Abstract
Staphylococcus aureus RN6390 presents a diauxic growth in milk, due to amino acid limitation. Inactivation of the oligopeptide permease Opp3 (dedicated to the nitrogen nutrition of the strain) not only affects the growth of the strain but also results in reduced expression levels of three major extracellular proteases.
Staphylococcus aureus is a ubiquitous gram-positive pathogen commonly found in the environment. Some S. aureus strains are able to produce enterotoxins that cause food poisoning (6). In France, contaminated dairy products are the main source of staphylococcal food-borne disease outbreaks (3). Thus, the development of this bacterium in milk is a major concern for the safety of dairy products. Our objective was to evaluate the growth of S. aureus in milk, with special attention focused on the role of the peptide transport systems.
Opp3 is involved in amino acid supply during growth of S. aureus RN6390 in milk.
S. aureus strain RN6390 (7) was grown at 37°C in reconstituted skim milk (Nilac; Duiven, The Netherlands) with shaking (200 rpm). The time courses of cell populations for seven independent replicate cultures were evaluated by plating culture dilutions after disrupting bacterial clumps with an Ultra Turrax (IKA, Staufen, Germany).
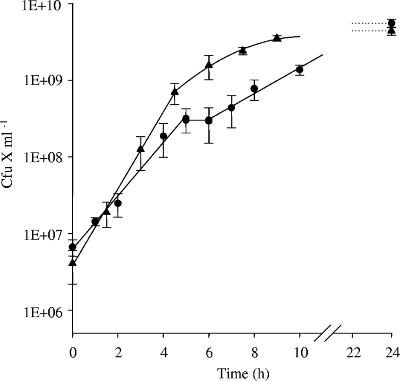
Linear regression analysis performed for each growth experiment discarded the hypothesis of a single exponential growth phase, with the standard errors of the estimate in the same range as the growth rates (from 0.70 to 1.19 h−1). S. aureus presented a diauxic growth in milk, with two distinct exponential growth phases separated by a short transition period (Fig. 1). The mean growth rates of the two exponential phases were 1.35 ± 0.03 h−1 and 0.60 ± 0.04 h−1 (means for seven determinations ± confidence limits at P values of 0.95), with standard errors of the estimate lower than 0.23 and 0.17 h−1, respectively.
FIG. 1.
Growth of the S. aureus RN6390 wild-type strain in milk (•) or in milk supplemented with a mixture of 18 amino acids (▴).
When milk was supplemented with a mixture of 18 amino acids (10), growth of S. aureus RN6390 was clearly modified (Fig. 1). Only one single exponential growth phase was observed (μ = 1.70 h−1), whereas the final population was not affected (ca. 5 × 109 CFU·ml−1). These results suggest that the amino acid composition, rather than the concentration of amino acids present in nonsupplemented milk, accounts for the growth limitation of the strain.
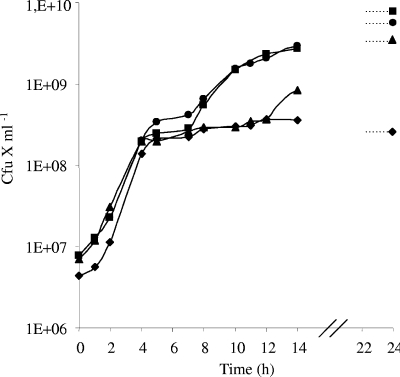
Five putative peptide transport systems are present in S. aureus: four ABC transporters, named Opp (oligopeptide permease), and one permease, DtpT. Among those, only Opp3 and DtpT exert a nutritional function, ensuring the import of tri- to octapeptides and di- and tripeptides, respectively (5). The role of these two peptide transport systems in growth in milk was evaluated by measuring the growth of opp3 and dtpT mutant strains. All deletion mutants were constructed by double crossover using the pMad vector as previously described (1, 5). While the dtpT mutant strain grew similarly to the wild-type strain, the growth of the opp3 mutant strain was altered, as the duration of the transition period was fivefold longer (Fig. 2). This suggests that oligopeptides are used as a source of amino acids at this stage of growth. The double opp3 dtpT mutant presented only the first exponential phase and a strongly reduced final population (about 10-fold), indicating that di- and tripeptides are a significant source of amino acids when the strain is unable to import oligopeptides. These results were further supported by growths in milk supplemented with a mixture of 18 amino acids in which all mutant strains grew as the wild-type strain (data not shown).
FIG. 2.
Growth of S. aureus RN6390 wild-type (•) and dtpT (▪), opp3 (▴), and opp3 dtpT (♦) mutant strains during growth in milk.
The growth in milk of the opp1 opp2 opp4 dtpT mutant strain was comparable to that for the wild type (data not shown), confirming the absence of significant nutritional function for Opp1, Opp2, and Opp4 systems, as reported previously (4).
Altogether, these results allow us to postulate the following three-stage scheme of nitrogen nutrition in S. aureus: (i) first, the bacteria utilize free amino acids from milk to reach a cell density of about 3 ×108 to 5 ×108 CFU ml−1; (ii) the pool of free amino acids in milk becomes too low, resulting in a transient growth arrest (this is supported by the fact that none of the amino acids required by S. aureus for an efficient growth, namely, Glu, Cys, Leu, Met, Gly, Val, Thr, Arg, and Lys [3], could be detected anymore by high-performance liquid chromatography in their free form in the milk cultures at this stage of growth [data not shown]); and (iii) a complementary source of amino acids supplied as peptides allows bacteria to restart growth and to reach high cell densities (about 5 × 109 CFU·ml−1).
Opp3 participates in extracellular protease regulation during growth in milk.
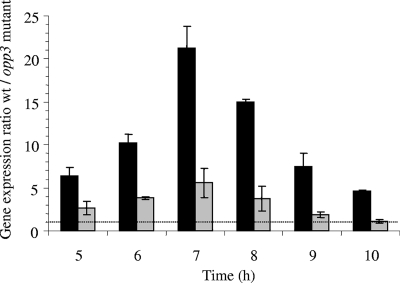
Milk coagulation was observed during the transition period of the wild-type strain. As the culture was stirred, S. aureus developed a respiratory metabolism that did not result in a decrease of pH. Consequently, milk clotting was most probably due to casein proteolysis. This phenomenon was delayed in the cultures of opp3 and opp3 dtpT mutant strains (1.5 h and 6 h, respectively). As the cell densities of the strains were in similar ranges at this growth stage (Fig. 2), this delay might be caused by reductions in the proteolytic activities of the mutant strains. To evaluate this hypothesis, the extracellular proteolytic activities of the wild-type and opp3 mutant strains were evaluated. Supernatants from transition period cultures were settled onto cellulose disks placed onto agar (1.5%) plates containing 3% casein. After incubation at 37°C, the halo of proteolysis (white precipitate of casein) observed with the opp3 mutant was smaller than that of the wild-type strain, indicating a lower proteolytic activity (data not shown). Ten extracellular proteases (SspA and B; Aur; ScpA; and SplA, B, C, D, E, and F) have been identified in S. aureus. To test a putative deregulation in the transcription of one or several of the proteases encoded by genes in the opp3 mutant strain, quantitative real-time PCR on the sspA, aur, scpA, and splA genes was performed (sspB and the splB to -F genes are cotranscribed with sspA and splA, respectively). RNAs and cDNA were prepared as previously described (5), and quantitative PCR was performed by using a SYBR green master mix (Applied Biosystems) with 2 ng of cDNA (primers are listed in Table 1). gyrB was validated as a stably expressed gene under our conditions and used to normalize the quantity of each cDNA tested. Inactivation of opp3 did not affect the expression levels of scpA and splA (data not shown). However, the levels of sspA and, to a lower extent, aur transcripts were reduced in the opp3 mutant (Fig. 3). As expected from the genetic organization of ssp genes, the sspB transcript level was also decreased (data not shown). We further observed a 1.5-h delay in milk coagulation in the culture of an sspA isogenic mutant strain (8), compared to the level for the parental RN6390 strain, confirming the implication of Ssp protease activity in the degradation of milk proteins (data not shown). Altogether, these results indicate that the delay in coagulation with the opp3 mutant strain results from a reduction in proteolytic activity caused at least by a decrease in ssp transcription. It is worth noting that no delay in coagulation could be observed between wild-type and opp3 mutant strains when milk was supplemented with a mixture of amino acids. This suggests that Opp3 could be involved in environmental sensing by modulating intracellular amino acid pools acting on pleiotropic regulators, as already demonstrated for other bacterial Opp systems (2, 4, 9). The amino acids and the regulator involved in this mechanism remain to be elucidated.
TABLE 1.
Oligonucleotide primers used for quantitative PCR
| Gene | Primer (5′-3′)
|
|
|---|---|---|
| First | Second | |
| gyrB | ATCGGTGGCGACTTTGATCT | CGGCATCAGTCATAATGACGATT |
| aur | TGAAGATGTCTACACACCTGGAAAA | AATTGTTCTGGGTTTGACATGCT |
| sspA | ACTTGTGAGTTCTCCAGCAGCAA | TGACTGCGTTTGTTGTGGAT |
| sspB | TGTAGGAATTATGATCCTTGCACAA | CAACTGCTAGCGCATGTCCTAA |
FIG. 3.
Effect of opp3 deletion on expression of sspA (black bars) and aur (gray bars) by S. aureus RN6390 grown in milk. Expression levels of sspA and aur in the S. aureus wild-type and opp3 mutant strains were estimated by quantitative PCR. For each strain and each time point, gene expression was normalized using gyrB as the stably expressed reference gene. Results are presented as ratios of gene expression (wild-type [wt] strain/opp3 mutant). Vertical bars indicate the standard deviations.
In conclusion, peptides transported by Opp3 play a dual role during growth of S. aureus in milk, supplying bacteria with nitrogenous nutrients and influencing expression of genes encoding three major extracellular proteases, most probably via an environmental sensing mechanism. Thus, Opp3 might represent a new target for controlling S. aureus development in dairy products.
Acknowledgments
We are grateful to Samira Makzhami and Claudia Bevilacqua (PICT, INRA Jouy en Josas, France) for their contribution to the quantitative-PCR analyses.
A. Hiron received a fellowship from the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche.
Footnotes
Published ahead of print on 13 March 2009.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkman, A. B., T. J. Ettema, W. M. de Vos, and J. van der Oost. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48:287-294. [DOI] [PubMed] [Google Scholar]
- 3.De Buyser, M. L., B. Dufour, M. Maire, and V. Lafarge. 2001. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int. J. Food Microbiol. 67:1-17. [DOI] [PubMed] [Google Scholar]
- 4.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227-1239. [DOI] [PubMed] [Google Scholar]
- 5.Hiron, A., E. Borezee-Durant, J. C. Piard, and V. Juillard. 2007. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 189:5119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Loir, Y., F. Baron, and M. Gautier. 2003. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2:63-76. [PubMed] [Google Scholar]
- 7.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rice, K., R. Peralta, D. Bast, J. de Azavedo, and M. J. McGavin. 2001. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203-207. [DOI] [PubMed] [Google Scholar]
- 10.Taylor, D., and K. T. Holland. 1989. Amino acid requirements for the growth and production of some exocellular products of Staphylococcus aureus. J. Appl. Bacteriol. 66:319-329. [DOI] [PubMed] [Google Scholar]