Abstract
Plant surfaces, colonized by numerous and diverse bacterial species, are often considered hot spots for horizontal gene transfer (HGT) between plants and bacteria. Plant DNA released during the degradation of plant tissues can persist and remain biologically active for significant periods of time, suggesting that soil or plant-associated bacteria could be in direct contact with plant DNA. In addition, nutrients released during the decaying process may provide a copiotrophic environment conducive for opportunistic microbial growth. Using Acinetobacter baylyi strain BD413 and transplastomic tobacco plants harboring the aadA gene as models, the objective of this study was to determine whether specific niches could be shown to foster bacterial growth on intact or decaying plant tissues, to develop a competence state, and to possibly acquire exogenous plant DNA by natural transformation. Visualization of HGT in situ was performed using A. baylyi strain BD413(rbcL-ΔPaadA::gfp) carrying a promoterless aadA::gfp fusion. Both antibiotic resistance and green fluorescence phenotypes were restored in recombinant bacterial cells after homologous recombination with transgenic plant DNA. Opportunistic growth occurred on decaying plant tissues, and a significant proportion of the bacteria developed a competence state. Quantification of transformants clearly supported the idea that the phytosphere constitutes a hot spot for HGT between plants and bacteria. The nondisruptive approach used to visualize transformants in situ provides new insights into environmental factors influencing HGT for plant tissues.
Despite the annually increasing acreage planted with genetically modified plants worldwide, the ongoing debate on their ecological safety is controversial and gave impetus to different studies of the putative horizontal gene transfer (HGT) of recombinant DNA from plant to bacteria (12, 30). Research regarding the fate of plant transgenes in environmental microbial communities is driven by practical societal concerns related to the potential dissemination of antibiotic resistance determinants in the environment and by fundamental evolution questions about gene transfer between species and kingdoms. Different parts of a plant (globally defined as the phytosphere) support the growth of numerous and diverse bacteria that colonize the surfaces or internal tissues and display advantageous, neutral, or pathogenic functions toward the plant (1, 9, 22). However, the plant as a whole is exposed to many environmental challenges and does not always provide the same favorable conditions for bacterial growth. The latter depends on several factors, such as the presence of nutrients, moisture, shelter from desiccation and UV, and shelter from grazing and predation, all of which fluctuate rapidly and are heterogeneously distributed in and on the plant. Hence, bacterial growth seems to occur mostly in nutrient-rich, few, and localized microhabitats on plant surfaces where bacteria would form aggregates (17, 20, 21, 22, 33). The presence of large clusters of bacteria at sites of relative nutrient abundance on plant surfaces might also increase the potential for metabolic and genetic exchange (19). For example, bacterial growth and relatively high rates of transfer for a conjugative plasmid were reported to occur on plant surfaces (2, 3, 5). Similarly, availability of growth substrates, high bacterial density, and the presence of solid leaf surfaces were thought to induce gene transfer by conjugation in the phyllosphere at significantly high rates (26).
Of the three mechanisms of bacterial HGT, natural transformation is considered to be the only one that could be effectively implicated in the transfer of DNA from transgenic plants to bacteria (4, 25). Although plants support bacterial growth, only putative evidence of DNA released by naturally degrading plant tissue being involved in a natural transformation process exists (23). Ceccherini et al. (6) showed, for example, that although most of the plant DNA was degraded within a short time by plant nucleases in planta during the process of plant decay, a measurable fraction escaped degradation and was still able to transform a recipient soil isolate in vitro (6).
In order to assess plant-to-bacteria gene transfer, some studies have been conducted with different plant compartments. For example, the possibility for Acinetobacter baylyi strain BD413 to grow opportunistically in Ralstonia solanacearum-infected plant tissues revealed a new niche for this soil bacterium: the pathosphere. Moreover, this bacterium could be naturally transformed therein by artificially added or indigenous transgenic DNA (10, 11). Yet, other plant compartments could be as propitious to HGT; for example, the residuesphere (i.e., the naturally degrading plant material at the interface with soil) has been shown to provide conditions for growth and conjugal gene transfer between indigenous soil bacteria (7, 34). The litter and the residues of annual crops represent an important amount of final plant production, which are often left in the field after harvest and, in most cases, account for up to 60% of the world's plant biomass (14).
The assessment of natural transformation events in the soil or the phytosphere has, however, revealed several methodological challenges and biases, since quantification of transformation events has often been conducted with a cultivation-based approach requiring the plating of recipient bacteria on selective media supplemented with antibiotics. Antibiotic resistance determinants are widespread in soil environments, providing a technical intricacy in the discrimination between recipients with newly acquired traits and indigenous antibiotic-resistant flora with naturally fitted analogous genes. In addition, discrepancies between transformation frequencies determined on plates and those determined by cell densitometry revealed that the latter were usually higher by 2 orders of magnitude (37), leading to an underestimation of the phenomenon in natural settings. In addition, the uncertainty of whether each colony enumerated on a plate belongs to a single independent transformation event or is one of many clones (from one event) extracted from the sample makes transformation frequency calculations imprecise. Another drawback of the plating step is the disruptive sampling of material to rescue transformants, which averages the frequency calculations over the entire sample. Due to the spatial heterogeneity of available nutrients and biologically active DNA, localized spots, which are most conducive for HGT, might be delimited at microscale. However, to date, knowledge about the effective topology is lacking, although the heterogeneity of bacterial growth on plant surfaces has been shown (15, 20, 21, 22).
The objectives of this study were to determine whether, during the natural or pathogen-induced decay of plant tissues, specific niches could be shown to foster bacterial growth, to develop a competence state, and to possibly acquire exogenous plant DNA using the Acinetobacter baylyi strain BD413 via natural transformation. Visualization of bacterial colonization plant material and detection of HGT events were performed at the leaf and bacterial scales using a cultivation-independent assay that relies upon a bioreporter tool (32). Microcosm-based experiments revealed that bacterial growth and competence development occur in different compartments of the plant. Isolation and direct visualization of transformants in situ suggest that some compartments of the phytosphere can be regarded as environmental hot spots for HGT.
MATERIALS AND METHODS
Plant material.
Wild-type and transplastomic tobacco plants (Nicotiana tabacum L. cv. PBD6) were grown in compost potting soil in a greenhouse at 23°C (±2°C), with a daily regimen of 16 h of light and 8 h of darkness. The relative humidity rate was, on average, 55% during the day and 72% at night. Transplastomic plants harbored the transgenic aadA gene (conferring resistance to spectinomycin and streptomycin) cloned between the chloroplastic genes rbcL and accD (11) and contained ca. 7,000 copies of the transgene per plant cell. For each experiment, entire leaves were sampled when plants were 9 to 10 weeks old.
Bacterial strains, plasmids, and culture media.
Escherichia coli strain DH5α(pCEA) harbored the pCEA plasmid which contained the aadA gene flanked with part of the rbcL and accD tobacco plastid sequences (11). The strain was grown at 37°C on Luria-Bertani modified (LBm) medium (Bacto tryptone extract, 10 g/liter; NaCl, 5 g/liter; yeast extract, 5 g/liter in 1 liter distilled water) supplemented with ampicillin (50 μg/ml) and spectinomycin (50 μg/ml) (Sigma, St. Louis, MO). The naturally transformable Acinetobacter baylyi strain BD413 was chosen as the model bacterium; a first strain harbored the recombinant plasmid pBAB2 that contained a recombinogenic site with plastidic tobacco sequences rbcL and accD (11) to favor homologous recombination (Fig. 1A). The strain was cultured at 28°C on LBm medium supplemented with ampicillin (50 μg/ml) and nalidixic acid (20 μg/ml) (Sigma, St. Louis, MO). Transformants were selected on LBm medium containing ampicillin (50 μg/ml), nalidixic acid (20 μg/ml), and spectinomycin (50 μg/ml) (Sigma, St. Louis, MO) after 2 days of incubation at 28°C. A. baylyi strain BD413(rbcL-aadA::gfp) and its nonfunctional counterpart, A. baylyi strain BD413(rbcL-ΔPaadA::gfp) (Fig. 1B), were used to quantify and visualize total bacterial populations and transformants in situ, respectively (32). The two strains were grown at 28°C on LBm medium supplemented with kanamycin (50 μg/ml) and rifampin (50 μg/ml), and for the A. baylyi strain BD413(rbcL-aadA::gfp) and rescued transformants, with spectinomycin (50 μg/ml) (Sigma, St. Louis, MO). Ralstonia solanacearum strain K60 was cultured in B solid medium supplemented with 12 μg/ml gentamicin at 28°C for 48 h or in B broth until reaching an optical density at 600 nm (OD600) of 0.8.
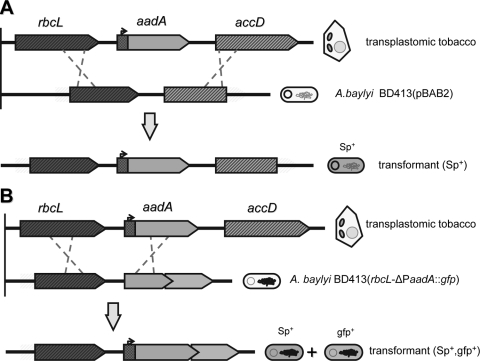
FIG. 1.
Schematic representation of homologous recombination events in A. baylyi reporter strains used in this study. (A) A. baylyi strain BD413(pBAB2) harbors the pBAB2 plasmid (11) which contains two neighboring chloroplastic gene sequences (accD and rbcL), favoring recombination with homologous sequences of the transplastomic tobacco DNA flanking the aadA transgene. After homologous recombination with plant DNA, the transgene is inserted in the plasmid and confers a spectinomycin-resistant phenotype (Sp+) to the transformed cell. (B) A. baylyi strain BD413(rbcL-ΔPaadA::gfp) carries a promoterless aadA::gfp fusion downstream of rbcL, cloned in the chromosome between the lipB and the lipA genes. The functionality of the aadA::gfp fusion is restored after homologous recombination with the transgenic DNA, conferring both antibiotic-resistant and green fluorescent (gfp+) phenotypes to the recombinant cells.
DNA extraction.
Plant genomic DNA was extracted from entire tobacco leaves. Sampled leaves were individually ground in liquid nitrogen after removal of the central vein. Plant DNA was extracted using the DNeasy plant kit (Qiagen, Mannheim, Germany) by following the manufacturer's instructions. Plasmid pCEA was isolated from E. coli DH5α(pCEA) using the QIAfilter plasmid midikit (Qiagen, Mannheim, Germany) by following the manufacturer's instructions. Total bacterial and plasmid DNA extractions from putative recombinants and recipients were performed using the NucleoSpin tissue and plasmid kits, respectively (Macherey-Nagel, Düren, Germany), according to the manufacturer's instructions.
Plant inoculation and incubation.
Intact and ground tobacco leaf tissues were inoculated with A. baylyi strain BD413. Leaf discs (2 cm in diameter) were cut from tobacco leaves and soaked twice in a 5% (wt/vol) bleach solution for 10 min, then rinsed twice in sterile distilled water, and dried on sterile Whatman paper (Whatman, Maidstone, United Kingdom). Half of the discs were ground in distilled sterile water in a proportion of 1 g/5 ml with an Ultra-Turrax T25 homogenizer operating at 25,000 rpm (Ystral GmbH, Dottingen, Germany) for 5 min. Intact leaf discs (ca. 0.1 g) and ground leaf tissues (ca. 0.2 g) were placed on 2.5-cm-diameter, 0.2-μm Isopore membrane filters (Millipore, Billerica, MA) lying in petri dishes containing a layer of agar supplemented with cycloheximide (200 μg/ml). A. baylyi strain BD413(pBAB2) was cultured overnight in LBm broth and then diluted 10-fold in fresh medium and left growing until it reached an OD corresponding to 2 × 108 CFU/ml. Cells were then centrifuged, washed, and resuspended in sterile saline solution (0.8% NaCl). One hundred microliters of a 1,000-fold dilution corresponding to a final inoculum of ca. 104 CFU was spotted in droplets of ca. 10 μl on ground or intact leaves and left to incubate at 28°C for up to 15 days, with three replicates per leaf. In parallel and as a negative control, 100 μl of sterile saline solution was also deposited on leaves. Visualization of the colonization pattern of plant tissues was performed after inoculating ground or intact wild-type tobacco leaf discs with A. baylyi strains BD413(rbcL-aadA::gfp) and BD413(rbcL-ΔPaadA::gfp). Inoculated leaf tissues were incubated at 28°C for up to 15 days.
In order to detect potential gene transfer following pathogen-induced plant decay, individual transplastomic leaves were inoculated with a mixture of Ralstonia solanacearum strain K60 and A. baylyi strain BD413(rbcL-ΔPaadA::gfp). Bacterial cells of the two strains were centrifuged and resuspended in 0.8% NaCl, and the cell concentration was adjusted to ca. 2 × 108 CFU/ml. The final inoculum was prepared by mixing 1 ml of the R. solanacearum K60 cell suspension with 2 ml of the A. baylyi cell suspension. Three hundred microliters of the final suspension was injected into the central vein of each tobacco leaf. Leaves were placed in petri dishes containing a layer of water agar supplemented with cycloheximide and incubated at 28°C until the appearance of severe symptoms of wilting (for up to 11 days).
Growth and competence development of A. baylyi strain BD413.
Growth and competence development of A. baylyi strain BD413(pBAB2) on wild-type tobacco leaf tissues was estimated by quantifying population size and transformation frequency in vitro, respectively. Three filters were sampled every 24 h for up to 7 days and individually washed in 5 ml of saline solution (NaCl, 0.8%) by vortexing the tubes for 5 min at maximum speed. Total and revertant population sizes were determined by serially diluting and plating suspensions on solid agar media supplemented with the appropriate antibiotics. Assessment of competence was determined by exposing 500 μl of bacterial suspensions obtained by washing leaf tissue to 1 μg of the pCEA plasmid in 50 μl of ultrapure water for 2 h at 28°C. Cell suspensions were serially diluted in 0.8% NaCl, plated on LBm agar medium supplemented with the appropriate antibiotics, and incubated for 2 days at 28°C before total population sizes and transformants were quantified.
Growth and competence development were also determined for cells grown in LBm medium. A. baylyi strain BD413(pBAB2) cells were grown overnight in LBm medium, diluted 25-fold with fresh LBm medium supplemented with the appropriate antibiotics, and cultured for 54 h. During this time interval, 350 μl of cells was periodically sampled and exposed to 1 μg of the pCEA plasmid in 50 μl of ultrapure water, incubated for 2 h at 28°C, and diluted and plated on LBm medium supplemented with the appropriate antibiotics.
Natural transformation of A. baylyi strain BD413 in vitro and in the phytosphere.
The transformation abilities of the different strains were initially determined in vitro in order to determine transformation frequencies under optimal conditions. Aliquots of exponentially growing A. baylyi strain BD413(pBAB2) and A. baylyi strain BD413(rbcL-ΔPaadA::gfp) (500 μl; OD600, 0.8) were exposed to pCEA plasmid DNA, purified transplastomic DNA, and leaf homogenate. Each experiment was done in triplicate for each assay. A. baylyi cells inoculated onto intact or ground leaf discs were recovered by washing each disc in a Falcon tube with 5 ml of sterile saline solution and vortexing the tubes for 5 min at maximum speed. Individual R. solanacearum-infected leaf segments (1 by 2 cm) were randomly cut per leaf, weighed, and used to quantify and visualize A. baylyi transformants. Quantification was performed after crushing infected segments in 5 ml sterile saline solution with the Ultra-Turrax T25 homogenizer. Population sizes were determined by serially diluting and plating suspensions on solid agar media supplemented with the appropriate antibiotics. In order to avoid ex planta transformants, suspensions were treated with DNase I (0.1 mg/ml) at 37°C for 1 h before plating all the remaining suspension on selective LBm solid media for the enumeration of transformant cells.
The stability of the different constructs for the bioreporter strains of A. baylyi was assessed by determining the number of revertants (spontaneous mutants resistant to spectinomycin) in vitro and in situ. Cells of the two strains were grown in LBm broth for up to 7 days or on ground leaf tissues of wild-type tobaccos for up to 15 days, and total population size and revertant population size were determined every 24 h by plating on LBm medium and spectinomycin-supplemented LBm solid medium. Reversion frequency was determined from the number of revertants (i.e., spontaneous mutants resistant to spectinomycin) observed divided by the total bacterial size.
Analysis of transformants.
The presence of the transgene in spectinomycin-resistant clones of A. baylyi strain BD413(pBAB2) was confirmed by PCR using previously published specific primers p1351cpl2up and p416 (6). These primers targeted an 853-bp sequence and were complementary to a part of the rbcL and the aadA gene. Confirmation of the rescue of the marker system in the A. baylyi strain BD413(rbcL-ΔPaadA::gfp) spectinomycin-resistant clones was performed by PCR on a template of 1 μl of genomic DNA. Primers Promo F (5′ ATCTTTCTATTGTTGTCTTGGAT-3′) and Promo R (5′-GGTCACCGTAACCAGCAAAT CAA-3′) were used to discriminate between the recombinant clones containing a functional promoter (size of the amplicon = 333 bp) and the recipients bearing a deletion in the promoter (size of the amplicon = 190 bp). The PCR consisted of a denaturation step at 95°C for 4 min, followed by 35 cycles consisting of 40 s at 95°C, 40 s at 56°C, and 40 s at 72°C, and a final extension step at 72°C for 5 min.
Visualization of bacterial cells on decaying or intact plant tissues.
The spatial localization of total and transformed bacterial cells was determined directly in situ by epifluorescence microscopy. Processed samples consisted of either intact or ground transplastomic tobacco leaves that had been inoculated with A. baylyi strains BD413(rbcL-ΔPaadA::gfp) and BD413(rbcL-aadA::gfp) or symptomatic leaves coinoculated with R. solanacearum K60 and A. baylyi strain BD413(rbcL-ΔPaadA::gfp). Three samples consisting of either intact or ground leaf tissues were selected for each treatment. Four segments of approximately 1 cm2 were cut from each intact leaf and placed on a glass slide. Ground leaf tissues incubated on a filter membrane were observed undisturbed under the microscope. A drop of Aqua-Poly/Mount mounting medium (Polysciences, Inc., Warrington, PA) was placed in the center of a coverslip, which was then gently pressed down onto the leaf material. To visualize total bacterial cells, a solution of ethidium bromide (final solution, 1 μg/ml) was mixed with Aqua-Poly/Mount (Polysciences, Inc., Warrington, PA) before mounting. Samples were then immediately observed using an Axioskop Zeiss microscope equipped with a 10×/0.30, 20×/0.50, 40×/0.75, or 100×/1.30 (objective/numerical aperture) Plan-Neofluar (Zeiss, Inc., Oberkochen, Germany). Filter set 10 (excitation bandpass [BP], 450/490 nm; beam splitter Farbteiler [FT], 510 nm; emission BP, 515/565 nm) was used to visualize green fluorescence emitted by cells expressing the green fluorescent protein (GFP). Total cells stained with ethidium bromide were visualized using filter set 15 (excitation BP, 534/558 nm; beam splitter FT, 580 nm; emission longpass [LP], 590 nm). Images were captured with an AxioCam MRc5 digital camera and the AxioVision version 4.3 software (Zeiss, Inc., Oberkochen, Germany).
RESULTS
Growth of Acinetobacter on plant tissues.
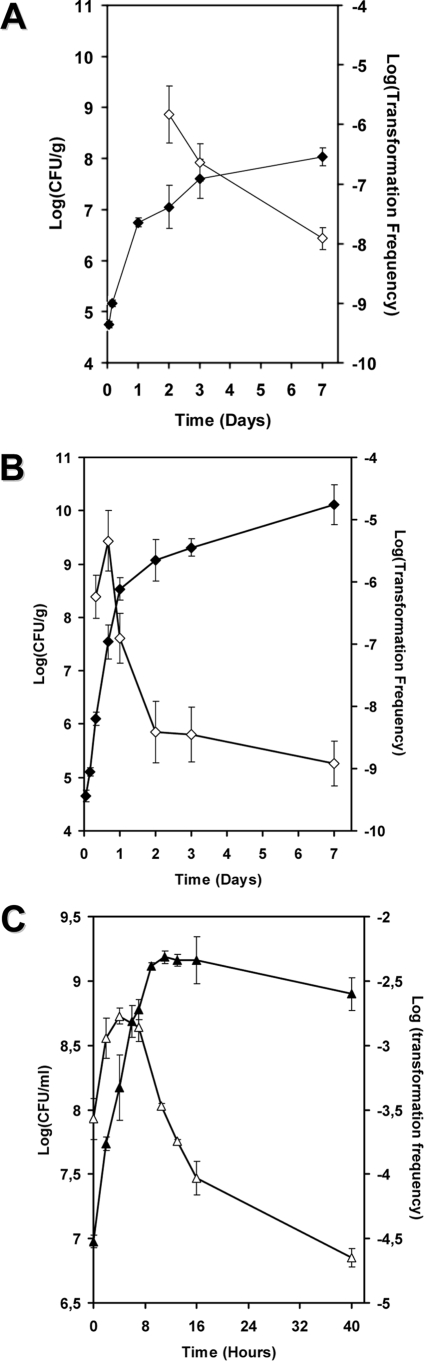
A. baylyi strain BD413(pBAB2) cells grew onto intact leaves. The initial amount of cells inoculated onto intact leaves was (2.3 ± 0.3) × 104 cells. Population sizes, which were determined at different intervals, indicated effective and consistent growth of the strain from the time of inoculation (Fig. 2A). The number of recovered cells increased from (6.0 ± 1.2) × 105 CFU/g (fresh weight) at approximately 4 h after inoculation to (1.1 ± 1.5) × 108 CFU/g (fresh weight) after 1 week (stationary phase). Likewise, when the same number of cells of A. baylyi was spotted on crushed leaf discs simulating naturally decaying tissues, the number of recovered cells indicated that consistent and rapid growth had occurred. After 24 h, they had increased by 4 orders of magnitude, reaching a density of (3.4 ± 1.1) × 108 CFU/g of fresh tissue (Fig. 2B). After 7 days of incubation, ground leaves were supporting a population size of (1.3 ± 2.4) × 1010 CFU/g (fresh weight). Kinetic growth of A. baylyi BD413 was determined in vitro as a control. The population size increased exponentially in the first 16 h and entered stationary phase thereafter (Fig. 2C).
FIG. 2.
Growth (filled diamonds) and competence development (open diamonds) kinetics of A. baylyi strain BD413(pBAB2) inoculated onto leaf surfaces (A) or ground leaf discs (B) of wild-type tobacco plants and incubated for 7 days or inoculated into LBm broth and grown for 40 h (C). Competence development is expressed as the log number of transformation frequencies (transformants/recipients) determined after exposure of the bacterial suspensions recovered from leaf tissues to the plasmid pCEA in vitro.
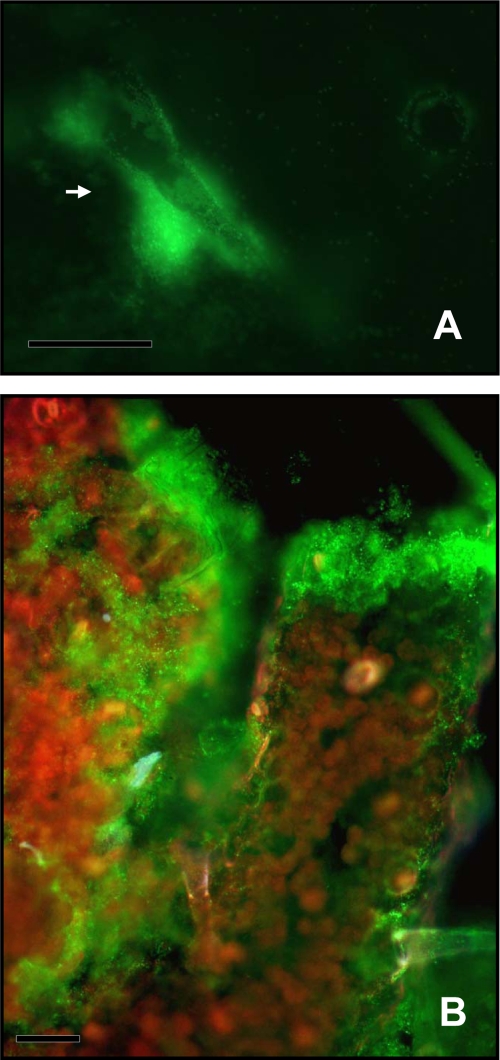
Total populations were visualized using epifluorescence microscopy either by tracking the constitutively GFP-expressing A. baylyi strain BD413 or after staining with ethidium bromide when the bioreporter bearing the promoterless aadA::gfp fusion was used. After 2 days of incubation, cells grown on intact leaf tissues were localized around trichomes and along veins with a scattered, uneven distribution all over their surface (Fig. 3A). Colonization on ground tissues was more important than that on intact leaves. Bacterial cells were heterogeneously distributed on the anatomical structures of the plant. Cells were spread on the surface of the mashed plant material and formed large aggregates either on the surface of the numerous clumps of debris (see Fig. S2 in the supplemental material) or on the margins of leaf residues (see Fig. S3 in the supplemental material).
FIG. 3.
Visualization of cells of A. baylyi strain BD413(rbcL-PaadA::gfp) constitutively expressing GFP after 7 days of incubation on intact tobacco leaf surfaces (A) or on ground leaf tissues (B). The arrow points at a large aggregate formed at the base of a trichome. Bars = 50 μm.
Competence development of A. baylyi BD413 cells grown in vitro and on plant tissues.
Competence development of A. baylyi strain BD413(pBAB2) cells grown in vitro and on leaf tissues was assessed as a function of time. After in vitro transformation, spectinomycin-resistant clones could be detected on intact leaf discs only after 48 h of incubation. The global transformation frequency was, on average, (1.5 ± 3.0) × 10−6 for samples taken after 2 days and decreased to (1.2 ± 1.5) × 10−8 after 7 days incubation on intact leaves (Fig. 2A). When suspensions of the A. baylyi strain BD413(pBAB2) were harvested from ground tobacco leaves, (5.7 ± 2.2) × 10−7 transformants per recipients were detected after 8 h of growth on leaves. After 16 h, competence reached a maximum, with transformation frequencies of (4.6 ± 3.1) × 10−6, corresponding to late exponential growth phase. Thereafter, transformation frequencies decreased with time (Fig. 2B). Transformation ability of A. baylyi strain BD413(pBAB2) was also determined in vitro for up to 40 h; competent bacteria were detected at all sampling times. The transformation frequencies determined at the start of the experiment were (2.7 ± 0.4) × 10−4. Maximum transformation frequency was reached after 4 h and decreased thereafter (Fig. 2C).
Natural transformation of A. baylyi BD413 in vitro and on plant tissues.
The transformation potential of A. baylyi BD413(pBAB2) and A. baylyi BD413(rbcL-ΔPaadA::gfp) was measured in vitro with different forms of transgenic DNA (Table 1). For both strains, transformation frequencies decreased as a function of the number of donor genes present in the transformation mixture. Transformation frequencies for the A. baylyi strain BD413(pBAB2) were, on average, higher than those for the A. baylyi BD413(rbcL-ΔPaadA::gfp) strain by 2 orders of magnitude, independent of the type of transforming DNA utilized (Table 1). When A. baylyi cells were used to determine if bacterial cells could be naturally transformed in situ by the DNA released from the plant, no transformants were detected from bacterial cells recovered from intact leaf tissues. However, transformants could be detected after incubation on crushed leaf discs for 8, 24, and 96 h at frequencies of (2.0 ± 3.1) × 10−10. PCR amplification of spectinomycin-resistant clones confirmed their recombinant nature (see Fig. S1 in the supplemental material). Transformants of A. baylyi strain BD413(rbcL-ΔPaadA::gfp) grown on ground leaf tissues were not detected on all samples and only after 48 h of incubation. Their frequencies were close to the detection limit and averaged 10−9. Fluorescence could be detected in all of the colonies recovered. When analyzed by PCR, spectinomycin-resistant clones could be discriminated from the amplification product on agarose gel, with the appearance of a band of 300 bp, characteristic of the restored promoter (see Fig. S1 in the supplemental material). Transformants were also detected after plating the suspension derived from experiments where A. baylyi was coinoculated with the plant pathogen R. solanacearum: the total population size of A. baylyi BD413(rbcL-ΔPaadA::gfp) averaged (2.9 ± 1.7) × 1011 CFU/g (fresh weight) at the sampling time of 7 days. Transformation frequencies for A. baylyi strain BD413(rbcL-ΔPaadA::gfp) were higher in the presence of the pathogen and were ca. (1.0 ± 5.4) × 10−8. No revertants were observed after sampling inoculated ground leaves and broth culture after 15 days and 7 days, respectively (data not shown).
TABLE 1.
Transformation experiments with A. baylyi strain BD413(rbcL-ΔPaadA::gfp) and A. baylyi strain BD413(pBAB2) exposed to plasmid pCEA and to different sources of transplastomic and wild-type plant DNA
| A. baylyi straina | Donor DNAb | Amt (material) | No. of transformants per assayc | Transformation frequencyc |
|---|---|---|---|---|
| BD413(rbcL-ΔPaadA::gfp) | Linear pCEA plasmid | 1 μg (DNA) | (1.3 ± 0.6) × 105 | (1.4 ± 0.4) × 10−5 |
| Total DNA from TP leaf tissue | 5 μg (DNA) | (1.3 ± 0.2) × 102 | (2.4 ± 0.4) × 10−8 | |
| TP ground leaves | 500 mg | (3.2 ± 1.9) × 101 | (8.4 ± 2.2) × 10−10 | |
| WT ground leaves | 500 mg | 0 | <10−10 | |
| BD413(pBAB2) | Linear pCEA plasmid | 1 μg (DNA) | (5.0 ± 0,1) × 105 | (1.7 ± 0,8) × 10−3 |
| TP total DNA from leaf tissue | 5 μg (DNA) | (6.0 ± 0.8) × 102 | (4.1 ± 2.3) × 10−6 | |
| TP ground leaves | 500 mg | (1.0 ± 0.1) × 101 | (1.5 ± 6.0) × 10−8 | |
| WT ground leaves | 500 mg | 0 | <10−10 |
Recipient concentrations were 3.4 × 107 CFU/ml and 7.0 × 108 CFU/ml for A. baylyi strain BD413(rbcL-ΔPaadA::gfp) and A. baylyi strain BD413(pBAB2), respectively.
TP, transplastomic; WT, wild-type.
Data are means ± standard deviations from three independent experiments.
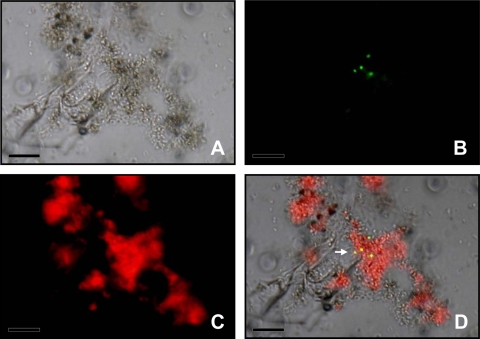
The use of the marker rescue transformation system allowed the visualization of HGT events at single-cell level directly on plant tissues without the need for selective pressure. Recombinants were visualized directly by using different filters. Superimposing images captured with identical fields of view, but different filters, localized single recombinant cells (Fig. 4) as well as cell aggregates (Fig. 5A and B). At higher magnification, microcolonies could be identified (Fig. 5C and D). When leaves infected by R. solanacearum were sampled, tissues showed the typical symptoms of bacterial wilt, such as chlorosis and necrosis. Leaf segments observed by microscopy revealed a heterogeneous disintegration of cellular plant components, while aggregates of transformants of A. baylyi strain BD413(rbcL-ΔPaadA::gfp) could be localized either in degraded tissue or inside leaf veins.
FIG. 4.
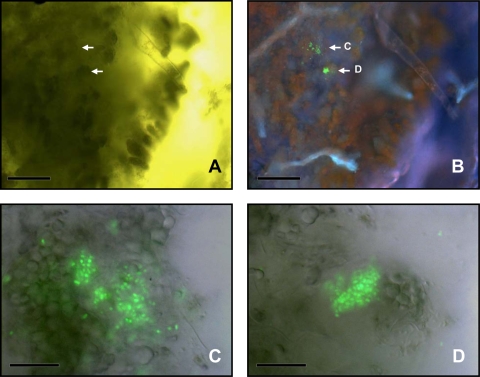
Visualization of A. baylyi transformants in the residuesphere of transplastomic tobacco plant. (A) Bright-field image showing plant debris and bacterial cells; (B) A. baylyi transformants expressing GFP after restoration of the promoter activity through HGT between the plant and the bacteria; (C) red fluorescence emitted by the chloroplasts; (D) images obtained with the different filter sets were combined to provide spatial context for transformant localization. The arrow points at four transformed bacterial cells. Bars = 10 μm.
FIG. 5.
Visualization of A. baylyi transformants in the residuesphere of transplastomic tobacco plant. (A) Bright-field image; arrows point at the localization of transformants. (B) Epifluorescence micrograph of the same field of view showing A. baylyi transformants expressing GFP (green), chloroplasts (red), and veins (cyan). (C, D) Details showing cell clusters of A. baylyi transformants expressing GFP after restoration of the promoter activity through HGT between the plant and the bacteria. Panel A and B bars, 50 μm; panel C and D bars, 20 μm.
DISCUSSION
The present study demonstrated that Acinetobacter baylyi BD413 can develop epiphytic populations, develop a competent state, and be naturally transformed on N. tabacum leaf tissues. Population sizes were 100-fold smaller on intact leaves than on leaf homogenates simulating plant decay, suggesting that conditions for microbial growth are more favorable on the latter due to enhanced availability of nutrients and moisture. When Acinetobacter baylyi BD413 was coinoculated with a vascular plant pathogen, it was able to grow in planta (10), indicating that the major factor for growth was the release of nutrients from damaged cells. Here, we observed that growth also occurred when the strain was inoculated onto the healthy plant surface tissues without an accompanying plant pathogen. Examination of leaf surface colonization patterns revealed that cells were preferentially associated with trichomes and interstices, supporting the general finding that these microhabitats provide a habitat for bacterial growth and survival (20, 21, 26). Acinetobacter baylyi BD413 growth on leaf surfaces probably occurs in part due to the consumption of leaf exudates, such as carbohydrates, amino acids, and organic acids, known to be emitted on intact leaf surface (15, 18). Moreover, such conditions were reported to stimulate gene transfer by conjugation in the phytosphere (13, 16, 36). The likelihood of this event occurring in natural systems cannot be neglected, since litter and naturally decaying plant tissues are closely associated with soil in the residuesphere.
Furthermore, we found that a significant proportion of Acinetobacter baylyi BD413 cells were able to develop a competent state in situ, as determined by exposure of bacterial cells recovered from leaves to a plasmid bearing a selectable marker. Competence development kinetics were consistent with those already observed (29) and correlated to exponential growth phase. While the fraction of cells that became competent in the phyllosphere and the residuesphere was 2 orders of magnitude less than the cells in vitro, these compartments are conducive to bacterial growth and HGT between plants and bacteria (39, 41). These “hot spots” for gene transfer, where bacteria can cluster and form microcolonies or even biofilms (e.g., on roots, aerial plant surfaces, and decaying organic matter and at the interfaces between organic phases and mineral particles), are characterized by their capacity to enhance bacterial metabolic activity, and hence, rates of genetic exchange processes (40, 41). Microscopic observations revealed that bacteria were distributed heterogeneously on the leaf surface, especially in the case of residual tissues; bacterial aggregates were concentrated on margins and extracellular interstices (see Fig. S3 in the supplemental material). We hypothesize that nutrients and DNA are more available in these locations. Thus, in addition, to bacterial recovery and transformation testing in vitro, we were able to visualize gene transfer in situ specifically and directly at these locations using the GFP reporter system.
In the phyllosphere, characterized by a higher degree of oligotrophy than the residuesphere, the probability of a physical encounter between DNA molecules and competent bacteria would be lower. This could explain why no transformants were detected in our microcosms simulating phyllosphere conditions. In contrast to the uninjured phyllosphere, assessment of the potential of natural transformation in the pathosphere confirmed the tremendous adaptability of A. baylyi to grow as an opportunistic bacterium (10). There, bacterial densities were extremely high, and so were transformation frequencies compared to those we obtained in the residuesphere, suggesting that of the three compartments investigated in this study, the pathosphere would be the most conducive for plant to bacteria gene transfer. Although considerations can be made about the possible competition for resources due to simultaneous growth of the two bacterial species in planta, our findings show that the carrying capacity of leaf tissues could allow coexistence of conspicuous bacterial populations.
Previous field studies attempting to detect transfer of engineered plant transgene by natural transformation into resident field bacterial population (8, 27) relied on detection strategies based on the cultivation of bacteria from soils on selective media and on the screening via various biomolecular techniques for the presence of transgenic DNA. Although they resulted in isolation of a high number of antibiotic-resistant colonies on transformant selective media, neither of them was able to confirm the incorporation of transgenic plant DNA into the bacterial isolates. Published results on this issue are, however, scarce and limited and, when critically reviewed, were described to rely on detection methods and sample sizes inadequate to identify rare bacterial transformants among the 108 bacteria ideally found in every gram of soil (24, 31). Furthermore, these 108 cells occupy only 0.02% of the colonizable volume available in soil (28). One of the more important stochastic factors influencing HGT in the environment is the proximity of donor DNA and recipient cells (41). By concentrating on visualizing the potential hotspots for gene transfer, we were able to observe transformants in situ.
Although visualization of transformants in situ in various hot spots was possible with our detection system, in some cases, the number of transformants that were detected by epifluorescence microscopy observation in situ seems to exceed the number of transformants recovered after plating. Most likely, biases result from mechanical processes. Removal of bacterial cells from vegetal samples by washing might have been only partially efficient, or aggregates might have remained intact, leading to an underestimation of potential cultivable transformants.
In addition, the number of transformants detected after plating suspensions harvested from infected leaf segments might have been representative of individual recombinants or the result of clonal multiplication of a few original single transformation events. During direct visualization of leaf samples, transformants were often found in clusters, such as microcolonies (Fig. 5), strongly suggesting clonal multiplication of single transformation events. Uncertainty on transformation frequencies could be resolved by coupling the reporter gene approach to new analytical methods not requiring cultivation like flow cytometry, which allows fine enumeration of recombinants (35, 38). Hence, the use of the bioreporter tool could be applied in natural environments, with new experiments relying on flow cytometry and differential genetic labeling of recombinant and recipient cells. From an ecological perspective, however, focusing strictly on transformation frequency quantification in situ might be limiting if we omit selective pressure and ecological considerations, since in theory even a single extremely rare transformation event may initiate a chain of ecological consequences depending on the transgene.
Our experiments were conducted in nearly sterile environments, and therefore, the transformation frequencies we observed are likely to be different than those that would be observed in the presence of the natural microflora of plant leaf surfaces due to the different possible interactions (e.g., competition or synergy). While our approach detected and visualized rare transformation events in laboratory experiments with microcosms simulating natural conditions, the application of this tool to field studies could provide insights into factors that affect HGT between prokaryotes or other biota.
Supplementary Material
Acknowledgments
We are grateful to Carolyne Freyssinet for technical help and Denis Desbouchage, in charge of the greenhouse facilities of the IFR 41 at the University Claude Bernard, Lyon I, for his assistance.
This work was supported by grant QLK3-CT-2001-02242 (TRANSBAC, 5th RTD Program, Quality of Life and Management of Living Resources) from the EU.
Footnotes
Published ahead of print on 27 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andrews, J. H., and R. F. Harris. 2000. The ecology and biogeography of microorganisms of plant surfaces. Annu. Rev. Phytopathol. 38:145-180. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, M. J., A. K. Lilley, and J. P. Diaper. 1996. Gene transfer between microorganisms in the phyllosphere, p. 102-123. In C. E. Morris, P. C. Nicot, and C. Nguy-The (ed.), Aerial plant surface microbiology. Plenum Press, New York, NY.
- 3.Bailey, M. J., P. B. Rainey, X.-X. Zhang, and A. K. Lilley. 2002. Population dynamics, gene transfer and gene expression in plasmids: the role of the horizontal gene pool in local adaptation at the plant surface, p. 173-192. In S. E. Lindow, E. I. Hecht-Poinar, and V. J. Elliot (ed.), Phyllosphere microbiology. APS Press, St. Paul, MN.
- 4.Bertolla, F., and P. Simonet. 1999. Horizontal gene transfers in the environment: natural transformation as a putative process for gene transfers between transgenic plants and microorganisms. Res. Microbiol. 150:375-384. [DOI] [PubMed] [Google Scholar]
- 5.Bjorklof, K., E. L. Nurmiaho-Lassila, N. Klinger, K. Haahtela, and M. Romantschuk. 2000. Colonization strategies and conjugal gene transfer of inoculated Pseudomonas syringae on the leaf surface. J. Appl. Microbiol. 89:423-432. [DOI] [PubMed] [Google Scholar]
- 6.Ceccherini, M. T., J. Pote, E. Kay, V. T. Van, J. Marechal, G. Pietramellara, P. Nannipieri, T. M. Vogel, and P. Simonet. 2003. Degradation and transformability of DNA from transgenic leaves. Appl. Environ. Microbiol. 69:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Lipthay, J. R., T. Barkay, and S. J. Sørensen. 2001. Enhanced degradation of phenoxyacetic acid in soil by horizontal transfer of the tfdA gene encoding a 2,4-dichlorophenoxyacetic acid dioxygenase. FEMS Microbiol. Ecol. 35:75-84. [DOI] [PubMed] [Google Scholar]
- 8.Gebhard, F., and K. Smalla. 1999. Monitoring field releases of genetically modified sugar beets for persistence of transgenic plant DNA and horizontal gene transfer. FEMS Microbiol. Ecol. 28:261-272. [Google Scholar]
- 9.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kay, E., F. Bertolla, T. M. Vogel, and P. Simonet. 2002. Opportunistic colonization of Ralstonia solanacearum-infected plants by Acinetobacter sp. and its natural competence development. Microb. Ecol. 43:291-297. [DOI] [PubMed] [Google Scholar]
- 11.Kay, E., T. M. Vogel, F. Bertolla, R. Nalin, and P. Simonet. 2002. In situ transfer of antibiotic resistance genes from transgenic (transplastomic) tobacco plants to bacteria. Appl. Environ. Microbiol. 68:3345-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keese, P. 2008. Risks from GMOs due to horizontal gene transfer. Environ. Biosafety Res. 7:123-149. [DOI] [PubMed] [Google Scholar]
- 13.Kidambi, S. P., S. Ripp, and R. V. Miller. 1994. Evidence for phage-mediated gene transfer among Pseudomonas aeruginosa strains on the phylloplane. Appl. Environ. Microbiol. 60:496-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lal, R. 2005. World crop residues production and implications of its use as a biofuel. Environ. Int. 31:575-584. [DOI] [PubMed] [Google Scholar]
- 15.Leveau, J. H., and S. E. Lindow. 2001. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 98:3446-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lilley, A. K., J. C. Fry, M. J. Day, and M. J. Bailey. 1994. In situ transfer of an exogenously isolated plasmid between Pseudomonas spp. in sugarbeet rhizosphere. Microbiology 140:27-33. [Google Scholar]
- 17.Lindow, S. E., and M. T. Brandl. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercier, J., and S. E. Lindow. 2000. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl. Environ. Microbiol. 66:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255-261. [DOI] [PubMed] [Google Scholar]
- 20.Monier, J.-M., and S. E. Lindow. 2003. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. USA 100:15977-15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monier, J.-M., and S. E. Lindow. 2004. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl. Environ. Microbiol. 70:346-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris, C. E., and J.-M. Monier. 2003. The ecological significance of biofilm formation by plant-associated bacteria. Annu. Rev. Phytopathol. 41:429-453. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen, K. M., P. J. Johnsen, D. Bensasson, and D. Daffonchio. 2007. Release and persistence of extracellular DNA in the environment. Environ. Biosafety Res. 6:37-53. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen, K. M., and J. P. Towsend. 2004. Monitoring and modeling horizontal gene transfer. Nat. Biotechnol. 22:1110-1114. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen, K. M., A. M. Bones, K. Smalla, and J. D. van Elsas. 1998. Horizontal gene transfer from transgenic plants to terrestrial bacteria—a rare event? FEMS Microbiol. Rev. 22:79-103. [DOI] [PubMed] [Google Scholar]
- 26.Normander, B., B. B. Christensen, S. Molin, and N. Kroer. 1998. Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris). Appl. Environ. Microbiol. 64:1902-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paget, E., M. Lebrun, G. Freyssinet, and P. Simonet. 1998. The fate of recombinant plant DNA in soil. Eur. J. Soil Biol. 34:81-88. [Google Scholar]
- 28.Pallud, C., A. Dechesne, J. P. Gaudet, D. Debouzie, and G. L. Grundmann. 2004. Modification of spatial distribution of 2,4-dichlorophenoxyacetic acid degrader microhabitats during growth in soil columns. Appl. Environ. Microbiol. 70:2709-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmen, R., B. Vosman, P. Buijsman, C. K. Breek, and K. J. Hellingwerf. 1993. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J. Gen. Microbiol. 139:295-305. [DOI] [PubMed] [Google Scholar]
- 30.Pontiroli, A., P. Simonet, A. Frostegard, T. M. Vogel, and J.-M. Monier. 2007. Fate of transgenic plant DNA in the environment. Environ. Biosafety Res. 6:15-35. [DOI] [PubMed] [Google Scholar]
- 31.Ray, J. L., and K. M. Nielsen. 2005. Experimental methods for assaying natural transformation and inferring horizontal gene transfer, p. 491-520. In E. A. Zimmer and E. H. Roalson (ed.), Molecular evolution: producing the biochemical data. Methods in enzymology. Academic Press, New York, NY. [DOI] [PubMed]
- 32.Rizzi, A., A. Pontiroli, L. Brusetti, S. Borin, C. Sorlini, A. Abruzzese, G. A. Sacchi, T. M. Vogel, P. Simonet, M. Bazzicalupo, K. M. Nielsen, J.-M. Monier, and D. Daffonchio. 2008. Strategy for in situ detection of natural transformation-based horizontal gene transfer events. Appl. Environ. Microbiol. 74:1250-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudrappa, T., M. L. Biedrzycki, and H. P. Bais. 2008. Causes and consequences of plant-associated biofilms. FEMS Microbiol. Ecol. 64:153-166. [DOI] [PubMed] [Google Scholar]
- 34.Sengelov, G., G. A. Kowalchuk, and S. J. Sørensen. 2000. Influence of fungal-bacterial interactions on bacterial conjugation in the residuesphere. FEMS Microbiol. Ecol. 31:39-45. [DOI] [PubMed] [Google Scholar]
- 35.Sørensen, S. J., M. J. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700-710. [DOI] [PubMed] [Google Scholar]
- 36.Sørensen, S. J., and L. E. Jensen. 1998. Transfer of plasmid RP4 in the spermosphere and rhizosphere of barley seedling. Antonie van Leeuwenhoek 73:69-77. [DOI] [PubMed] [Google Scholar]
- 37.Sørensen, S. J., S. Musovic, and G. Oregaard. 2004. Culture independent detection of horizontal gene transfer in natural environments, abstr. 125. Abstr. 10th Int. Symp. Microb. Ecol. ISME, Cancun, Mexico.
- 38.Sørensen, S. J., A. H. Sørensen, L. H. Hansen, G. Oregaard, and D. Veal. 2003. Direct detection and quantification of horizontal gene transfer by using flow cytometry and gfp as a reporter gene. Curr. Microbiol. 47:129-133. [DOI] [PubMed] [Google Scholar]
- 39.van Elsas, J. D., and M. J. Bailey. 2002. The ecology of transfer of mobile genetic elements. FEMS Microbiol. Ecol. 42:187-197. [DOI] [PubMed] [Google Scholar]
- 40.van Elsas, J. D., J. C. Fry, P. Hirsch, and S. Molin. 2000. Ecology of plasmid transfer and spread, p. 175-206. In C. M. Thomas (ed.), The horizontal gene pool, bacterial plasmids and gene spread. Harwood Scientific Publishers, Amsterdam, The Netherlands.
- 41.van Elsas, J. D., S. Turner, and M. J. Bailey. 2003. Horizontal gene transfer in the phytosphere. New Phytol. 157:525-537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.