Abstract
Estuarine sediments are the location for significant bacterial removal of anthropogenically derived inorganic nitrogen, in particular nitrate, from the aquatic environment. In this study, rates of benthic denitrification (DN), dissimilatory nitrate reduction to ammonium (DNRA), and anammox (AN) at three sites along a nitrate concentration gradient in the Colne estuary, United Kingdom, were determined, and the numbers of functional genes (narG, napA, nirS, and nrfA) and corresponding transcripts encoding enzymes mediating nitrate reduction were determined by reverse transcription-quantitative PCR. In situ rates of DN and DNRA decreased toward the estuary mouth, with the findings from slurry experiments suggesting that the potential for DNRA increased while the DN potential decreased as nitrate concentrations declined. AN was detected only at the estuary head, accounting for ∼30% of N2 formation, with 16S rRNA genes from anammox-related bacteria also detected only at this site. Numbers of narG genes declined along the estuary, while napA gene numbers were stable, suggesting that NAP-mediated nitrate reduction remained important at low nitrate concentrations. nirS gene numbers (as indicators of DN) also decreased along the estuary, whereas nrfA (an indicator for DNRA) was detected only at the two uppermost sites. Similarly, nitrate and nitrite reductase gene transcripts were detected only at the top two sites. A regression analysis of log(n + 1) process rate data and log(n + 1) mean gene abundances showed significant relationships between DN and nirS and between DNRA and nrfA. Although these log-log relationships indicate an underlying relationship between the genetic potential for nitrate reduction and the corresponding process activity, fine-scale environmentally induced changes in rates of nitrate reduction are likely to be controlled at cellular and protein levels.
Estuaries are major conduits for the transport of anthropogenically derived nitrogen (e.g., from fertilizer runoff and from wastewater treatment plants) from land to sea (18, 20). Estuarine sediments are now recognized as being an important location for the removal of inorganic nitrogen from this environment via benthic nitrate reduction to nitrogenous gases (21). Previously, we have utilized the isotope-pairing technique (22) to investigate rates of denitrification (DN) of nitrate to N2O and N2 along the nitrate and salinity gradients in the hypernutrified Colne estuary, United Kingdom, and demonstrated that DN rates are highest at the estuary head, where nitrate levels are also at their highest (7, 9). However, DN represents only one of three key pathways relevant to nitrate reduction, with a recent review suggesting that the importance of DN in aquatic systems may be overstated (3). A substantial proportion of nitrate may alternatively be converted to ammonium (1, 16, 17) via dissimilatory nitrate reduction to ammonium (DNRA), in which case inorganic nitrogen is retained within the aquatic environment. Nitrite, usually derived from nitrate reduction, can also be converted to N2 via the anammox (AN) pathway, involving the simultaneous conversion of nitrite and ammonium to N2 (19, 47). All three nitrate/nitrite reduction pathways are catalyzed by benthic microbial (primarily bacterial) assemblages, and the fate of nitrate is dependent upon the composition and activity of these organisms. DN and DNRA are mediated by diverse polyphyletic taxa (2, 37); conversely, nitrite reduction via AN is currently considered to be conducted only by members of the Planctomycetes (41). DN and DNRA pathways are catalyzed by a series of reductase enzymes encoded by genes that can be used as proxies in determining the potential for nitrate reduction processes within environments (26), although the enzyme proteins themselves remain the nearest proxy of activity. Briefly, narG and napA genes encode subunits of two distinct nitrate reductases (NAR and NAP, which are present in the cell membrane and in the periplasm, respectively), which mediate the reduction of nitrate to nitrite in both DN and DNRA. Subsequent commitment to the DN pathway to generate nitric oxide, with the eventual formation of either nitrous oxide or N2, is mediated by nitrite reductases encoded by nirS or nirK (reviewed in reference 49). In contrast, in DNRA, nitrite is reduced to ammonium by the NrfA enzyme encoded by nrfA (reviewed in reference 37). Consequently, molecular analyses of nirS and nrfA genes can be used to investigate the genetic potential in the environment for commitment to either DN or DNRA within the benthic nitrogen flow. We have previously investigated the presence of some of these key functional genes along the Colne estuary, together with their corresponding RNA transcripts (23, 38, 39), and subsequent sequence analyses have revealed a complex and diverse community of nitrate-reducing bacteria along the estuarine nitrate and salinity gradients. While our subsequent quantitative PCR (Q-PCR) and reverse transcription (RT)-Q-PCR analyses suggested a decline in the abundance of nitrate and nitrite reductase genes and transcripts from the head to the mouth of the estuary (38), no attempt has been made to investigate direct linkages between the distributions of such key functional genes and nitrate reduction process rate measurements. Moreover, to provide a complete assessment of nitrate/nitrite reduction within the estuarine environment, it is also essential to determine the rates of, and the potential for, AN. The occurrence of AN in a number of estuarine and coastal sediments has now been reported (13, 30, 31, 44, 45). At this time, a diagnostic functional gene marker for AN has yet to be confirmed, and consequently, the presence of AN bacteria is currently assessed using PCR assays that target AN bacterium-specific 16S rRNA sequences (25, 28).
The aims of this research were, first, to determine whether there was variation in the rates of DN, DNRA, and AN along the estuary; second, to determine whether there was variation in the presence and abundance of gene markers serving as proxies for bacteria responsible for DN, DNRA, and AN along the estuary; and third, to investigate interrelationships between the rates of these processes and the abundances of the gene markers for these processes.
MATERIALS AND METHODS
Field sampling.
Field sampling was conducted in February and March 2005 at three sites along the Colne estuary, United Kingdom, at Hythe (the head), Alresford Creek (the middle), and Brightlingsea (the mouth), as described previously (8, 9). At each site, replicate small (10-cm-long) cores of sediment were collected into core tubes (internal diameter, 3.4 cm, and length, 22 cm) for process rate measurements (n, 5 per treatment). The use of cores preserves vertical (nitrate) concentration gradients and, hence, is the sampling method best suited to avoiding experimental effects when determining nitrate reduction rates. In addition, in February 2005, triplicate sediment samples were taken from the top 1 cm of sediment for molecular analysis. Nitrate reduction occurs only in the top 0 to 1 cm (i.e., surface sediment), into which nitrate penetrates from the overlying water. These samples were transported on ice and returned to the laboratory within 1 h of sampling. Aliquots (0.5 g [wet weight]) of homogenized sediment from each replicate were then stored at −70°C prior to their use for nucleic acid isolation.
Measurement of DN and DNRA rates.
Rates of DN and DNRA in both February and March 2005 were determined. The sediment cores (see above) were processed as described by Dong et al. (7, 9). 15N-labeled nitrate solution (1 ml) was added to the site water (120 ml) above each core to give a final 15N-nitrate concentration of 20% of the in situ nitrate. The isotope was allowed to equilibrate for 30 min after the addition, and then core tubes were closed and incubated in the dark at an in situ temperature for 3 h. (Preliminary experiments showed that DN and DNRA responses were linear with respect to changes in 15NO3− concentrations in the overlying water and the incubation time) (see Fig. S1 in the supplemental material). During this incubation period, dissolved oxygen depletion was <20% of air equilibration. At the end of the incubation, the sediment core and water were mixed to form a slurry and then samples (12.5 ml) were removed to enable the quantification of DN gaseous products (N2O and N2). Subsamples (10 ml) of the slurried sediment cores were taken for the subsequent recovery of 15NH4+ to determine rates of DNRA. The pH of each slurry sample for DNRA was adjusted to >12 via the addition of 0.5 ml of 2 M NaOH, and then the sample was steam distilled to carry over NH3 into two acid traps, each containing 5 ml of 0.01 M HCl, in sequence. Ammonia was effectively retained in the first trap, but any accidental carryover of alkali into the first trap might liberate NH3, which would then be retained in the second trap. Preliminary experiments showed that the first 120 ml of distillate contained >98% of the NH4+ in the sediment sample, normally in the first trap. After distillation, the NH4+ concentration in each trap was measured colorimetrically using a 1-ml subsample and then the remaining NH4+ was absorbed onto 100 mg of zeolite Y (reference no. D-958; Caltel International Ltd., Sheffield, United Kingdom), which absorbed 95% of the NH4+. Preliminary experiments with known 15/14NH4+ ratio standards showed that there was no selective isotope fractionation by the zeolite during adsorption, with measured 15NH4+/14NH4+ ratios not being significantly different from the ratios in the standards (P < 0.05; t test) (data not shown). The zeolite was then filtered through a glass fiber filter (Whatman, Maidstone, United Kingdom), and the filter was air dried. The trapped powdered zeolite was then carefully scraped away from the glass fiber filter, and ∼20 mg was weighed into 6- by 4-mm tin cups (Elemental Microanalysis Ltd., Cambridge, United Kingdom) prior to being loaded into the carousel of an NA1500 carbon/nitrogen elemental analyzer (Carlo Erba, Milan, Italy) which was interfaced with an isotope ratio mass spectrometer (Dennis Leigh Technologies, Manchester, United Kingdom). Samples were then combusted at >1,000°C and passed through reducing copper turnings, magnesium perchlorate, and Carbosorb prior to entering the ion source of the isotope ratio mass spectrometer via continuous flow. Beam data for N2 at masses with m/z values of 28, 29, and 30 were then recorded for each sample. Reference standards and blanks were run prior to analysis and after every 10th sample. Blanks of zeolite alone showed that nitrogen was below detectable levels.
The rate of DNRA was calculated as follows: rate of DNRA (in micromoles of N per square meter per hour) = 15N excess × amount of NH4+ (in micromoles of N) in 1 ml of slurry × V (in milliliters)/15N enrichment/incubation time (in hours) × R, where 15N excess = 2 × [(sample 30N/28N ratio) − (reference 30N/28N ratio)] +[(sample 29N/28N ratio) − (reference 29N/28N ratio)], amount of NH4+ in 1 ml of slurry = NH4+ concentration (in micromolars) × volume of distillate (in liters)/10 ml of slurry, V is the total slurry volume per square meter of sediment (V = height of core [in centimeters] × 100 cm × 100 cm), 15N enrichment = 15NO3−/(15NO3− + 14NO3−) ratio, and R is the 14NO3−/(15NO3− + 14NO3−) ratio in the water column.
Measurement of AN rates.
AN (together with DN and DNRA) in sediment samples obtained along the estuary in March 2005 was measured by established methods (31) by incubating two series of sediment cores with the addition of 100 μM or 200 μM 15NO3− (final concentration) in the water above the cores.
Determination of nitrate reduction potential.
In order to examine the potential for nitrate reduction along the estuary, a series of sediment slurry experiments was established. During March, samples of sediment (0 to 2 cm) at each of the three sample sites along the estuary were collected into glass jars completely filled to exclude oxygen and the jars were returned to the laboratory. Slurries (50%, vol/vol) of sediment in deoxygenated artificial seawater (Tropic Marin Centre, Rickmansworth, United Kingdom) at salinities appropriate to the sites were dispensed into 120-ml serum bottles under a continual stream of oxygen-free N2, and the bottles were sealed with Suba-Seals. Sodium nitrate solution (2 ml of 50 mM) was added to each flask to give an elevated initial concentration of approximately 2,000 μM nitrate. Half of the flasks had 2 ml of 500 mM sodium chlorate solution added (final concentration, 20 mM); sodium chlorate is a selective inhibitor of the NAR but not of the NAP nitrate reductase (33). Half of each series of flasks (both those with and without chlorate) had acetylene (10%, vol/vol) injected into the headspace to inhibit the reduction of N2O to N2 and thus provide a measurement of DN by comparing N2O accumulation levels in the presence and absence of acetylene (24). Each of the treatments was done in triplicate. The flasks were incubated with gentle shaking at 15°C for 4 h, and then 50-μl samples of the headspaces were taken for subsequent analysis of N2O by injection into a gas chromatograph with an electron capture detector (32). Samples of the slurry were also analyzed for residual nitrate, nitrite, and ammonium by using an autoanalyzer [Scalar (UK) Ltd., York] and standard colorimetric methods. Finally, a slurry subsample was taken for the evaluation of residual nitrate and nitrite, and residual ammonium was recovered from each flask by steam distillation (recovering both soluble and exchangeable ammonium) as described above.
Nucleic acid extraction, Q-PCR, and RT-Q-PCR.
DNA and RNA were extracted from 0.5-g sediment samples by using Lysing Matrix B tubes (Bio-101; QBiogene, Cambridge, United Kingdom) as described previously (38). Nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and gene transcripts from triplicate sediment samples collected from each site were quantified using a series of (RT)-Q-PCR TaqMan assays described previously (38). narG, napA, and nrfA primer and probe sets were designed to target subgroups of these genes identified within clone libraries amplified by PCR from DNA isolated from Colne estuary sediments (38). The suite of nirS primers and probes was designed to target sequences related to nirS mRNA clones amplified by RT-PCR from mRNA isolated from Colne estuary sediments (23).
PCR detection and sequence analysis of AN-related bacterial 16S rRNA genes.
16S rRNA genes related to those from AN bacteria were amplified from DNA extracted from sediments collected in February 2005 by using the primers Brod541F (5′-GAG CAC GTA GGT GGG TTT GT-3′) and Brod1260F (5′-GGA TTC GCT TCA CCT CTC GG-3′) under the amplification conditions stated previously (25). Clone library construction and subsequent sequencing of amplified 16S rRNA genes were carried out as described previously (38). Sequence alignments were constructed using ClustalX (42), and distance matrices were calculated using the DNADIST program in PHYLIP (10). Phylogenetic trees were created from the distance matrices by the neighbor-joining method (35) using the Kimura substitution algorithm (15) in PHYLIP. Consensus trees were calculated after bootstrapping (1,000 replicate trees).
Statistical analyses.
Variation in nitrate reduction rates and in gene numbers among sites were analyzed using a one-way analysis of variance (ANOVA), followed by a post hoc Tukey test (46), in SPSS version 14. Data were first log(n + 1) transformed and then used for regression analyses of the process rate and gene abundance data sets. Compositional changes in the bacterial nitrate- and nitrite-reducing guilds present at the three sites along the estuary were investigated by comparing the abundances of the four genes (including representatives of nine phylotypes) at the three sites. A Bray-Curtis resemblance matrix (6) was created from the log(n + 1)-transformed data and then used to generate two-dimensional multidimensional scaling (MDS) plots using PRIMER-6 (PRIMER-E Ltd., Plymouth Marine Laboratory, United Kingdom), with variation assessed using a one-way analysis of similarities (ANOSIM) (5).
Nucleotide sequence accession numbers.
Nucleotide sequences were deposited in the GenBank database with accession numbers EU394239 to EU394279.
RESULTS
DN, DNRA, and AN rate measurements along the estuary.
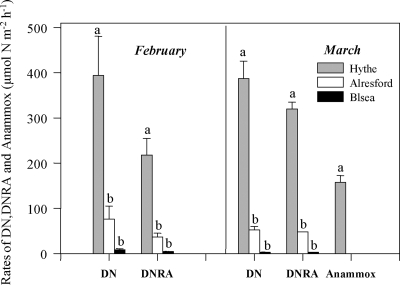
Rates of DN to both N2O and N2 and of DNRA decreased from the head (Hythe) toward the mouth (Brightlingsea) of the estuary (Fig. 1). Rates of DN and DNRA at the Hythe site were significantly higher (P < 0.05; repeated-measures ANOVA) than those at the other two sites, which were not significantly different from each other. Rates of AN in sediments sampled in March 2005 were also determined, and AN was detected only at the Hythe site (mean AN activity ± standard error, 157 ± 15 μmol of N m−2 h−1), where it accounted for 30% of N2 formation (Fig. 1).
FIG. 1.
Rates of DN, DNRA, and AN at three sites along the Colne estuary in February and March 2005. AN activity was not detected at Alresford or Brightlingsea (Blsea) in March 2005. Standard errors (n = 5) are represented by error bars, and for each nitrate reduction process, statistically significant differences (P = 0.05) between sites are indicated by different letters.
Nitrate reduction potential along the Colne estuary.
Using slurry experiments, we investigated the potential for nitrate removal along the Colne estuary (Table 1). Nitrate removal potentials were highest at the Hythe site and were significantly lower (P < 0.05; ANOVA) at Alresford and Brightlingsea. At the Hythe site, slightly higher levels of nitrate reduction were due to the activity of NAP, while at Alresford and Brightlingsea, NAR activity was greater. The proportion of nitrate denitrified (to either N2O or N2) decreased from the head to the mouth of the estuary, while the proportion of DNRA was highest at the intermediate Alresford site. DN was the dominant potential removal pathway at the Hythe site, while DNRA activity was greater than DN activity at Alresford and Brightlingsea. However, while there was good agreement (∼97%) between the moles of nitrate removed and the N products formed at Alresford, much smaller proportions of the reduced nitrate at the Hythe and Brightlingsea sites (∼55 and 44.5%, respectively) could be accounted for directly (Table 1).
TABLE 1.
Determination of nitrate removal potentials and end product formation in sediment slurry experiments with sediments from Hythe, Alresford, and Brightlingsea collected in March 2005a
| Site | Treatment | NO3− removal rate | % Removal by NAR:NAP | N2 production rate | NO2− production rate | NH4+ production rate | (N2 + N2O)/ NO3− ratio | NH4+/NO3− ratio |
|---|---|---|---|---|---|---|---|---|
| Hythe | No chlorate | 1.725 (0.099) | 43:57 | 0.837 (0.012) | 0.001 (0.000) | 0.103 (0.025) | 0.485 (0.025) | 0.061 (0.017) |
| Chlorate | 0.990 (0.144) | 0.510 (0.030) | 0.000 (0.000) | 0.194 (0.015) | ||||
| Alresford | No chlorate | 0.710 (0.145) | 66:33 | 0.265 (0.011) | 0.057 (0.013) | 0.391 (0.017) | 0.373 (0.094) | 0.594 (0.107) |
| Chlorate | 0.233 (0.026) | 0.147 (0.003) | 0.000 (0.000) | 0.254 (0.021) | ||||
| Brightlingsea | No chlorate | 0.847 (0.215) | 58:42 | 0.103 (0.002) | 0.135 (0.027) | 0.139 (0.007) | 0.121 (0.049) | 0.199 (0.069) |
| Chlorate | 0.355 (0.206) | 0.093 (0.003) | 0.000 (0.001) | 0.055 (0.005) |
The initial NO3− concentration was 2 mM, slurries were incubated for 4 h, and N2, N2O, and NH4+ production was measured. All rates are means (±standard errors; n = 3) in micromoles of N per gram (dry weight) of sediment per hour. The last two columns show the moles of N2 plus N2O or of NH4+ produced per mole of NO3− removed. Chlorate at 20 mM was used to inhibit NAR and estimate nitrate reduction by either NAR or NAP.
(RT)-Q-PCR quantification of nitrate and nitrite reductase genes and transcripts.
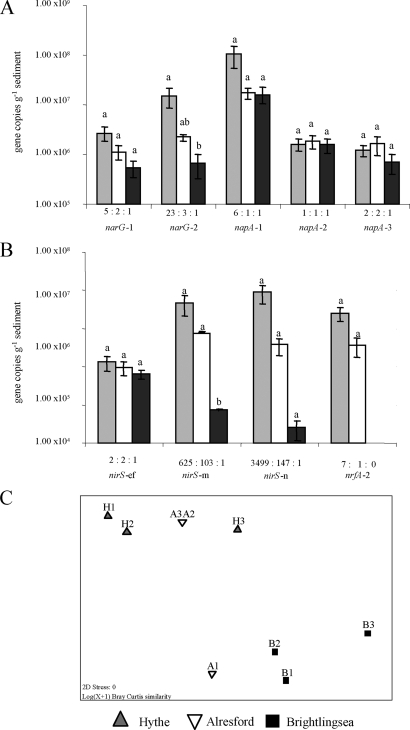
Q-PCR assays were used to quantify variation in the abundance of nitrate and nitrite reductase genes representing nine phylotypes (38) in sediments along the estuary sampled in February 2005 (Fig. 2). Nitrate reductase (narG and napA) and nitrite reductase (nirS and nrfA) gene numbers generally decreased along the estuary. Q-PCR analysis of narG genes of two phylotypes showed a general decline in gene numbers from the head (Hythe) to the mouth (Brightlingsea) of the estuary, although this decline yielded a significance difference only between the Hythe and Brightlingsea sites for the narG-2 gene (P < 0.05). Numbers of napA-1 were greater than those of the genes of the other two napA phylotypes at all three sites. No statistically significant decline in the napA genes of any phylotype along the estuary was observed, although napA-1 gene numbers were highest at the Hythe site. A decline in numbers of nirS genes (representing three phylotypes) was also seen, with a significant decline of nirS-m numbers along the estuary from the Hythe and Alresford sites to Brightlingsea (P < 0.05) but no significant decline in nirS-e&f and nirS-n gene numbers. However, it should be noted that although there was no significant difference in nirS-n gene copy numbers along the estuary (F2,6 = 4.78; P = 0.057), the P value was very close to the critical 0.05 level, indicating that there may be variation in gene numbers among sites. The subsequent post hoc test indicated that the P value for the difference in nirS-n gene numbers between Hythe and Brightlingsea was close to the critical 0.05 level (P = 0.058). Abundances of the nrfA-2 gene also declined along the estuary, with sequences of this phylotype not detected at the Brightlingsea site.
FIG. 2.
Variation in abundance (number of copies per gram of sediment) of nitrate and nitrite reductase genes along the Colne estuary in February 2005. (A and B) Data are shown for nitrate reductase genes narG and napA (A) and nitrite reductase genes nirS and nrfA (B). Details of the subgroups targeted by the Q-PCR primers and probes are given by Smith et al. (38). Standard errors (n = 3) for each separate Q-PCR assay are shown, and for each phylotype, statistically significant differences (P < 0.005) between sites are indicated by different letters. The ratios of gene numbers at the three sites are displayed for each phylotype. nrfA-2 genes were not detected below the CT cutoff value at the Brightlingsea site. Gene numbers were calculated from the following standard curves: narG-1, r2 = 0.995, y intercept = 40.52, and E = 100.9% (NTC undetected); narG-2, r2 = 0.999, y intercept = 44.57, E = 90.9%, and CT cutoff = 32.3; napA-1, r2 = 0.998, y intercept = 44.99, and E = 86.9% (NTC undetected); napA-2, r2 = 0.997, y intercept = 39.00, E = 89.6%, and CT cutoff = 32.83; napA-3, r2 = 0.996, y intercept = 47.60, and E = 89.2% (NTC undetected); nirS-e&f, r2 = 0.998, y intercept = 35.89, E = 86.0%, and CT cutoff = 35.00; nirS-m, r2 = 0.996, y intercept = 41.13, and E = 86.1% (NTC undetected); nirS-n, r2 = 0.998, y intercept = 42.84, and E = 82.1% (NTC undetected); and nrfA-2, r2 = 0.997, y intercept = 39.89, E = 96.06%, and CT cutoff = 28.51. (C) MDS plot of Bray-Curtis similarities from log(n + 1)-transformed gene numbers showing overall variation in nitrate and nitrite reductase gene abundances among sites (H, Hythe; A, Alresford; and B, Brightlingsea). ANOSIM R statistics were as follows: for the comparison of data from all sites, 0.564; for data from Hythe and Alresford, 0.037; for data from Hythe and Brightlingsea, 0.963; and for data from Alresford and Brightlingsea, 0.704. R values range between 0 and 1 (0, identical; 1, no similarity).
To investigate variation in the overall abundance of the nitrate and nitrite reductase genes along the estuary, an MDS plot of Bray-Curtis log(n + 1)-transformed gene numbers at each site (Fig. 2C), which separated the Hythe and Alresford sites from Brightlingsea, was generated. Pairwise ANOSIM indicated that there was no difference in the overall numbers of nitrate and nitrite reductase genes at the Hythe and Alresford sites but that reductase gene numbers at both sites were significantly different from those at Brightlingsea (R = 0.963 for Hythe numbers versus Brightlingsea numbers and R = 0.704 for Alresford numbers versus Brightlingsea numbers; ANOSIM).
RT-Q-PCR was then used to target and quantify mRNA transcripts of the narG, napA, nirS, and nrfA phylotypes in RNA extracts from the Colne estuary sediment samples obtained in February 2005 from each site (Table 2). RT-Q-PCR detection threshold cycle (CT) values for gene transcripts were low. CT cutoff values for each RT-Q-PCR assay were set at 3.3 cycles fewer than that for the no-template control (NTC), if detected, to ensure that the background signal was not contributing to transcript quantification (39). No gene expression above the detection limits was measured for any of the targeted genes at the Brightlingsea site. napA transcripts for two of the three phylotypes (those of napA-1 and napA-3) were detected at both the Hythe and Alresford sites, with no significant difference in transcript numbers for the two phylotypes between sites. The presence of both of the narG phylotypes, with similar transcript numbers, was detected only at the Hythe site. Of the three nirS phylotypes, only the nirS-e&f phylotype had transcripts detected at both the Hythe and Alresford sites, with transcript numbers greater at the Hythe site.
TABLE 2.
Nitrate and nitrite reductase gene transcript numbers in surface sediments at three sites along the Colne estuary in February 2005a
| Site | No. of transcripts of napA gene:
|
No. of transcripts of narG gene:
|
No. of transcripts of nirS gene:
|
No. of transcripts of nrfA-2 | |||||
|---|---|---|---|---|---|---|---|---|---|
| napA-1 | napA-2 | napA-3 | narG-1 | narG-2 | nirS-e&f | nirS-n | nirS-m | ||
| Hythe | 1.14 × 105 ± 1.14 × 105 | ND | 4.00 × 103 ± 4.00 × 103 | 3.08 × 105 ± 8.71 × 104 | 5.79 × 105 ± 1.66 × 104 | 2.42 × 106 ± 5.90 × 105 | ND | ND | ND |
| Alresford | 1.77 × 105 ± 1.77 × 105 | ND | 3.3 × 103 ± 3.3 × 103 | ND | ND | 9.77 × 105 ± 9.77 × 105 | ND | ND | ND |
| Brightlingsea | ND | ND | ND | ND | ND | ND | ND | ND | ND |
Gene transcript numbers are expressed as mean numbers of transcripts per gram of sediment. Surface sediments were those from depths of 0 to 1 cm. ND, not determined (no transcripts detected). Standard errors are indicated (n = 3). RT-Q-PCR standard curve descriptors and detection levels were as follows: napA-1, r2 = 0.997, y intercept = 44.94, and E = 92.7% (NTC undetected); napA-3, r2 = 0.997, y intercept = 37.85, and E = 88.6% (NTC undetected); narG-1, r2 = 0.997, y intercept = 38.15, E = 89.2%, and CT cutoff = 33.18; narG-2, r2 = 0.998, y intercept = 45.51, and E = 83.0% (NTC undetected); and nirS-e&f, r2 = 0.997, y intercept = 42.54, E = 88.6%, and CT cutoff = 34.13.
Distribution and diversity of AN bacteria along the Colne estuary.
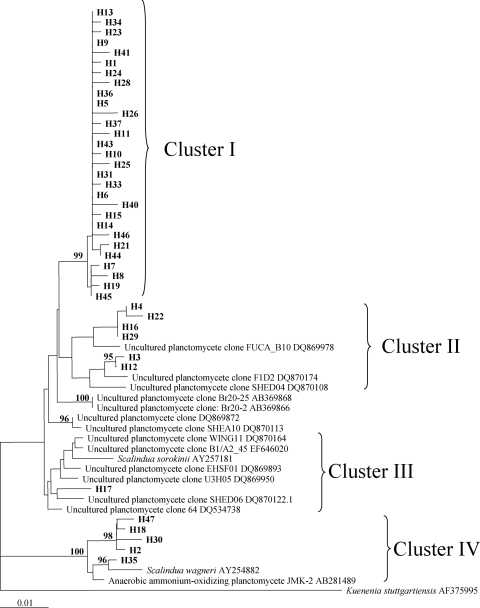
AN-related bacterial 16S rRNA genes were amplified from DNA isolated from the Hythe site but could not be detected at Alresford or Brightlingsea (data not shown). Subsequent sequence analyses of cloned 16S rRNA genes amplified from the Hythe sediments showed the genes to be highly similar (95 to 99% identical) to those from other uncultured planctomycetes from marine and freshwater sediments and from members of the “Candidatus Scalindua” genus (36). Phylogenetic analysis grouped the Hythe sequences into four clusters, with the majority (70% of these sequences) forming a cluster (Fig. 3, cluster I) with high bootstrap support (99%) that was distinct from other sequences from known AN bacteria and environmental clones in the GenBank database (∼97% identical to “Candidatus Scalindua sorokinii” sequences and ∼94% identical to “Candidatus Scalindua wagneri” sequences). A further 12% of the sequences from the Hythe site were most closely related to the 16S rRNA gene from “Candidatus Scalindua wagneri” (Fig. 3, cluster IV).
FIG. 3.
Neighbor-joining phylogenetic tree of 16S rRNA genes of planctomycete-related bacteria amplified from Hythe sediment samples (obtained in February 2005) by using the AN bacterium-specific primers Brod541F-Brod1260R. Evolutionary distances were calculated using the Kimura substitution algorithm. The 16S rRNA gene sequence from “Candidatus Kuenenia stuttgartiensis” was used as the out-group. Sequences from this study are indicated in bold. The scale bar indicates 10% sequence divergence. Bootstrap values above 70% are indicated.
Regression analyses of nitrate reduction process rates and nitrate and nitrite reductase gene abundances.
Regression analyses of the log(n + 1)-transformed process rate data compared against the log(n + 1) mean numbers of individual gene copies were conducted to investigate possible relationships between the rates of DN and DNRA and their corresponding genetic determinants. For DN, there was a significant regression relationship between DN rates and nirS gene numbers [for log (DN rate + 1) and log (nirS-n copy number + 1), r2 = 0.79 and P < 0.01; for log (DN rate + 1) and log (nirS-m copy number + 1), r2 = 0.71 and P < 0.01; and for log (DN rate + 1) and log (nirS-e&f copy number + 1), r2 = 0.85 and P < 0.01)]. For DNRA, there was also a significant regression relationship between DNRA rates and nrfA-2 numbers [log (DNRA rate + 1) and log (nrfA-2 copy number + 1): r2 = 0.94; P < 0.01]. Finally, a significant regression relationship between the rates of DN and DNRA [log (DN rate + 1) and log (DNRA rate + 1): r2 = 0.89; P < 0.01] along the estuary was also found.
DISCUSSION
Previously, we have described, separately, variation in the rates of DN (8, 9) and variation in the distribution and diversity of key nitrate and nitrite reductase genes driving nitrate reduction pathways (23, 38, 39) in sediments at three sites along the nutrient gradient of the Colne estuary. In the present study, we undertook an integrated investigation of the three main nitrate/nitrite reduction pathways by simultaneously measuring rates of DN and, for the first time in the Colne estuary, rates of DNRA and AN, with subsequent comparison to molecular data sets describing variation in the abundance of reductase and 16S rRNA genes serving as genetic markers for nitrate reduction, DN, DNRA, and AN within the estuarine sediments. The preliminary experiments that were conducted (see Fig. S1 in the supplemental material) confirmed that the conditions conformed to the requirements for the isotope-pairing technique, namely, that rates of 15NO3− reduction were related to the concentrations of 15NO3− added to core tubes, while the time course experiment confirmed that 15NO3− reduction was linear over the time period used.
Rates of DN declined from the estuary head (Hythe) to the mouth (Brightlingsea) (Fig. 1) as nitrate concentrations declined downstream, as shown previously for the Colne (7, 9) and other (43) estuaries. DNRA rates were also seen to decline along the Colne estuary, with the lowest rates at the marine site. This finding contrasts with those of a study of estuaries in Texas, in which potential DNRA activity was found to increase with increasing salinity (11), although in the latter study not all of the 15NH4+ generated was recovered. AN activity contributed about 30% of N2 formation at the Hythe site but was not detected at Alresford or Brightlingsea (Fig. 1). The proportion of N2 formation due to AN at the Hythe site was higher than any previous estimates of AN in estuarine sediments: Risgaard-Petersen et al. (31) suggested that AN accounted for up to ∼26% of N2 production in fjord sediments in Denmark, while Trimmer et al. (44) reported AN′s contributing only 1 to 8% of N2 production along the Thames estuary, United Kingdom. Recently, in the Chesapeake Bay in the United States, the proportion of N2 production due to AN in homogenized surface (0- to 1-cm-deep) sediments was found to range from 0 to 22%, with the highest proportions of N2 production via AN observed in freshwater sediments, where nitrate concentrations in the water column were greatest (28), as found along the Colne in our present study. It is perhaps not surprising that the level of AN activity is high at the Hythe site, which has highly organic sediments with high ammonium and nitrite concentrations in pore waters (8, 34).
Sediment slurry experiments showed that the potential for nitrate reduction declined markedly along the estuary (Table 1) and that the potential for N2 formation was greatest at the Hythe site. The ratio of the moles of N2 produced to the moles of NO3− removed decreased down the estuary, while the ratio of the moles of NH4+ produced to the moles of NO3− removed increased (Table 1), indicating a decrease in the potential for DN and an increase in the potential for DNRA down the estuary. This change in the relative significance of DN and DNRA has been attributed previously (4, 12, 16, 48) to increases in the ratio of electron donors to electron acceptors in soils or sediments, which stimulate DNRA relative to DN, and in the present case is due probably to the decrease in nitrate concentrations in the water column toward the mouth of the estuary. There is currently no way of directly measuring available organic matter within sediments. Recently, Strohm et al. (40) have shown that the levels of ATP synthesis in DN in laboratory cultures of denitrifying and nitrate-ammonifying bacteria are far lower than would be expected from free energy changes and are lower than those in DNRA. Thus, when competition for nitrate increases down the estuary, as nitrate concentrations decrease, nitrate-ammonifying bacteria are likely to be competitively more efficient than denitrifiers, as is indicated by the proportional increase in the importance of DNRA.
Interestingly, for both the Hythe and Brightlingsea sites, a large proportion (up to ∼55%) (Table 1) of the reduced nitrate (in terms of moles of nitrate removed) within the sediment slurries could not be accounted for by the formation of products of DN (N2O and N2) or DNRA (NH4+). For the Hythe site, the missing ∼45% of reduced nitrate may be accounted for partly by conversion by AN, as N2 formed via AN would not have been quantified using the acetylene-inhibited accumulation of N2O; Jensen et al. (14) have shown that acetylene inhibits AN but that N2O is not a product of AN. Indeed, in situ rate measurements (Fig. 1) showed AN to be responsible for ∼30% of the formation of N2 at the Hythe site in March 2005. It was also noticeable that during the slurry experiment, where large amounts of nitrate were present, nitrite never accumulated in the Hythe sediment slurry, although nitrite did accumulate in sediment slurries from Alresford and Brightlingsea, commensurate with AN′s removing nitrite at the Hythe site but not at Alresford and Brightlingsea. For the Brightlingsea site, we are not able to account for the missing ∼55% of reduced nitrate, although this proportion represents a much smaller amount of nitrate reduction within these sediments than that represented by the missing ∼45% within sediments from Hythe.
Results from slurry experiments in which sediments were either unamended or amended with chlorate to selectively inhibit NAR but not NAP (Table 1) suggested that NAP was proportionately more important than NAR at the Hythe site, where nitrate concentrations and reduction potentials were greatest, but that NAR activity increased proportionately, albeit at lower rates, at both Alresford and Brightlingsea. Richardson (29) has argued that NAR is associated with anaerobic energy conservation, expressed under anaerobic conditions, and that NAP is involved in redox balancing of chemoheterotrophic growth on reduced carbon sources under more oxidized conditions. In a nitrate-limited chemostat, a strain of Escherichia coli expressing only NAR was outcompeted by a second strain expressing only NAP, while under C-limited and nitrate-sufficient conditions, the strain expressing only NAR outcompeted the strain expressing only NAP (27). Hence, periplasmic NAP, which has a higher affinity for nitrate than NAR, was suggested to be more effective than NAR for nitrate scavenging and subsequent reduction at low nitrate concentrations and in oxidized environments. However, the increased importance of NAR activity at both Alresford and Brightlingsea, where nitrate concentrations are lower than those at the Hythe site, tends to contradict this model. Although the balance between electron acceptors and electron donors in the Colne estuary sediments is not known because it is not possible to directly measure the latter, the work of Potter et al. (27) might tend to suggest that the sediments at the Hythe site, where nitrate concentrations are highest, are more C limited than the sediments at sites lower down the estuary, which are more nitrate limited.
Variation in nitrate and nitrite reductase gene and transcript abundance along the estuary in February, 2005, was investigated using (RT)-Q-PCR. While declines in numbers of narG, nirS, and nrfA genes from the Hythe site to Brightlingsea were found (Fig. 2), napA genes of two of the three napA phylotypes (napA-2 and napA-3) were present in similar numbers at all three sites along the estuary. The results from our previous investigation of sediments sampled in October 2005 (38) had also shown a general decline in reductase gene numbers along the estuary, again with the exception of napA-3, the numbers of which remained similar. Together, these studies suggest some temporal stability in the numbers of the nitrate- and nitrite-reducing functional guilds within the Colne estuary. Quantification of nitrate and nitrite reductase gene transcripts showed these to be present only at the Hythe and Alresford sites (Table 2), consistent with the greater abundances of the corresponding genes at these sites (Fig. 2) and the higher nitrate reduction activity at these sites than at Brightlingsea (Fig. 1). While it should be recognized, as in any PCR-based study, that the use of specific primers based on a priori knowledge may not allow the targeting of the entire nitrate- and nitrite-reducing communities, this study has nevertheless attempted to quantify the abundance and expression of gene sequences that have been identified previously as being present or dominant within the Colne estuary (23, 38).
The detection of AN-related bacterial 16S rRNA genes by PCR only at the Hythe site in February 2005 was in agreement with the process rate measurements taken in March 2005, when AN activity occurred only at this site (Fig. 1). Sequence analyses of 16S rRNA genes from the Hythe sediments showed high similarities (95 to 99%) to those from other uncultured planctomycetes and to members of the taxa “Candidatus Scalindua wagneri” and “Candidatus Scalindua sorokinii” (Fig. 3, clusters III and IV). Interestingly, one dominant clade was present at the Hythe site (Fig. 3, cluster I), accounting for 77% of clones, and was distinct from the two “Candidatus Scalindua” spp., although more closely related to “Candidatus Scalindua sorokinii.” Previous 16S rRNA-based analyses of putative AN bacterial diversity have similarly identified only “Candidatus Scalindua”-related sequences in freshwater or marine sediments (25, 28), suggesting that this is the dominant AN taxon in nonwastewater environments and/or that the different primer sets used in these studies are potentially biased toward such sequences.
A general decline in both the rates of the three nitrate/nitrite reduction pathways and the abundances of nitrate and nitrite reductase genes (and transcripts) along the estuary was observed (see above). A correlation analysis of the data sets could not be used, as the samples for rate and gene abundance measurements were not paired. Significant regression relationships (P < 0.01) between the log(n + 1) rates of DN and the log(n + 1) abundances of the nirS genes of the three phylotypes and, similarly, between the log(n + 1) rates of DNRA and the log(n + 1) abundances of an nrfA gene of a single phylotype at sites along the estuary were found, showing that rates of DN and DNRA were broadly related at a log-log level to the abundances of genes encoding these different pathways. AN-related bacterial 16S rRNA genes were detected only at the Hythe site, concurrent with the detection of AN activity only at this site, where concentrations of ammonium and nitrite are greatest (8, 34). Moreover, the significant regression relationship of DN with DNRA (P < 0.01) suggests that generally similar environmental factors will simultaneously influence both processes along the estuary, the most likely of these being the decline in concentrations of nitrate and of electron donors in the surface sediments along the estuary.
While we have demonstrated significant broad (log-scale) relationships between nitrate reduction process rates and subgroups of corresponding functional genes, it is nevertheless important to recognize that more subtle transcriptional regulation of gene operons is occurring, together with further regulation of the proteins involved by the environment (e.g., temperature and oxygen and substrate concentrations). Such environmental regulation of these processes means that any tighter linkage than a broad log-log relationship between nitrate reduction process rates and abundances of corresponding reductase genes and/or transcripts is unlikely.
Supplementary Material
Acknowledgments
We thank John Green for excellent technical support in field sampling and in processing stable isotope samples.
This work was supported by the Natural Environment Research Council of the United Kingdom through research grant NER/A/S/2002/00962, awarded to D.B.N. and A.M.O. S.P. was supported by Marie-Curie intra-European fellowship 024108 from the European Commission.
Footnotes
Published ahead of print on 20 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.An, S., and W. S. Gardiner. 2002. Dissimilatory nitrate reduction to ammonium (DNRA) as a nitrogen link, versus denitrification as a sink in a shallow estuary (Laguna Madre/Baffin Bay, Texas). Mar. Ecol. Prog. Ser. 237:41-50. [Google Scholar]
- 2.Bothe, H., G. Jost, M. Schloter, B. B. Ward, and K.-P. Witzel. 2000. Molecular analysis of ammonification and denitrification in natural environments. FEMS Microbiol. Rev. 24:673-690. [DOI] [PubMed] [Google Scholar]
- 3.Burgin, A. J., and S. K. Hamilton. 2007. Have we over emphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front. Ecol. Environ. 5:89-96. [Google Scholar]
- 4.Christensen, P. B., S. Rysgaard, N. P. Sloth, T. Dalsgaard, and S. Schwaerter. 2000. Sediment mineralization, nutrient fluxes, denitrification, and dissimilatory reduction to ammonium in an estuarine fjord with sea cage trout farms. Aquat. Microb. Ecol. 21:73-84. [Google Scholar]
- 5.Clarke, K. R. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117-143. [Google Scholar]
- 6.Clarke, K. R., P. J. Somerfield, and M. G. Chapman. 2006. On resemblance measures for ecological studies, including taxonomic dissimilarities and zero-adjusted Bray-Curtis coefficient for denuded assemblages. J. Exp. Mar. Biol. Ecol. 330:55-80. [Google Scholar]
- 7.Dong, L. F., D. B. Nedwell, and A. Stott. 2006. Sources of nitrogen used for denitrification and nitrous oxide formation in sediments of the hypernutrified Colne, the nutrified Humber, and the oligotrophic Conwy estuaries, United Kingdom. Limnol. Oceanogr. 50:545-557. [Google Scholar]
- 8.Dong, L. F., D. B. Nedwell, G. J. C. Underwood, D. C. O. Thornton, and I. Rusmana. 2002. Nitrous oxide formation in the Colne estuary, England: the central role of nitrite. Appl. Environ. Microbiol. 68:1240-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, L. F., D. C. O. Thornton, D. B. Nedwell, and G. J. C. Underwood. 2000. Denitrification in sediments of the River Colne estuary, England. Mar. Ecol. Prog. Ser. 203:109-122. [Google Scholar]
- 10.Felsenstein, J. 1989. PHYLIP-Phylogeny Inference package. Cladistics 5:164-166. [Google Scholar]
- 11.Gardner, W. S., M. J. McCarthy, S. An, D. Sobolev, K. S. Sell, and D. Brock. 2006. Nitrogen fixation and dissimilatory nitrate reduction to ammonium (DNRA) support nitrogen dynamics in Texas estuaries. Limnol Oceanogr. 51:558-568. [Google Scholar]
- 12.Herbert, R. A., and D. B. Nedwell. 1990. Role of environmental factors in regulating nitrate respiration in intertidal sediments, p. 77-90. In N. P. Revsbech and J. Sorensen (ed.), Denitrification in soils and sediments. Plenum Press, New York, NY.
- 13.Hietanen, S. 2007. Anaerobic ammonium oxidation (anammox) in sediments of the Gulf of Finland. Aquat. Microb. Ecol. 48:197-205. [Google Scholar]
- 14.Jensen, M. M., B. Thamdrup, and T. Dalsgaard. 2007. Effects of specific inhibitors on anammox and denitrification in marine sediments. Appl. Environ. Microbiol. 73:3151-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, United Kingdom.
- 16.King, D., and D. B. Nedwell. 1987. The adaptation of nitrate-reducing bacterial communities in estuarine sediments in response to overlying nitrate load. FEMS Microbiol. Ecol. 45:15-20. [Google Scholar]
- 17.Koike, A., and A. Hattori. 1978. Denitrification and ammonium formation in anaerobic coastal sediments. Appl. Environ. Microbiol. 35:278-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer, B., E. W. Boyer, C. Goodale, N. A. Jaworski, N. V. Breeman, R. W. Howarth, S. Seitzinger, G. Billen, K. Lajtha, K. Nadelhoffer, D. V. Dam, L. J. Hetling, M. Nosal, and K. Paustian. 2002. Sources of nitrate in rivers draining sixteen watersheds in the northern U.S.: isotopic constraints. Biogeochemistry 57-58:171-197. [Google Scholar]
- 19.Mulder, A. January 1992. Anoxic ammonium oxidation. U.S. patent 5,078,884.
- 20.Nedwell, D. B., L. F. Dong, A. Sage, and G. J. C. Underwood. 2002. Variations of the nutrient loads to the mainland UK estuaries: correlations with catchment areas, urbanisation and coastal eutrophication. Estuar. Coast. Shelf Sci. 54:951-970. [Google Scholar]
- 21.Nedwell, D. B., T. Jickells, M. T. Trimmer, and R. Sanders. 1999. Nutrients in estuaries, p. 43-92. In D. B. Nedwell and D. Raffaelli (ed.), Estuaries. Academic Press, London, United Kingdom.
- 22.Nielsen, L. P. 1992. Denitrification in sediment determined from nitrogen isotope pairing. FEMS Microbiol. Ecol. 86:357-362. [Google Scholar]
- 23.Nogales, B., K. N. Timmiss, D. B. Nedwell, and A. M. Osborn. 2002. Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Appl. Environ. Microbiol. 68:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogilvie, B., D. B. Nedwell, R. M. Harrison, A. Robinson, and A. Sage. 1997. High nitrate, muddy estuaries as nitrogen sinks: the nitrogen budget of the River Colne estuary (United Kingdom). Mar. Ecol. Prog. Ser. 150:217-228. [Google Scholar]
- 25.Penton, R. C., A. H. Devon, and J. M. Tiedje. 2006. Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl. Environ. Microbiol. 72:6829-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philippot, L., and S. Hallin. 2005. Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr. Opin. Microbiol. 8:234-239. [DOI] [PubMed] [Google Scholar]
- 27.Potter, L. C., P. Millington, L. Griffiths, G. H. Thomas, and J. A. Cole. 1999. Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem. J. 344:77-84. [PMC free article] [PubMed] [Google Scholar]
- 28.Rich, J. J., O. R. Dale, B. Song, and B. B. Ward. 2008. Anaerobic ammonium oxidation (anammox) in Chesapeake Bay sediments. Microb. Ecol. 55:311-320. [DOI] [PubMed] [Google Scholar]
- 29.Richardson, D. J. 2000. Bacterial respiration: a flexible process for a changing environment. Microbiology 146:551-571. [DOI] [PubMed] [Google Scholar]
- 30.Risgaard-Petersen, N., R. L. Meyer, M. Schmid, M. S. M. Jetten, A. Enrich-Prast, S. Rysgaard, and N. P. Revsbech. 2004. Anaerobic ammonium oxidation in an estuarine sediment. Aquat. Microb. Ecol. 36:293-304. [Google Scholar]
- 31.Risgaard-Petersen, N., L. P. Nielsen, S. Rysgaard, T. Dalsgaard, and R. L. Meyer. 2003. Application of the isotope pairing technique in sediments where anammox and denitrification coexist. Limnol. Oceanogr. Methods 1:63-73. [Google Scholar]
- 32.Robinson, A. D., D. B. Nedwell, R. M. Harrison, and B. G. Ogilvie. 1998. Hypernutrified estuaries as sources of N2O emission to the atmosphere: the estuary of the River Colne, Essex, UK. Mar. Ecol. Prog. Ser. 164:59-71. [Google Scholar]
- 33.Rusmana, I., and D. B. Nedwell. 2004. Use of chlorate as a selective inhibitor to distinguish membrane-bound nitrate reductase (Nar) and periplasmic nitrate reductase (Nap) of dissimilative nitrate reducing bacteria in sediment. FEMS Microbiol. Ecol. 48:379-386. [DOI] [PubMed] [Google Scholar]
- 34.Sage, A. S. 1995. Removal of nitrate from estuarine water and its reduction in the bottom sediments. Ph.D. thesis. University of Essex, Colchester, United Kingdom.
- 35.Saitou, N., and M. Nei. 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Schmid, M., K. Walsh, R. Webb, W. I. C. Rijpstra, K. van de Pas-Schoonen, M. J. Verbruggen, T. Hill, B. Moffet, J. Fuerst, S. Schouten, J. S. S. Damste, J. Harris, P. Shaw, M. Jetten, and M. Strous. 2003. Candidatus “Scalindua brodae,” sp. nov., Candidatus “Scalindua wagneri,” sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 26:529-538. [DOI] [PubMed] [Google Scholar]
- 37.Simon, J. 2002. Enzymology and bioenergetics or respiratory nitrate ammonification. FEMS Microbiol. Rev. 26:285-309. [DOI] [PubMed] [Google Scholar]
- 38.Smith, C. J., D. B. Nedwell, L. F. Dong, and A. M. Osborn. 2007. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl. Environ. Microbiol. 73:3612-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, C. J., D. B. Nedwell, L. F. Dong, and A. M. Osborn. 2006. Evaluation of quantitative polymerase chain reaction-based approaches for determining gene copy number and gene transcript numbers in environmental samples. Environ. Microbiol. 8:804-815. [DOI] [PubMed] [Google Scholar]
- 40.Strohm, T. O., B. Griffin, W. G. Zumft, and B. Schink. 2007. Growth yields in bacterial denitrification and nitrate ammonification. Appl. Environ. Microbiol. 73:1420-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strous, M., J. A. Fuerst, E. H. M. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. M. Jetten. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trimmer, M., D. B. Nedwell, D. B. Sivyer, and S. J. Malcolm. 2000. Seasonal benthic organic matter mineralisation measured by oxygen uptake and denitrification along a transect of the inner and outer River Thames estuary, U.K. Mar. Ecol. Prog. Ser. 197:103-119. [Google Scholar]
- 44.Trimmer, M., J. C. Nicholls, and B. Deflandre. 2003. Anaerobic ammonium oxidation measured in sediments along the Thames estuary, United Kingdom. Appl. Environ. Microbiol. 69:6447-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trimmer, M., N. Risgaard-Petersen, J. C. Nicholls, and P. Engström. 2006. Direct measurement of anaerobic ammonium oxidation (anammox) and denitrification in intact sediment cores. Mar. Ecol. Prog. Ser. 326:37-47. [Google Scholar]
- 46.Tukey, J. W. 1953. The collected works of John W. Tukey, vol. 3. Multiple comparisons, p. 1948-1983. Chapman and Hall, New York, NY. [Google Scholar]
- 47.van de Graaf, A. A., A. Mulder, P. de Bruijn, M. S. Jetten, L. A. Robertson, and J. G. Kuenen. 1995. Anaerobic oxidation of ammonium is a biologically mediated process. Appl. Environ. Microbiol. 61:1246-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin, S. X., D. Chen, L. M. Chen, and R. Edis. 2002. Dissimilatory nitrate reduction to ammonium and responsible microorganisms in two Chinese and Australian paddy soils. Soil Biol. Biochem. 34:1131-1137. [Google Scholar]
- 49.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.