU per square centimeter and t is time in hours. The AUPPC values for the strains were compared by analysis of variance and subsequent separation of sample means by the Waller-Duncan multiple-range test with SAS version 9.0 (SAS Inc., Cary, NC). Each experiment was conducted three times.
Plant disease severity assays.
Disease severity assays were conducted on plants in the greenhouse to determine the effect of X. perforans WT 91-118 and mutant strains on symptom development. In each test, four young (four-true-leaf stage) plants were inoculated with each strain by being dipped into bacterial suspensions of the 91-118, 91-118ΔbcnB, 91-118ΔopgH, and 91-118ΔopgHΔbcnB strains and X. euvesicatoria strain 91-106 (3 × 106 CFU/ml amended with the surfactant Silwet L-77 [Loveland Industries, Inc., Greeley, CO]) at 0.025% for 15 s. Plants were maintained in the greenhouse during the evaluation period. The plants were assessed for disease severity by visual estimation of the percentage of leaf tissue with lesions at 14 to 21 days after inoculation. Disease severity assessments were made based on leaf and stem ratings compiled from three separate greenhouse inoculation tests.
Antagonism assays.
Assays were performed to determine the antagonistic effects of the WT and mutant X. perforans strains on bacteriocin-sensitive X. euvesicatoria strain E3-1. Internal and external (phyllosphere) populations of WT and individual mutant X. perforans strains were evaluated separately by using two assays to monitor antagonism by the mutants.
Internal plant antagonism assay.
An inoculum was prepared as described above. Leaflets of 6-week-old seedlings of the tomato cultigen Florida 47 (Asgrow, Oxnard, CA) were inoculated with X. perforans and X. euvesicatoria strains (15 leaflets per strain) at 5 × 107 and 5 × 106 CFU/ml, respectively, with a hypodermic syringe as described previously (12). The mutant and WT X. perforans strains were inoculated into leaflets by infiltration 12 h prior to inoculation with the sensitive strain (E3-1). Untreated controls were inoculated with 1 ml of sterile tap water. Each treatment consisted of three replications. Following inoculation, plants were incubated in a growth room on a 12-h photoperiod of cool, white fluorescent light at 24 to 28°C. Leaf tissue samples were assayed at 24-h intervals for 96 h. Populations of the X. euvesicatoria sensitive strain (E3-1SmNal) in leaflets were determined as already described, and statistical analyses were performed. Each experiment was conducted three times.
Phyllosphere antagonism assays.
Phyllosphere antagonism assays were conducted on plants in growth chambers to determine if the gene deletion affected the level of antagonism toward external leaf populations of X. euvesicatoria strain E3-1 by comparing the antagonistic effect of the mutants with that of parent strain 91-118. Six-week-old Bonny Best tomato seedlings were dipped into an inoculum containing 5 × 107 CFU/ml WT or mutant X. perforans amended with 0.025% Silwet L-77 7 days prior to spray inoculation with a 5 × 107-CFU/ml suspension of X. euvesicatoria strain E3-1. For the untreated checks, water was inoculated at the initial inoculation. Following spray inoculation, plants were incubated at 24 to 28°C. Three leaflets per treatment were sampled every 24 h for 96 h for quantification of E3-1 populations. Each leaflet was weighed, placed in a polyethylene bag containing 10 ml of sterile tap water, and shaken vigorously on a wrist action shaker (Burrel Co., Oakland, CA) for 20 min to dislodge surface bacteria. The leaflet weights were used to calculate bacterial populations per gram of leaf tissue. The leaf wash was then serially diluted 10-fold and plated onto NANalSm medium to selectively determine the bacterial population of strain E3-1. Populations were determined, and statistical analysis was performed as described above. Each experiment was conducted three times.
Field experiments.
Field experiments were set up in a completely randomized block design consisting of four replications. Raised beds were 0.91 m wide and were covered with white plastic mulch. Plots were arranged in paired beds that were 1.83 m from center to center, and each set of paired beds was 7.32 m apart. Plots within the paired beds were spaced 6.1 m apart. Each plot contained 20 plants spaced 457 cm apart.
Field experiments were performed with X. perforans mutant strain 91-118ΔopgHΔbcnB to evaluate antagonism to X. euvesicatoria strain E3-1. An inoculum was prepared as already described, and the bacterial suspensions were adjusted to 5 × 107 CFU/ml containing 0.025% Silwet L-77. Plants were dipped into suspensions of the 91-118ΔopgHΔbcnB strain 24 h prior to spray inoculation with a 5 × 107-CFU/ml suspension of strain E3-1. Inoculated plants were transplanted in the field 24 h after the second inoculation. Six-week-old seedlings of tomato genotype Florida 47 were used in all experiments.
Incidence of strains in lesions.
In 2004, field experiments were conducted at the North Florida Research and Education Center (Quincy) to evaluate the relative incidence of WT X. euvesicatoria strain E3-1 and 91-118ΔopgHΔbcnB after recovery from symptomatic leaf tissue compared to that obtained with the growers’ standard (described below). The experiment consisted of six treatments, (i) an uninoculated control, (ii) E3-1 plus the growers’ standard, (iii) E3-1 alone, (iv) 91-118ΔopgHΔbcnB alone, (v) E3-1 and 91-118ΔopgHΔbcnB applied every 2 weeks, and (vi) E3-1 and 91-118ΔopgHΔbcnB applied weekly. Plants in the growers’ standard group were treated on weekly rotations with acibenzolar-S-methyl (0.055 g/liter) (Actigard 50WG; Syngenta Crop Protection Inc., Greensboro, NC) or copper hydroxide (3.6 g/liter) (Kocide 2000; E. I. du Pont de Nemours and Company, Wilmington, DE) plus mancozeb (2.5 g/liter) (Manzate 75DF; E. I. du Pont de Nemours and Company, Wilmington, DE). Symptomatic leaf tissue was collected every 2 weeks beginning 35 days after transplantation (DAT). Ten to 20 leaflets were randomly collected from each plot, and bacteria were isolated from 30 lesions. Individual lesions were macerated in 75 μl of sterile deionized water, and the suspensions were streaked onto NA medium amended with 134 μg/ml pentachloronitrobenzene (28) and 50 μg/ml cycloheximide to eliminate fungal contaminants from the samples. After 4 to 5 days, individual colonies were plated onto two media to differentiate X. perforans and X. euvesicatoria (NA medium amended with the appropriate antibiotics for 91-118ΔopgHΔbcnBRif Km and E3-1SmNal). After incubation at 28°C for 4 to 5 days, colonies typical of Xanthomonas were counted and populations were calculated. The overall incidence of strains was expressed by calculating the area under the incidence progress curve (AUIPC). The AUIPC is a modification of the area under the disease progress curve: standardized AUIPC = Σ [(xi + xi − 1)/2](ti − ti −1), where x is the arcsin of the percent recovery (to normalize the data) and t is the time (in days) after inoculation.
Incidence of phyllosphere populations.
In 2005, field experiments were conducted at two locations, the Northwest Florida Research and Education Center (Quincy) and the North Central Florida Citra Research Farm of the University of Florida (Citra), to evaluate the recovery of E3-1 and 91-118ΔopgHΔbcnB from the surface of asymptomatic leaf tissue. Asymptomatic leaf tissue was sampled every 2 weeks beginning at 20 to 25 DAT. Seven leaflets were collected from each plot. Each sample was weighed, placed into a polyethylene bag (Becton Dickinson, Rutherford, New Jersey) containing 5 to 10 ml of sterile tap water, and shaken at 200 rpm for 30 to 45 min. Serial 10-fold dilutions were made in sterile tap water. A 50-μl aliquot of each dilution was plated onto two NA medium plates, one amended with 134 μg/ml pentachloronitrobenzene and/or 50 μg/ml cycloheximide with addition of antibiotics for selection of E3-1 (Smr Nalr) and the other for the 91-118ΔopgHΔbcnB mutant (Rifr Kmr). After incubation at 28°C for 4 to 5 days, colonies typical of Xanthomonas were counted and populations were calculated. The AUIPC was calculated, and data were analyzed for statistical significance as described above. Each experiment was conducted three times.
RESULTS
Comparison of WT and mutant strains for population dynamics and pathogenicity.
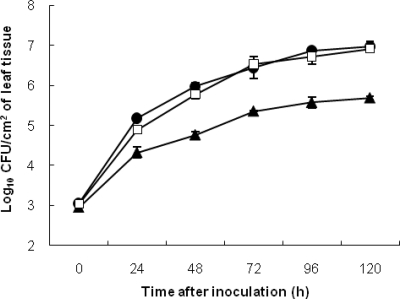
The WT 91-118 strain and strain 91-118ΔbcnB exhibited similar growth curves during the 120-h sampling period and based on the AUPPC calculations (Fig. 1). Internal populations of 91-118ΔopgHΔbcnB and 91-118ΔopgH were 1 to 1.5 log units lower than those of WT strain 91-118 and strain 91-118ΔbcnB (Fig. 1). In greenhouse experiments, the disease caused by 91-118ΔopgHΔbcnB (12%) was significantly less severe than that caused by 91-118 (39%) (Fig. 2). The 91-118::ΔopgHΔbcnB and 91-118::ΔopgH strains were compared for internal population size and the severity of the disease they caused. No significant differences were observed (data no shown).
FIG. 1.
Growth rates of WT and mutant X. perforans strains after infiltration of tomato genotype Bonny Best with 91-118ΔbcnB (□), 91-118ΔopgHΔbcnB (▴), and WT 91-118 (•). Error bars indicate standard errors.
FIG. 2.
Disease severity in tomato Bonny Best leaflets 2 weeks after dip inoculation at 5 × 106 CFU/ml with X. perforans strains 91-118 (WT; left) and 91-118ΔopgHΔbcnB (right). All dip treatments contained 0.025% Silwet L-77 in the suspension.
Antagonism assays.
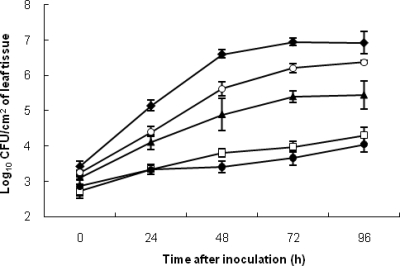
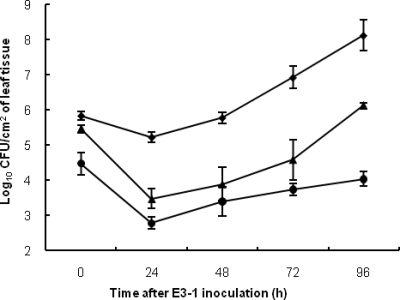
Both internal leaf tissue (Fig. 3) and phyllosphere (Fig. 4) antagonism assays performed under growth room conditions found that mutants 91-118ΔopgH and 91-118ΔopgHΔbcnB were moderately antagonistic, whereas WT 91-118 and 91-118ΔbcnB provided the greatest reduction in E3-1 populations. However, the actual differences between the WT and mutant strains were much less in the phyllosphere assay. The water control treatment consistently had significantly higher population levels and AUPPC than all of the other treatments (Fig. 3 and Table 2). When a bacterial suspension of the WT X. euvesicatoria strain (91-106) was injected into the leaflet prior to injection of a suspension of X. euvesicatoria strain E3-1SmNal cells, strain E3-1 populations were only reduced by approximately 0.5 log10 CFU/ml in both assays compared to those in the water control.
FIG. 3.
Growth rates of X. euvesicatoria strain E3-1 in leaflets of tomato genotype Bonny Best plants infiltrated at 5 × 107 CFU/ml with strain 91-118ΔbcnB (□), 91-118ΔopgHΔbcnB (▴), or 91-118 (WT; •), with WT X. euvesicatoria strain 91-106 (○), or with water (⧫), followed 18 h later by infiltration with strain E3-1 at 5 × 106 CFU/ml. Error bars indicate standard errors.
FIG. 4.
Phyllosphere antagonism assay in a growth room measuring X. euvesicatoria strain E3-1 in tomato genotype Bonny Best leaflets dip inoculated with 5 × 107-CFU/ml suspensions of strain 91-118ΔopgHΔbcnB (▴) or 91-118 (WT; •) or X. euvesicatoria strain 91-106 (⧫), followed 7 days later by spray inoculation of strain E3-1 at 5 × 107 CFU/ml. Error bars indicate standard errors.
TABLE 2.
Results of internal leaf tissue and phyllosphere antagonism experiments measuring X. euvesicatoria strain E3-1 populations when coinoculated with water, X. euvesicatoria strain E3-1, or WT or mutant X. perforans strain 91-118, measured as the AUPPC
| Treatment | AUPPCa
|
|
|---|---|---|
| Internal antagonism | Phyllosphere antagonism | |
| Water control | 115.3 (a)b | |
| X. euvesicatoria 91-106 | 101.7 (b) | 120.8 (a) |
| X. perforans 91-118 | 67.0 (d) | 69.0 (c) |
| X. perforans 91-118::ΔopgHΔbcnB | 90.8 (c) | 86.3 (b) |
AUPPC values are from assays of antagonism over a 96-h period based on recovered populations of X. euvesicatoria strain E3-1.
Values followed by the same letter are not significantly different based on the Waller-Duncan multiple-range test (P = 0.05).
Field study.
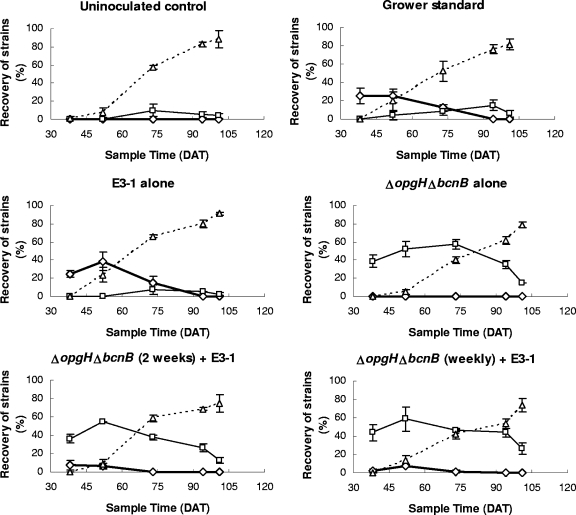
The attenuated mutant, 91-118ΔopgHΔbcnB, significantly reduced E3-1 populations in leaf tissue in the weekly and biweekly treatments in the field (Fig. 5). E3-1 was recovered from less than 5% of the samples of plants in 91-118ΔopgHΔbcnB-treated plots, compared to 26% of the plants in plots where E3-1 was applied alone (Table 3).
FIG. 5.
Quincy 2004 field experiment. Percent recovery of X. perforans strain 91-118ΔopgHΔbcnB (□), native WT strains (▵), and X. euvesicatoria strain E3-1 (⋄) from lesions from plants after the following treatments: (i) uninoculated control, (ii) E3-1 followed by the growers’ standard (copper hydroxide plus mancozeb and acibenzolar-S-methyl), (iii) E3-1 alone, (iv) 91-118ΔopgHΔbcnB alone, (v) E3-1 followed by 91-118ΔopgHΔbcnB applied every 2 weeks, and (vi) E3-1 followed by 91-118ΔopgHΔbcnB applied weekly. Error bars indicate standard errors.
TABLE 3.
Incidence and percent recovery of T1 X. euvesicatoria strain E3-1 isolates from lesions on tomato plants that received various treatments in three field experimentsa
| Treatment | AUIPC,a % recoveryb
|
||
|---|---|---|---|
| 2004, Citra | 2005
|
||
| Citra | Quincy | ||
| Uninoculated control | 0 (c),c 0 | 427 (b), 13 | 157 (b), 7 |
| Growers’ standardd | 563.8 (a), 21 | 1,271 (a), 37 | 757 (ab), 15 |
| T1 (E3-1) alone | 614.5 (a), 26 | 1,614 (a), 54 | 1,430 (a), 30 |
| 91-118::ΔopgHΔbcnB alone | 0 (c), 0 | 238 (b), 5 | 92 (b), 2 |
| 91-118::ΔopgHΔbcnB (2) + T1e | 181.0 (b), 5 | 469 (b), 12 | 890 (ab) 25 |
| 91-118::ΔopgHΔbcnB (1) + T1f | 158.2 (b), 4 | 333 (b), 8 | 477 (b), 11 |
AUIPC values are from fields evaluating E3-1 populations for each season for each treatment.
Percent recovery is the average percent recovery of E3-1 populations for the season for each treatment.
Values followed by the same letter are not significantly different based on the Waller-Duncan multiple-range test (P = 0.05).
Growers’ standard plots were treated with copper hydroxide plus mancozeb and acibenzolar-S-methyl every 2 weeks throughout the season.
The 91-118ΔopgHΔbcnB (2) + T1 plots were treated with the 91-118::ΔopgHΔbcnB strain every 2 weeks throughout the season.
The 91-118ΔopgHΔbcnB (1) + T1 plots were treated with the 91-118::ΔopgHΔbcnB strain weekly throughout the season.
In 2005 at Citra (Fig. 6) and Quincy (data not shown), the percent incidence of phyllosphere populations was determined. At both locations, the 91-118ΔopgHΔbcnB mutant produced significant reductions in E3-1 populations on plant leaf surfaces when it was applied weekly throughout the growing season (Fig. 6), and these results were equivalent to or better than those attained with the growers’ standard in both trials. At Citra, both 91-118ΔopgHΔbcnB treatments significantly reduced the recovery of E3-1 populations compared to that in plots with E3-1 alone (Fig. 6). The weekly and biweekly treatments with 91-118ΔopgHΔbcnB significantly reduced the incidence of E3-1 to 12 and 8%, respectively, in comparison with that in plots treated with the growers’ standard (37%) and E3-1 alone (54%) (Table 3).
FIG. 6.
Citra 2005 field experiment. Percent recovery of X. perforans strain 91-118ΔopgHΔbcnB (□) and X. euvesicatoria strain E3-1 (⋄) from asymptomatic leaves from plants that received the following treatments: (i) uninoculated control, (ii) E3-1 followed by the growers’ standard (copper hydroxide plus mancozeb and acibenzolar-S-methyl), (iii) E3-1 alone, (iv) 91-118ΔopgHΔbcnB alone, (v) E3-1 followed by 91-118ΔopgHΔbcnB applied every 2 weeks, and (vi) E3-1 followed by 91-118ΔopgHΔbcnB applied weekly. Error bars indicate standard errors.
At Quincy, E3-1 was recovered from 30% of the samples from plants in plots where E3-1 was applied alone (Table 3). In the treatment where 91-118ΔopgHΔbcnB was applied every 2 weeks, the frequency of recovery of E3-1 populations was not significantly different from that where E3-1 alone was applied. Weekly application of 91-118ΔopgHΔbcnB, however, significantly reduced the recovery of E3-1 populations compared to that achieved by treatments where the E3-1 strain was applied alone (an approximately 65% reduction) (Table 3).
DISCUSSION
We demonstrated that the pathogenically attenuated X. perforans mutant maintained sufficient populations on the leaf surface to suppress X. euvesicatoria while causing minimal disease. We selected a mutant with the previously described osmoregulated periplasmic glucan gene opgHXcv for our model attenuated mutant (22) because of its antagonism toward X. euvesicatoria (36) and reduced virulence when inoculated into susceptible plants. Our greenhouse experiments confirmed that both 91-118ΔopgH and 91-118ΔopgHΔbcnB were attenuated based on their reduced growth curves in plant tissue versus that of the WT strain and the inoculation experiment in which 91-118ΔopgHΔbcnB also caused significantly less disease. Although mutants 91-118ΔopgH and 91-118ΔopgHΔbcnB were effective at reducing X. euvesicatoria populations, there were no significant differences between these mutants in the in planta or phyllosphere assays conducted in the greenhouse. However, based on the work of Hert et al. (9), bcnB was deleted in the attenuated strain because in that work it was observed in field experiments that larger populations of the 91-118ΔbcnB mutant than of the WT strain persisted in the field.
It is important to note that although 91-118ΔopgHΔbcnB was not effective at reducing internal populations, it did reduce external populations of the sensitive strain. Lindemann et al. (17) observed a direct correlation between phyllosphere populations and the occurrence of disease. This correlation suggests that a reduction in X. euvesicatoria phyllosphere populations could result in a reduction in disease incidence in the field. The phyllosphere antagonism experiments suggest that a strategy for the biological control of X. euvesicatoria by 91-118ΔopgH (A. P. Hert and J. B. Jones, unpublished data) or 91-118ΔopgHΔbcnB may be effective in controlling disease by reducing phyllosphere populations below the threshold level necessary for lesion development. No additional antagonism was observed with the additional ΔbcnB mutant in the 91-118ΔopgH background (9); however, the benefit in terms of the apparent competitive advantage of the ΔbcnB mutant observed in the field was the rationale for using the double mutant, although no definitive proof in a field study was available at the time of the experimentation.
The field experiments, although less definitive, corroborated the greenhouse data in terms of the ability of the attenuated mutant to suppress X. euvesicatoria. In 2004, two hurricanes during the season introduced high populations of WT X. perforans into the field plots, which interfered with the interaction of E3-1 and 91-118ΔopgHΔbcnB. X. perforans was previously shown to be insensitive to the bacteriocins produced by other X. perforans strains (36). Introduction of nonattenuated X. perforans strains during the hurricanes resulted in predominance of the newly introduced X. perforans strain. Data collected in the early stages of the experiment suggested that weekly and biweekly treatments with 91-118ΔopgHΔbcnB significantly reduced X. euvesicatoria populations. These findings suggest that the use of this control method would be limited to locations where limited introductions of X. perforans exist. Disease severity was not measured due to the mid-season ingression of native X. perforans populations; however, it was observed that all treatments containing 91-118ΔopgHΔbcnB exhibited lower disease severity early in the season.
In the 2005 field experiments, phyllosphere populations were taken due to low disease severity (<10%) within the trials. In both experiments, 91-118ΔopgHΔbcnB effectively reduced the incidence of X. euvesicatoria and weekly application of 91-118ΔopgHΔbcnB reduced X. euvesicatoria populations significantly more than did the growers’ standard in two of the three trials and was equivalent in the third trial. This reduction in X. euvesicatoria populations is similar to previous field data obtained with 91-118ΔbcnB (9). Hert et al. (9) found that coinoculation of 91-118ΔbcnB and an X. euvesicatoria strain in the field yielded less than 5% recovery of the X. euvesicatoria strain over all of the seasons tested. These results suggest that the 91-118ΔopgHΔbcnB attenuated mutant has potential as a biological control agent for reducing X. euvesicatoria populations.
The reduction in X. euvesicatoria populations by 91-118ΔopgHΔbcnB in antagonism assays does indicate that it is less inhibitory than 91-118ΔbcnB. This suggests that bacteriocin expression could have also been affected by the ΔopgH mutant as a result of reduced internal populations. Weekly application of strain 91-118ΔopgHΔbcnB was more effective at reducing X. euvesicatoria populations than previously observed in similar experiments with hrp mutant X. perforans (18). The 91-118ΔopgHΔbcnB mutant internally colonizes leaf tissue more effectively than nonpathogenic strains do (22) and produced results similar to those observed by Etchebar et al. (6). Etchebar found that an hrcV mutant of R. solanacearum (6) colonized the stems of tomato plants more effectively than nonpathogenic mutants did.
Although the opgHXcv mutant was effective at suppressing X. euvesicatoria populations, there is potential for identifying other gene targets that can help improve biological control efficacy. Several other pathogenicity factors and associated genes have previously been described in X. euvesicatoria, including genes associated with the hrp system (hpaA, hpaB, hpaC), avirulence genes (avrBs2, xopA, xopD), and pathogenicity factors such as gum genes (1, 2, 10, 24, 25, 38). Previously, we tested these pathogenicity-associated genes for the phenotypes they produce in X. perforans and their potential use for creating further biocontrol agents with attenuated pathogenicity (9). When several of the genes were mutated in X. perforans strain 91-118, there were associated pathogenicity and growth curve reductions in planta. These mutants or mutants with changes in other pathogenicity-associated genes may improve our attenuated-pathogenicity biological control model system by allowing better internalization and subsequent competition between X. euvesicatoria and X. perforans populations without effects detrimental to the plant.
Sequencing of the X. euvesicatoria genome has provided significant new possibilities for developing candidates with attenuated pathogenicity. In 2005, Thieme et al. (34) published the X. euvesicatoria genome sequence and estimated more than 480 putative pathogenicity factors and associated genes. These genes were grouped into six categories, (i) secretion systems, (ii) flagellum, (iii) secreted proteins (via type III secretion system), (iv) detoxification, (v) surface structure and adhesion, and (vi) quorum sensing. Genomic sequencing of bacteria provides an opportunity to exploit these genes for our use.
Further research is needed to help us understand the interactions between pathogens. In this study, we attempted to create a bacterial antagonist with decreased virulence that is able to maintain a level of antagonism similar to that of the WT. Recent information from related bacteria and genomic sequences can be used to provide opportunities to improve our understanding of how pathogenic bacteria colonize and subsequently infect the host. Continued exploration of new innovative ideas will help us to use this knowledge in effective ways.
Acknowledgments
This research was supported by the Florida Agricultural Experiment Station and by a USDA T-STAR grant (2005-34135-16285-S, to J. B. Jones and M. T. Momol).
Footnotes
Published ahead of print on 13 March 2009.
REFERENCES
- 1.Büttner, D., C. Lorenz, E. Weber, and U. Bonas. 2006. Targeting of two effector protein classes to the type III secretion system by a HpaC- and HpaB-dependent protein complex from Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 59:513-527. [DOI] [PubMed] [Google Scholar]
- 2.Büttner, D., D. Gurlebeck, L. D. Noël, and U. Bonas. 2004. HpaB from Xanthomonas campestris pv. vesicatoria acts as an exit control protein in type III-dependent protein secretion. Mol. Microbiol. 54:755-768. [DOI] [PubMed] [Google Scholar]
- 3.Conover, R. A., and N. A. Gerhold. 1981. Mixtures of copper and maneb or mancozeb for control of bacterial spot of tomato and their compatibility for control of fungus diseases. Proc. Fla. State Hort. Soc. 94:154-156. [Google Scholar]
- 4.Daniels, M. J., C. E. Barber, D. C. Turner, W. G. Cleary, and M. Sawage. 1984. Isolation of mutants of Xanthomonas campestris pv. campestris showing altered pathogenicity. J. Gen. Microbiol. 130:2447-2455. [Google Scholar]
- 5.Dianese, A. C., P. Ji, and M. Wilson. 2003. Nutritional similarity between leaf-associated nonpathogenic bacteria and the pathogen is not predictive of efficacy in biological control of bacterial spot of tomato. Appl. Environ. Microbiol. 69:3484-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etchebar, C., D. Trigalet-Demery, G. Gijsegem, J. Vasse, and A. Trigalet. 1998. Xylem colonization by a hrcV mutant of Ralstonia solanacearum is a key factor for the efficient biological control of tomato bacterial wilt. Mol. Plant-Microbe Interact. 11:869-877. [Google Scholar]
- 7.Flaherty, J. E., J. B. Jones, B. K. Harbaugh, G. C. Somodi, and L. E. Jackson. 2000. Control of bacterial spot of tomato in the greenhouse and field with h-mutant bacteriophages. Hortscience 35:882-884. [Google Scholar]
- 8.Frey, P., P. Prior, C. Marie, A. Kotoujansky, D. Trigalet-Demery, and A. Trigalet. 1994. Hrp− mutants of Pseudomonas solanacearum as potential biocontrol agents of tomato bacterial wilt. Appl. Environ. Microbiol. 60:3175-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hert, A. P., P. D. Roberts, M. T. Momol, G. V. Minsavage, S. M. Tudor-Nelson, and J. B. Jones. 2005. Relative importance of bacteriocin-like genes in antagonism of Xanthomonas perforans tomato race 3 to Xanthomonas euvesicatoria tomato race 1 strains. Appl. Environ. Microbiol. 71:3581-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huguet, E., K. Hahn, K. Wengelnik, and U. Bonas. 1998. hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiol. 29:1379-1390. [DOI] [PubMed] [Google Scholar]
- 11.Ji, P., and M. Wilson. 2003. Enhancement of population size of a biological control agent and efficacy in control of bacterial speck of tomato through salicylate and ammonium sulfate amendments. Appl. Environ. Microbiol. 69:1290-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, J. B., H. Bouzar, G. C. Somodi, R. E. Stall, K. Pernezny, G. El-Morsy, and J. W. Scott. 1998. Evidence for the preemptive nature of tomato race 3 of Xanthomonas campestris pv. vesicatoria in Florida. Phytopathology 88:33-38. [DOI] [PubMed] [Google Scholar]
- 13.Jones, J. B., G. H. Lacy, H. Bouzar, R. E. Stall, and N. W. Schaad. 2004. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 27:755-762. [DOI] [PubMed] [Google Scholar]
- 14.Jones, J. B., R. E. Stall, and H. Bouzar. 1998. Diversity among xanthomonads pathogenic on pepper and tomato. Annu. Rev. Phytopathol. 36:41-58. [DOI] [PubMed] [Google Scholar]
- 15.Jones, J. B., R. E. Stall, J. W. Scott, G. C. Somondi, H. Bouzar, and N. C. Hodge. 1995. A third tomato race of Xanthomonas campestris pv. vesicatoria. Plant Dis. 79:395-398. [Google Scholar]
- 16.Koller, W. 1998. Chemical approaches to managing plant pathogens, p. 337-376. In J. R. Ruberson (ed.), Handbook of integrated pest management. Dekker, New York, NY.
- 17.Lindemann, J., D. C. Arny, and C. D. Upper. 1984. Use of an apparent infection threshold population of Pseudomonas syringae to predict incidence and severity of brown spot of bean. Phytopathology 74:1334-1339. [Google Scholar]
- 18.Liu, T. 1998. Biological control of tomato bacterial spot with a Hrp− mutant of Xanthomonas campestris pv. vesicatoria. M.S. thesis. University of Florida, Gainesville.
- 19.Louws, F. J., M. Wilson, H. L. Campbell, D. A. Cuppels, J. B. Jones, P. B. Shoemaker, F. Sahin, and S. A. Miller. 2001. Field control of bacterial spot and bacterial speck of tomato using a plant activator. Plant Dis. 85:481-488. [DOI] [PubMed] [Google Scholar]
- 20.Marco, G. M., and R. E. Stall. 1983. Control of bacterial spot of pepper initiated by strains of Xanthomonas campestris pv. vesicatoria which differ in sensitivity to copper. Plant Dis. 67:779-781. [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 22.Minsavage, G. V., M. B. Mudgett, R. E. Stall, and J. B. Jones. 2004. Importance of opgHXcv of Xanthomonas campestris pv. vesicatoria in host-parasite interactions. Mol. Plant-Microbe Interact. 17:152-161. [DOI] [PubMed] [Google Scholar]
- 23.Moss, W., J. M. Byrne, and M. Wilson. 1997. Interactions between Xanthomonas axonopodis pv. vesicatoria hrp mutants and the pathogenic parent. Phytopathology 87:S68. [Google Scholar]
- 24.Noël, L., F. Thieme, D. Nennstiel, and U. Bonas. 2002. Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noël, L., F. Thieme, D. Nennstiel, and U. Bonas. 2001. cDNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 41:1271-1281. [DOI] [PubMed] [Google Scholar]
- 26.Obradovic, A., J. B. Jones, M. T. Momol, B. Balogh, and S. M. Olson. 2004. Management of tomato bacterial spot in the field by foliar applications of bacteriophages and SAR inducers. Plant Dis. 88:736-740. [DOI] [PubMed] [Google Scholar]
- 27.Pohornezny, K., and R. B. Volin. 1983. The effect of bacterial spot on yield and quality of fresh market tomatoes. Hortscience 18:69-70. [Google Scholar]
- 28.Ribeiro, O. K. 1978. A source book of the genus Phytophthora, p. 55-57. J. Cramer, Stuttgart, Germany.
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Shaner, G., and R. Finney. 1997. The effect of nitrogen fertilization on the expression of slow mildewing resistance in Knox wheat. Phytopathology 67:1051-1056. [Google Scholar]
- 31.Stall, R. E., and P. L. Thayer. 1962. Streptomycin resistance of the bacterial spot pathogen and control with streptomycin. Plant Dis. Rep. 46:389-392. [Google Scholar]
- 32.Stall, R. E., D. C. Loschke, and J. B. Jones. 1986. Linkage of copper resistance and avirulence loci on a self-transmissible plasmid in Xanthomonas campestris pv. vesicatoria. Phytopathology 76:240-243. [Google Scholar]
- 33.Tagg, J. R., and A. R. McGiven. 1971. Assay system for bacteriocins. Appl. Microbiol. 21:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thieme, F., R. Koebnik, T. Bekel, C. Berger, J. Boch, D. Büttner, C. Caldana, L. Gaigalat, A. Goesmann, S. Kay, O. Kirchner, C. Lanz, B. Linke, A. C. McHardy, F. Meyer, G. Mittenhuber, D. H. Nies, U. Niesbach-Klösgen, T. Patschkowski, C. Rückert, O. Rupp, S. Schneiker, S. C. Schüster, F.-J. Vorhölter, E. Weber, A. Pühler, U. Bonas, D. Bartels, and O. Kaiser. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trigalet, A., and D. Trigalet-Demery. 1990. Use of avirulent mutants of Pseudomonas solanacearum for the biological control of bacterial wilt of tomato plants. Physiol. Mol. Plant Physiol. 36:27-38. [Google Scholar]
- 36.Tudor-Nelson, S. M., G. V. Minsavage, R. E. Stall, and J. B. Jones. 2003. Bacteriocin-like substances from tomato race 3 strains of Xanthomonas campestris pv. vesicatoria. Phytopathology 93:1421. [DOI] [PubMed] [Google Scholar]
- 37.Wang, J. F., J. B. Jones, J. W. Scott, and R. E. Stall. 1990. A new race of the tomato group of strains of Xanthomonas campestris pv. vesicatoria. Phytopathology 80:1070. [Google Scholar]
- 38.Wilson, M., H. L. Campbell, J. B. Jones, and D. A. Cuppels. 2002. Biological control of bacterial speck of tomato under field conditions at several locations in North America. Phytopathology 92:1284-1292. [DOI] [PubMed] [Google Scholar]