Abstract
Corynebacterium glutamicum accumulates up to 300 mM of inorganic polyphosphate (PolyP) in the cytosol or in granules. The gene products of cg0488 (ppx1) and cg1115 (ppx2) were shown to be active as exopolyphosphatases (PPX), as overexpression of either gene resulted in higher exopolyphosphatase activities in crude extracts and deletion of either gene with lower activities than those of the wild-type strain. PPX1 and PPX2 from C. glutamicum share only 25% identical amino acids and belong to different protein groups, which are distinct from enterobacterial, archaeal, and yeast exopolyphosphatases. In comparison to that in the wild type, more intracellular PolyP accumulated in the Δppx1 and Δppx2 deletion mutations but less when either ppx1 or ppx2 was overexpressed. When C. glutamicum was shifted from phosphate-rich to phosphate-limiting conditions, a growth advantage of the deletion mutants and a growth disadvantage of the overexpression strains compared to the wild type were observed. Growth experiments, exopolyphosphatase activities, and intracellular PolyP concentrations revealed PPX2 as being a major exopolyphosphatase from C. glutamicum. PPX2His was purified to homogeneity and shown to be active as a monomer. The enzyme required Mg2+ or Mn2+ cations but was inhibited by millimolar concentrations of Mg2+, Mn2+, and Ca2+. PPX2 from C. glutamicum was active with short-chain polyphosphates, even accepting pyrophosphate, and was inhibited by nucleoside triphosphates.
Inorganic polyphosphate (PolyP), a linear polymer made of up to hundreds of orthophosphate residues (Pi), has been found in all organisms tested for its presence (3, 4, 7, 12, 20, 22, 48). In nature's phosphorus cycle, diatom-derived PolyP has recently been shown to be critically important for marine phosphorus sequestration (6). In cells, PolyP may function as a means of storage of phosphorus and/or energy, may substitute ATP in kinase reactions, and was shown to be important in response to many stresses. Mutants of Escherichia coli, Pseudomonas aeruginosa, Shigella spp., Salmonella spp., Vibrio cholerae, and Helicobacter pylori with a low PolyP content showed defects in environmental stress responses and/or virulence (2, 14, 17, 38). In amino acid-starved E. coli, PolyP accumulates and is bound by Lon protease, which degrades ribosomal proteins to liberate amino acids (23).
The presence of PolyP granules is used as a diagnostic criterion to distinguish the pathogenic Corynebacterium diphtheriae from nonpathogenic corynebacteria, such as Corynebacterium glutamicum (54). However, these metachromatic granules have recently been shown to be present also in nonpathogenic C. glutamicum (33). When sufficient phosphate is available, C. glutamicum accumulates up to 300 mM of PolyP (24) either soluble in the cytosol or in volutin granules (18, 33). During growth of C. glutamicum on glucose, intracellular PolyP concentrations peaked in the early exponential growth phase and at the entry to stationary phase (18). Soluble PolyP prevailed in the stationary growth phase, while PolyP occurred in granules in the early exponential growth phase (18). C. glutamicum is widely used for the biotechnological production of about 2,200,000 tons of amino acids per year, mainly l-glutamate and l-lysine (50, 58), while the related Corynebacterium ammoniagenes is used for the production of the flavor-enhancing purine nucleotides IMP and XMP (30). As it is conceivable that engineering corynebacterial PolyP metabolism affects overproduction of amino acids or of the phosphorus-containing compounds IMP and XMP, the study of PolyP metabolism and the enzymes involved has recently received increasing attention.
PolyP formation in C. glutamicum was shown to be stimulated by MgCl2 (33), probably due to the magnesium dependence of PolyP synthesizing enzymes (27). In microorganisms, PolyP may be synthesized by PolyP kinases belonging to three distinct families (PPK1, PPK2, and PPK3; EC 2.7.4.1) from ATP or other nucleoside triphosphates (NTPs) in a reversible reaction (12). C. glutamicum possesses two PPK2 genes (ppk2A and ppk2B) (27). Purified PPK2B of C. glutamicum is active as a homotetramer and shows higher catalytic efficiency in the PolyP-forming direction than in the reverse direction, forming NTPs from PolyP. The intracellular PolyP content was increased by overexpression of ppk2B and decreased in the absence of PPK2B (27). Besides PPK2B, no other PolyP-dependent enzyme has been characterized in C. glutamicum, although the cg2091 gene product, a putative PolyP-dependent glucokinase (EC 2.7.1.63), was found to be associated with PolyP granules (33).
Degradation of PolyP by hydrolysis may be catalyzed by exopolyphosphatases (PPX) (EC 3.6.1.11) and/or endopolyphosphatases (PPN) (EC 3.6.1.10) (1, 49). Exopolyphosphatases hydrolyze PolyP from the chain's termini, liberating Pi. The C. glutamicum genome contains two genes encoding putative exopolyphosphatases (ppx1-cg0488 and ppx2-cg1115) (15), but their functions have not yet been characterized. The corresponding proteins are distinct from each other as they share only 25% identical amino acids. Both proteins show 25% amino acid identity to E. coli PPX (1), which possesses 200 additional C-terminal amino acids (56). Here, we have analyzed PolyP degradation in C. glutamicum and show that both cg0488 (ppx1) and cg1115 (ppx2) gene products are functional exopolyphosphatases. Growth experiments, determination of exopolyphosphatase activities, and intracellular PolyP concentrations in strains lacking or overexpressing these genes revealed that cg1115 (ppx2) encodes the major exopolyphosphatase of C. glutamicum, which was characterized enzymatically.
MATERIALS AND METHODS
Microorganisms and cultivation conditions.
All strains of Corynebacterium glutamicum used are based on the wild-type (WT) strain ATCC 13032. Deletion mutants lacking ppx1 or ppx2, the Δppx1 and Δppx2 strains, respectively, were used as well as the strains overexpressing ppx1 and ppx2, WT(pVWEx1-ppx1) and WT(pVWEx1-ppx2), respectively, which are based on the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible vector pVWEx1 (34). Luria-Bertani complex medium (44) was used for precultivation of C. glutamicum strains. When appropriate, kanamycin (50 μg/ml) and 1 mM IPTG were added. Growth experiments with C. glutamicum were carried out on CgXII medium (8) with glucose as the carbon source and were inoculated to an initial optical density at 600 nm (OD600) of 1 with washed LB precultures (10 min at 3,220 × g, washed once in an equal volume of 0.9% sodium chloride).
For a growth comparison of the WT, the Δppx1 and Δppx2 strains, and WT(pVWEx1), WT(pVWEx1-ppx1), and WT(pVWEx1-ppx2) with various phosphate concentrations, CgXII medium containing 13 mM, 0.13 mM, or no phosphate was used. For inoculation of these media, cells were harvested from LB precultures (containing 50 μg/ml and 1 mM IPTG for overexpression strains) by centrifugation (10 min at 3,220 × g), washed in phosphate-free medium, and centrifuged again. Fifty milliliters of the medium was inoculated to a final OD600 of 1 and incubated for 26 h at 30°C in 500-ml flasks. For enzyme activity determination in crude extracts, cells were grown on LB medium to the mid-exponential phase (OD600 of 4), harvested by centrifugation (10 min at 3,220 × g and 4°C), and washed in 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8, twice. Cells were stored at −20°C until use.
E. coli DH5α was used for cloning (11), and BL21(DE3) (52) was used for protein purification. BL21(DE3) carrying pET16b derivates was cultured on LB medium containing ampicillin (50 μg/ml). Biomass concentrations of C. glutamicum were calculated from OD600 values using the correlation of 0.25 g dry weight per OD600 (57).
Homologous overexpression of ppx1 and ppx2 from C. glutamicum.
For overexpression of ppx1, the following primers designed for amplification of the gene from genomic DNA of WT C. glutamicum were used: ppx1-OE-fw (5′-AAGGAGATATAGATGTGAGATTAGGTGTA-3′; the start codon is underlined; the ribosomal binding site is in italics) and ppx1-OE-rv (5′-TGTTACTCGAGTCCTTTGTCGATCCTG-3′; the stop codon is underlined). Similarly, for overexpression of ppx2, the gene was amplified via PCR from genomic DNA of WT C. glutamicum, which was prepared as described previously (9). PCR analysis was carried out using the following oligonucleotide primers: ppx2-OE-fw (5′-GCCTGCAGAAGGAGATATAGATATGACCCGTTACGCGGCC-3′; the start codon is underlined; the ribosome binding site is in italics; the PstI restriction site is bold) and ppx2-OE-rv (5′-GCTCTAGACTATTTCTTCAAAGAGTC-3′; the stop codon is underlined; the XbaI restriction site is bold). Amplified products were cloned into vector pGEM-T (Promega, Mannheim, Germany). DNA sequence analysis of the resulting plasmids pGEM-T-ppx1 and pGEM-T-ppx2 confirmed that the cloned PCR product did not contain mutations. Subsequently, ppx1 was cloned as a 984-bp fragment obtained by restriction of pGEM-T-ppx1 by NcoI, followed by treatment with the E. coli DNA polymerase I Klenow fragment and SalI restriction into the expression vector pVWEx1, which was restricted by XbaI restriction, followed by Klenow treatment and SalI restriction, to yield pVWEx1-ppx1. Similarly, the PstI-XbaI fragment of pGEM-T-ppx2 was cloned into PstI- and Xba-restricted pVWEx1, resulting in pVWEx1-ppx2. The vectors pVWEx1-ppx1 and pVWEx1-ppx2 allow IPTG-inducible expression of ppx1 and ppx2 in C. glutamicum.
Construction of the deletion mutants C. glutamicum Δppx1 and Δppx2.
In-frame deletions of ppx1 and ppx2 were constructed in WT C. glutamicum using pK19mobsacB (47) for two-step homologous recombination (8). Flanking regions of ppx1 were amplified by PCR analysis using the primer pairs Δppx1-A (5′-CATATCTAGACACGAATCGATGCCGCCGCTGGCGAAGACTCG-3′; the XbaI recognition site is bold) and Δppx1-B (5′-CCCATCCACTAAACTTAAACAGACGCGGTTTCCTTCTCCACGTGGTCG-3′; the 21-bp linker sequence is in italics) as well as Δppx1-C (5′-TGTTTAAGTTTAGTGGATGGGTCACATCAGATCGTGGCAGGTGCGCTAGTTGCGG-3′; the 21-bp linker sequence is in italics) and Δppx1-D (5′-GATATCTAGAGCTCAGGCTTGGGGGCCTCAACAACCTCAGGTTCTGCTG-3′; the XbaI recognition site is bold). Both amplified flanking regions were joined in a crossover PCR using primers Δppx1-A and Δppx1-D, and the resulting product was cloned into pK19mobsacB via its primer-attached XbaI recognition sites. The same strategy was used for the construction of a ppx2 deletion strain using primers Δppx2-A (5′-GCGGATCCCGAAGAGCAATTGGGAAGGGCC-3′; the BamHI restriction site is bold), Δppx2-B (5′-CCCATCCACTAAACTTAAACAGGCCGCGTAACGGGTCATC-3′; the 21-bp linker sequence is in italics), Δppx2-C (5′-TGTTTAAGTTTAGTGGATGGGCTTGGCCTGGTAGAAGCC-3′; the 21-bp linker sequence is in italics), and Δppx2-D (5′-GCAAGCTTTAGTGGACGATGAAGCAATCATC-3′; the HindIII restriction site is bold). The crossover PCR product was cloned into pK19mobsacB via the BamHI and HindIII recognition sites. Gene deletion mutagenesis with pK19mobsacBΔppx1 or pK19mobsacBΔppx2 was carried out as described previously (40). To verify deletion of ppx1 or ppx2, PCR amplifications using the primer pairs Δppx1-verif-A (5′-CCAATTAGACTCAAGCCACGTTAAATC-3′) and Δppx1-verif-B (5′-GCCCTCCACCGAAGCCACTTC-3′) or Δppx2-verif-A (5′-ACCAACTGAGGAAGCAACTGTG-3′) and Δppx2-verif-B (5′-CTTTGACATCGCAACTGCCCAT-3′) were performed, and deletion mutations were designated Δppx1 and Δppx2.
Heterologous expression of ppx2 from C. glutamicum, protein purification, and molecular weight determination.
For expression of ppx2 (cg1115) in E. coli BL21(DE3) (52), ppx2 was amplified via PCR from the genomic DNA of WT C. glutamicum using ppx2-EC-for (5′-GACCCATATGACCCGTTACGCGGCC-3′; the start codon is underlined; the NdeI recognition site is bold) and ppx2-EC-rev (5′-CTATTTCTTCAAAGAGTCGGCTTCTACC-3′; the stop codon is underlined). The amplified product was cloned into vector pGEM-T (Promega, Mannheim, Germany), resulting in vector pGEM-T-ppx2EC, and the absence of mutations was confirmed by sequencing. A ppx2 fragment of 999 bp obtained by NdeI restriction of pGEM-T-ppx2EC was ligated to NdeI-restricted pET16b (Novagen, Madison, WI). The vector, pET16b-ppx2, allows production of PPX2 carrying an N-terminal decahistidyl tag in E. coli BL21(DE3). LB medium was inoculated with a single colony from fresh transformation and grown overnight. For protein production, 500 ml of LB medium was inoculated with 5 ml of the overnight culture and incubated at 37°C. Induction with 0.5 mM IPTG was started at an OD600 of 0.5 to 0.6 and incubated at room temperature. The cells were harvested 4 h after induction, washed in 20 mM Tris, 300 mM NaCl, 5 mM imidazole, and 5% (vol/vol) glycerol (TNI 5) and stored at −20°C until protein purification. Protein purification was performed as described previously (27). The desalted protein was buffered in 50 mM PIPES, pH 6.8. The molecular weight of the purified protein was measured by gel filtration in 50 mM PIPES, pH 6.8, with 2 mM MgCl2 and 25 mM KCl as described previously (27).
Exopolyphosphatase assay.
Exopolyphosphatase activity was measured discontinuously by taking samples of 10 μl from a 1-ml reaction mixture containing 50 mM PIPES, pH 6.8, 25 mM KCl, and 2 mM MgCl2 at defined times and by using quantification of PolyP with 1 ml toluidine blue solution (6 mg/liter) containing 40 mM acetic acid (31). The ratio of 530 nm to 630 nm was measured spectrophotometrically after calibration using PolyP20 in concentrations of up to 10 mM Pi units. Alternatively, the EnzChek phosphate assay kit (Molecular Probes, Göttingen, Germany) was used according to the instructions of the supplier to measure exopolyphosphatase activity in a continuous spectrophotometric assay. This assay was carried out at 30°C in 1-ml cuvettes containing 50 mM PIPES, pH 6.8, 25 mM KCl, 2 mM MgCl2. Conditions were optimized using concentrations of MgCl2 (0 to 20 mM) and KCl (0 to 200 mM), temperatures from 10 to 67°C, a pH range from 5.6 to 8.3, and PolyP concentrations of up to 50 mM (Pi residues). To characterize the substrate spectrum, PolyPs of the following different chain lengths were used: PolyP3, PolyP5, PolyP15, PolyP25, PolyP45, PolyP65, PolyP75 (Sigma-Aldrich, Taufkirchen, Germany), and PolyP20 (B. K. Giulini, Ladenburg, Germany). All PolyP concentrations are indicated in millimolar Pi units.
31P NMR spectroscopy.
Intracellular PolyP contents were analyzed by 31P nuclear magnetic resonance (NMR) spectroscopy as described earlier (27).
Statistical analysis.
To determine whether changes in biomass formation or exopolyphosphatase activity were statistically significant, the t test was applied; data were marked by an asterisk for a P value of <0.05 and by two asterisks for a P value of <0.005.
Sequence comparison and alignment.
Sequence homologues of the examined proteins were obtained from NCBI using BLAST and aligned using ClustalW, and phylogenetic trees were constructed using the neighbor-joining method (43) with 1,000 bootstrap replicates. Accession numbers (gene identifiers) of proteins used for comparison were as follows: Cg0488 (PPX1), Cg1115 (PPX2), and Cg2988 (PPA) of C. glutamicum; DIP0920 of Corynebacterium diphtheriae; ro02065 and ro05780 of Rhodococcus sp. RHA1; MSMEG_0939 and MSMEG_5413 of Mycobacterium smegmatis strain MC2 155; Mmcs_0664 and Mmcs_4237 of Mycobacterium sp. MCS; Arth_3352 and Arth_1150 of Arthrobacter sp. FB24; BlinB01001419 and BlinB01002403 of Brevibacterium linens BL2; CE0428 and CE1046 of Corynebacterium efficiens YS-314; cu0217 and cu0592 of Corynebacterium urealyticum DSM 7109; nfa51810 and nfa48560 of Nocardia farcinica IFM 10152; Rv0496 and Rv1026 of Mycobacterium tuberculosis H37Rv; ML2434 and ML0258 of Mycobacterium leprae TN; SCO3348 and SCO3093 of Streptomyces coelicolor A3(2); AAur_3332 and AAur1_1266 of Arthrobacter aurescens TC1; Npun_R4665 of Nostoc punctiforme PCC 73102; Ava_3530 of Anabaena variabilis; jk1480 and jk1908 of Corynebacterium jeikeium K411; b2502 (PPX), b4226 (PPA), and b3779 (guanosine pentaphosphate phosphohydrolase [GPP]) of Escherichia coli strain K12 substrain MG1655; SSO1193 of Sulfolobus solfataricus P2; aq_891 of “Aquifex aeolicus” VF5; AAK69116 of Serratia marcescens; AAO48270 of Trypanosoma cruzi; YDR452W (PPN1), YBR011C (PPA), and YHR201C (PPX1) of Saccharomyces cerevisiae; PA5241 of Pseudomonas aeruginosa PAO1; MAP0993 and MAP3987 of Mycobacterium avium subspecies paratuberculosis K-10; and VC0395_A2696 (GPP) of Vibrio cholerae O395l217.
RESULTS
Overexpression and deletion of genes coding for putative exopolyphosphatases.
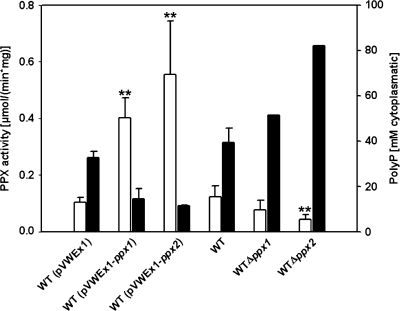
In order to determine whether the gene products of ppx1-cg0488 and ppx2-cg1115 carry exopolyphosphatase activity and to characterize their influence on growth and PolyP accumulation by C. glutamicum, these genes were cloned into the IPTG-inducible expression vector pVWEx1 and the resulting plasmids were transformed into WT C. glutamicum. Determination of exopolyphosphatase activity in crude extracts of C. glutamicum WT(pVWEx1), WT(pVWEx1-ppx1), and WT(pVWEx1-ppx2) revealed that overexpression of ppx1 increased the specific activity of exopolyphosphatase by fourfold (0.43 ± 0.01 μmol min−1 mg−1) compared to that of the empty vector control (0.11 ± 0.01 μmol min−1 mg−1) (Fig. 1). Crude cell extracts of C. glutamicum WT(pVWEx1-ppx2) showed sixfold-higher exopolyphosphatase activity (0.80 ± 0.29 μmol min−1 mg−1) than those of WT(pVWEx1) (0.11 ± 0.01 μmol min−1 mg−1) (Fig. 1). Thus, both ppx1 and ppx2 appear to code for enzymes with exopolyphosphatase activity. Accordingly, analysis of the constructed deletion mutations Δppx1 and Δppx2 revealed reduced exopolyphosphatase activities in the absence of ppx1 (0.08 ± 0.03 μmol min−1 mg−1) and ppx2 (0.04 ± 0.02 μmol min−1 mg−1) compared to those of WT C. glutamicum (0.12 ± 0.04 μmol min−1 mg−1) (Fig. 1).
FIG. 1.
Exopolyphosphatase activity and PolyP accumulation in various strains of C. glutamicum. Open columns, exopolyphosphatase activity; filled columns, PolyP content expressed in millimolar Pi. Average values and standard deviations of at least three independent determinations are shown. Enzyme activity was measured spectrophotometrically using the EnzChek phosphate assay kit in 1 ml containing 50 mM PIPES, pH 6.8, 25 mM KCl, 2 mM MgCl2, and 40 mM PolyP20. *, P < 0.05; **, P < 0.005.
The cellular PolyP contents of WT C. glutamicum, strains Δppx1 and Δppx2, and WT(pVWEx1), WT(pVWEx1-ppx1), and WT(pVWEx1-ppx2) were analyzed by 31P NMR (Fig. 1). While the deletion of ppx1 and ppx2 resulted in an increase in intracellular PolyP content (a slight increase for Δppx1; a twofold increase for Δppx2) compared to that of WT C. glutamicum, the overexpression of these genes reduced the cellular PolyP content by more than twofold for WT(pVWEx1-ppx1) and WT(pVWEx1-ppx2) compared to the empty vector control WT(pVWEx1).
Taken together, as overexpression and deletion of ppx2 had a greater influence on cellular PolyP content and exopolyphosphatase activity than overexpression and deletion of ppx1, PPX2 appears to be the major exopolyphosphatase of C. glutamicum and was chosen for purification and enzymatic characterization.
Biochemical characterization of exopolyphosphatase PPX2His.
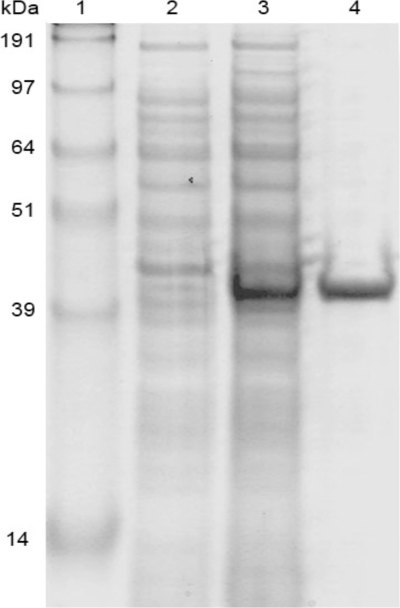
In order to produce PPX2 with an N-terminal decahistidyl tag, ppx2 was cloned into pET16b and expressed in E. coli BL21(DE3). PPX2His was purified from the soluble fraction of the cell extract as shown in Fig. 2. A potential effect of including the His tag on PPX2 activity could not be assessed, as attempts to cleave the His tag were not successful. As judged from gel filtration chromatography, PPX2His is active as a monomer (data not shown). Exopolyphosphatase activity could either be monitored discontinuously by following the decrease of toluidine blue-detectable PolyP or continuously by following phosphate formation in a purine ribonucleoside phosphorylase-coupled assay.
FIG. 2.
Purification of PPX2His of C. glutamicum from E. coli BL21(DE3) (pET16b-ppx2). (Lane 1) SeaBlue Plus2 prestained standard (Invitrogen) containing proteins of the indicated molecular masses. Also shown is Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of protein from extracts before (lane 2) and 4 h after (lane 3) (both 150 μl OD600−1) induction with 0.5 mM IPTG and 12 μg PPX2His purified by Ni-nitrilotriacetic acid chromatography (lane 4).
Optimal conditions for the exopolyphosphatase activity of PPX2His were determined. Of the bivalent cations tested at a concentration of 2 mM, Mg2+ showed the highest stimulation of PPX2His activity. Whereas CaCl2, BaCl2, CuSO4, and SnCl2 did not show any activating effect on PPX2His, the addition of 2 mM MnCl2, FeCl2, and ZnSO4 also stimulated PPX2His but to lesser extents (0.86-fold, 0.26-fold, and 0.11-fold compared to that of MgCl2) (data not shown). When various concentrations of MgCl2 and MnCl2 were tested, a gradual increase in activity was observed up to concentrations of about 2 mM. Higher concentrations of MgCl2 and MnCl2 reduced PPX2His activity, e.g., twofold at 3.5 mM MnCl2 and twofold at 10 mM MgCl2. At a pH of 6.8, the highest activity of PPX2His was reached in 50 mM of PIPES buffer (tested in a pH range of 6 to 8.3). PPX2His activity was further increased in the presence of KCl, with 25 mM KCl resulting in a threefold increase in the specific activity.
To determine the stability of PPX2His against irreversible thermal denaturation, the enzyme was preheated in 50 mM PIPES buffer, pH 6.8, containing 2 mM MgCl2 and 25 mM KCl, before measuring the activity at 30°C, the optimal temperature for growth of C. glutamicum. After preincubation for 60 min at 25°C, 30°C, and 40°C, no significant loss of activity was observed (data not shown), while at temperatures above 40°C PPX2His, activity decreased. After preheating at 45°C for 30 and 60 min, respectively, 70% and 40% of the PPX activity remained. When preheated at 50°C, PPX2His lost its activity quickly, and after incubation for 60 min, no activity remained. Preheating at 60°C inactivated PPX2His completely within 5 min (data not shown).
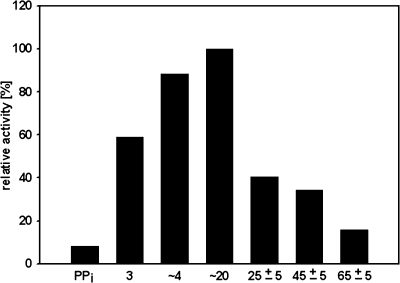
When various short- to medium-chain PolyPs were tested, PPX2His was most active with PolyPs of 3 to 20 phosphate residues (Fig. 3). As the activity with PPX2His with long-chain PolyPs could not be tested because these were not available commercially, it could not be excluded that PPX2His is active with long-chain PolyPs. While comparable maximal activities of PPX2His were observed with PolyP3, PolyP4, and PolyP20 as substrates, the substrate affinity was highest for PolyP3. Thus, PPX2His showed the highest catalytic efficiency with PolyP3 as the substrate (a kcat/Km ratio of 15.5 s−1 mM−1) (Table 1). Because PPX2His is able to hydrolyze the inorganic diphosphates and triphosphates PolyP3 and pyrophosphate, it was tested whether NTPs were substrates of PPX2His. However, PPX2His did not show detectable activity with ATP or GTP.
FIG. 3.
Influence of PolyP chain length on the activity of PPX2His. For PolyP standards, see Materials and Methods. Enzyme activity was measured spectrophotometrically using the EnzChek phosphate assay kit in 1 ml containing 50 mM PIPES, pH 6.8, 25 mM KCl, 2 mM MgCl2, up to 15 μg PPX2His, and up to 40 mM PolyP.
TABLE 1.
Kinetic parameters of PPX2His from C. glutamicuma
| Substrate | Km (mM Pi) | Vmax (μmol min−1 mg−1 of protein) | kcat (s−1) | kcat/Km(s−1 mM−1) |
|---|---|---|---|---|
| PolyP3 | 0.04 | 1.0 | 0.6 | 15.5 |
| PolyP4 | 0.11 | 1.5 | 0.9 | 8.5 |
| PolyP20 | 9.70 | 1.7 | 1.0 | 0.1 |
Enzyme activity was measured spectrophotometrically using the EnzChek phosphate assay kit in 1 ml reaction mixture containing 50 mM PIPES, pH 6.8, 25 mM KCl, and 2 mM MgCl2; PolyP concentrations of up to 1, 10, and 40 mM for PolyP3, PolyP4, and PolyP20, respectively; and up to 15 μg of purified PPX2His.
Interestingly, NTPs, deoxynucleoside triphosphates (dNTPs), and ADP were identified as inhibitors of PPX2His that showed half-maximal inhibition (Ki) at 0.4 mM for dGTP, 0.6 mM for ADP and dATP, 0.8 mM for GTP and ITP, and 2.6 mM for ATP. While glucose, fructose, and the central carbon metabolites glucose 6-phosphate, fructose 6-phosphate, fructose 1,6-bisphosphate, fructose 1-phosphate, 6-phosphogluconate, ribose 5-phosphate, and α-ketoglutarate did not inhibit PPX2His (data not shown), concentrations of 3.8 mM CaCl2 and 8.2 mM pyrophosphate led to half-maximal inhibition of PPX2His. Thus, PPX2 activity appears to be controlled by the “energy charge” of the C. glutamicum cell.
Role of exopolyphosphatases for growth of C. glutamicum after transfer to phosphate-limiting conditions.
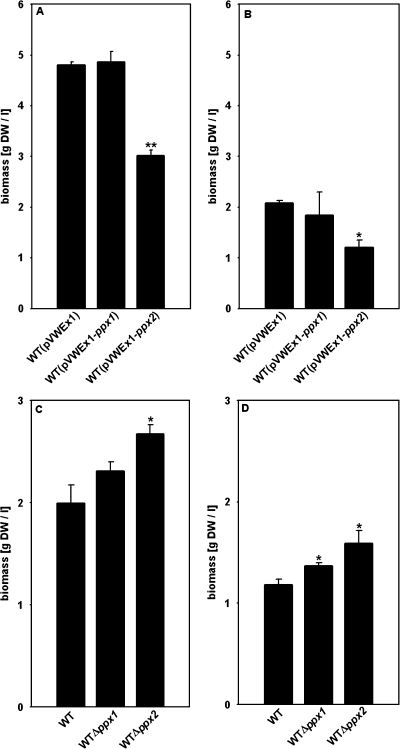
With sufficient phosphate supply, overexpression of ppx1 and ppx2 mildly slowed growth of C. glutamicum on CgXII minimal medium containing 200 mM glucose as the carbon source. While C. glutamicum WT(pVWEx1) reached a growth rate of 0.39 ± 0.01 h−1, C. glutamicum WT(pVWEx1-ppx1) and WT(pVWEx1-ppx2) grew slightly slower (growth rates of 0.37 ± 0.01 h−1 and 0.31 ± 0.01 h−1, respectively), but all three strains grew to comparable biomass concentrations (data not shown). The ppx1 and ppx2 deletion strains Δppx1 and Δppx2 grew as fast as the WT (growth rates of 0.39 ± 0.02 h−1, 0.40 ± 0.01 h−1, and 0.41 ± 0.01 h−1, respectively) and formed comparable biomass concentrations to WT C. glutamicum under phosphate-sufficient conditions (data not shown). When transferred from phosphate-sufficient to phosphate-limiting conditions, C. glutamicum WT(pVWEx1-ppx1) and WT(pVWEx1-ppx2) exhibited a growth disadvantage and formed less biomass than C. glutamicum WT(pVWEx1) (Fig. 4B). Accordingly, the Δppx1 and Δppx2 deletion mutants formed more biomass and showed higher growth rates than WT C. glutamicum (0.28 ± 0.01 h−1 and 0.27 ± 0.01 h−1, respectively, compared to 0.24 ± 0.01 h−1) when shifted to phosphate-limiting conditions (Fig. 4D and data not shown).
FIG. 4.
Biomass formation (grams dry weight per liter) of C. glutamicum strains WT(pVWEx1), WT(pVWEx1-ppx1) and WT(pVWEx1-ppx2) (A and B), the WT, and the Δppx1 and Δppx2 strains (C and D), after transfer to minimal medium with different phosphate concentrations. Cells cultured overnight on LB medium were used to inoculate CgXII minimal medium with 4% (wt/vol) glucose and 0.13 mM (A and C) or 0 mM (B and D) phosphate. Cultivations shown in panels A and B were performed in the presence of 25 μg/ml kanamycin and 1 mM IPTG. Biomass formation was determined after 12 to 13 h of incubation. Averages and standard deviations of at least three independent cultivations are shown. *, P < 0.05; **, P < 0.005.
Phylogenetic analysis of PPX1 and PPX2.
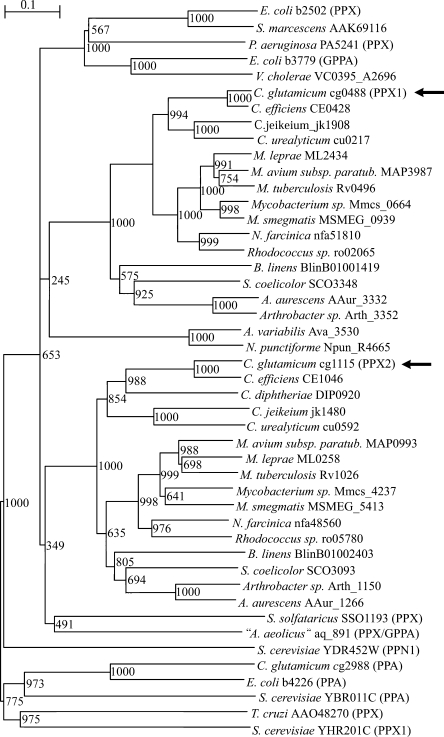
Databank searches with the amino acid sequences for PPX1 and PPX2 of C. glutamicum revealed similarities to the biochemically characterized exopolyphosphatases, e.g., those of E. coli, “Aquifex aeolicus,” and S. cerevisiae, as well as to a number of putative or hypothetical proteins from other bacteria. A phylogenetic tree of the exopolyphosphatase protein family was constructed using the deduced amino acid sequences of the biochemically characterized exopolyphosphatases from E. coli (1), P. aeruginosa (61), S. solfataricus (5), T. cruzi (10), S. cerevisiae (PPX1 and PPN1) (49, 60), and “A. aeolicus” (PPX/GPP) (21) and of 32 putative exopolyphosphatases deposited in public databases. In addition, the amino acid sequences of the biochemically characterized inorganic pyrophosphatases (PPA) from S. cerevisiae, E. coli, and C. glutamicum (36), the guanosine pentaphosphate phosphohydrolases (GPP) of E. coli (16), and the putative GPP of V. cholerae were included in this analysis. The resulting phylogenetic tree is depicted in Fig. 5. Proteins homologous to PPX2 from C. glutamicum are encoded in the genomes of actinomycetes. The PPX2 homologues can be clearly distinguished from the cluster of proteins homologous to PPX1 from C. glutamicum as well as from the cluster of enterobacterial exopolyphosphatases such as PPX of E. coli and from the exopolyphosphatases of the archaea “A. aeolicus” and Sulfolobus solfataricus. Neither exopolyphosphatase PPN1 nor PPX1 from S. cerevisiae nor the pyrophosphatases from S. cerevisiae, E. coli, and C. glutamicum clustered with the other proteins. The phylogenetic analysis suggests that, e.g., the M. tuberculosis homologues of PPX1 (Rv0496) and PPX2 (Rv1026), which are annotated as conserved hypothetical proteins, might be active as exopolyphosphatases. The genomes of the actinomycetes typically encode homologues of both PPX1 and PPX2, as is the case for the corynebacteria C. efficiens, C. jeikeium, and C. urealyticum, whereas C. diphtheriae lacks a PPX1 homologue.
FIG. 5.
Phylogenetic tree of polyphosphatase (PPX, PPN), pyrophosphatase (PPA), and guanosine pentaphosphate phosphohydrolase (GPP) proteins and putative homologues. Numbers at the nodes represent bootstrap values. Full names of organisms are listed in Materials and Methods. Gene identifiers are shown after the names of organisms. Names of biochemically characterized enzymes are in parentheses. The positions of PPX1 and PPX2 of C. glutamicum are pointed out by arrows. M. smegmatis, Mycobacterium smegmatis; M. avium subsp. paratub., M. avium subspecies paratuberculosis.
A structure-based sequence comparison based on the crystal structure of PPX from E. coli (37) was performed using PPX protein sequences from P. aeruginosa (61), S. solfataricus (5), PPX1 and PPX2 from C. glutamicum, and their homologues from M. tuberculosis, N. farcinica, S. coelicolor, and C. diphtheriae. All analyzed protein sequences contained the putative catalytic Glu and Arg residues corresponding to Glu121 and Arg93 from E. coli (37). The probable Mg2+ coordinating sites (Asp143 and Glu150 in PPX from E. coli) are present in all sequences except for PPX from S. solfataricus (Glu instead of Asp143). Also, the glycine-rich phosphate-binding loop (P-loop Gly145-Ser148) (37) is present in all sequences examined, while the polyphosphate-binding canyon (Fig. 6) is not conserved in the examined proteins. Several regions distinguish C. glutamicum PPX1 and homologues from C. glutamicum PPX2 and homologues (Fig. 6) while, e.g., C. glutamicum PPX1 and its homologues contain Glu261-Ile-Cys-Pro-Phe-Ala-Leu-Arg268 (numbers from C. glutamicum PPX1) in the region α helix α11; this region is less conserved in C. glutamicum PPX2 and its homologues (Fig. 6). On the other hand, α helix α12 contains Asp304-Ile-Leu-Asp-Gly308 (numbers from C. glutamicum PPX2) in C. glutamicum PPX2 and its homologues, whereas the corresponding region in C. glutamicum PPX1 and its homologues contain Leu/Val-Ile-Leu-Arg/Thr-Arg/Lys (Fig. 6).
FIG. 6.
Structure-based sequence alignment of the characterized exopolyphosphatases from E. coli, P. aeruginosa, and S. solfataricus with PPX1 and PPX2 from C. glutamicum and their homologues from M. tuberculosis, N. farcinica, S. coelicolor, and C. diphtheriae. The alignment was based on the crystal structure of E. coli PPX (37), and secondary structural motifs are highlighted above the alignment. Probable Mg2+ coordinating Asp143 and Glu150 are marked with asterisks. Putative catalytic Glu121 and Arg93 are highlighted with a circle and a square, respectively. Triangles indicate putative PolyP-binding canyons and diamonds indicate P-loop residues Gly145-Ser148. Gene identifiers are shown after the names of organisms. Sequence alignment was carried out using ClustalW, and the alignment was formatted using BoxShade.
DISCUSSION
As shown here, the genome of C. glutamicum codes for two functional exopolyphosphatases which are distinct from each other and from other known exopolyphosphatases. The phylogenetic analysis of PPX1 and PPX2 from C. glutamicum and of other characterized and putative exopolyphosphatases (Fig. 5) indicated that at least six different groups of exopolyphosphatase enzymes exist, i.e., those similar to (i) enterobacterial PPX (e.g., E. coli PPX) (1), (ii) S. cerevisiae PPN1 (49), (iii) S. cerevisiae PPX1 (60), (iv) archaeal PPX (e.g., S. solfataricus PPX) (5), (v) C. glutamicum PPX1, and (vi) C. glutamicum PPX2. The diversity of polyphosphatase enzymes known currently exceeds that of the three classes of PolyP kinases (4). Exopolyphosphatase genes from other members of the suborder Corynebacterineae of the Actinomycetales have not yet been characterized. Based on the sequence similarity of deduced gene products, a number of homologues of PPX1 and PPX2 from C. glutamicum (Fig. 5) were identified in Corynebacterium, Rhodococcus, Nocardia, and Mycobacterium species (and also in representatives of other suborders of the Actinomycetales, such as Streptomyces coelicolor, Arthrobacter aurescens, or Brevibacterium linens), and it is proposed that these proteins are also active as exopolyphosphatases. Although gene-directed mutagenesis has not been used to study the function of the PPX1 and PPX2 homologues in other species of the Corynebacterineae, some information about the Rv0496 and Rv1026 genes from M. tuberculosis is available from a transposon mutant screen, in which Rv0496 and Rv1026 were not predicted to be required for in vivo survival in the M. tuberculosis transposon mutant screen (45). Both genes were implied in M. tuberculosis pathogenicity, as Rv0496 was identified as a novel T-cell antigen (42) and Rv1026 was shown to be induced when M. tuberculosis infects macrophages (51). Whether the PPX1 and PPX2 homologues encoded by Rv0496 and Rv1026 in M. tuberculosis are indeed active as exopolyphosphatases and, if so, whether their enzymatic activity is relevant to pathogenicity remain to be studied. It is conspicuous that genomes of representatives of the Actinomycetales in general contain both a PPX1 and a PPX2 homologue, while the genome of C. diphtheriae contains a PPX2 gene homologue but lacks a PPX1 gene homologue. This observation likely reflects the fact that gene loss played an important role in the evolution of the C. diphtheriae genome (32), which comprises 2,320 genes compared to 3,002 genes in C. glutamicum.
The occurrence of multiple exopolyphosphatases within a species is not restricted to members of the Actinomycetales but is also known, e.g., for E. coli and S. cerevisiae. In E. coli, PolyP may be hydrolyzed by PPX (1), by SurE, a stationary-phase survival protein (35), as well as by GPP, the guanosine pentaphosphate phosphohydrolase involved in the stringent response (16). Currently, it is not known whether the bifunctional GPP/synthetase from C. glutamicum encoded by the rel gene (55) is active as an exopolyphosphatase. In S. cerevisiae, two genes encoding exopolyphosphatases are known, PPX1 (60) and PPN1 (49). However, the finding that a PPX1 PPN1 double mutant is still able to hydrolyze PolyP suggests that an additional, as-yet-unknown polyphosphatase is active in S. cerevisiae (26). Nonoverlapping substrate specificity may be a reason for the occurrence of two or more exopolyphosphatases within one species. In E. coli, PPX and GPP hydrolyze long-chain PolyPs (1, 16) and SurE hydrolyzes short-chain PolyPs (35). Human PPX and the PPX of Leishmania major also prefer short-chain PolyPs (41, 53). In S. cerevisiae, subcellular localization and substrate specificity distinguish the PPX1 and PPN1 gene products (25). PPX1 is a ∼40-kDa exopolyphosphatase which is active in the cytosol and soluble mitochondrial fraction and preferably hydrolyzes short-chain PolyPs. High-molecular-mass polyphosphatase (120 to 830 kDa) activities hydrolyzing long-chain PolyPs are present in the nucleus and the mitochondrial membrane fraction and are dependent on PPN1. PPX2 from C. glutamicum was active with short-chain PolyPs (Fig. 3). Based on the finding that the chain length of PolyP extracted from C. glutamicum cells was 800 to 1,000 (C. Lambert and S. M. Schoberth, unpublished data) and the observation that C. glutamicum PPX2 is active with short-chain PolyPs, it is tempting to speculate that PPX1 from C. glutamicum might hydrolyze long-chain PolyPs. However, currently it remains unknown whether PPX2 and PPX1 differ with respect to substrate specificity.
The cation requirement of PPX2 from C. glutamicum is typical for exopolyphosphatases. Potassium ions stimulated the activity of PPX2 from C. glutamicum, which requires Mg2+ or Mn2+ like PPX from E. coli (1). Higher concentrations of these divalent cations and of Ca2+ inhibited PPX activity, which is in accordance with the finding that PolyP storage formation is increased with high magnesium concentrations in the medium (33) and with the proposal of intracellular sequestration of metal cations by PolyP (20). Instead of intracellular sequestration, the metal tolerance of other microorganisms, e.g., tolerance against divalent copper ions found in Sulfolobus metallicus, involves hydrolysis of PolyP, stimulation of PPX activity by copper ions, and efflux of cupric phosphate (39). The substrate specificity of PPX2 from C. glutamicum toward short-chain PolyPs (Fig. 3) was similar to that of SurE from E. coli and of 28-kDa exopolyphosphatase from S. cerevisiae (29), while PPX1 from S. cerevisiae (59) and PPX (1) and GPP (16) from E. coli preferentially hydrolyze long-chain PolyPs. PPX2 from C. glutamicum showed the highest catalytic efficiency with PolyP3, with a kcat/Km value of 15.5 s−1 mM−1 (Table 1). Although pyrophosphate inhibits PPX2 with a Ki value of 8.2 mM, pyrophosphate is also a substrate of PPX2 from C. glutamicum (Fig. 3), as has been described for S. cerevisiae polyphosphatase (29). Typically, pyrophosphate is hydrolyzed by PPA enzymes, which are clearly distinct from the bacterial and archaeal exopolyphosphatases (Fig. 5). C. glutamicum possesses PPA, which is encoded by cg2988 and which requires Mg2+ for pyrophosphate hydrolysis (36). Interestingly, PPA from C. glutamicum was shown to interact with the cell division protein FtsZ and was postulated to be essential because attempts to disrupt ppa were unsuccessful (36). As PPX2 from C. glutamicum also hydrolyzes pyrophosphate (Fig. 3), it is proposed that PPA from C. glutamicum has an essential function other than its enzymatic activity as pyrophosphatase.
The activity of PPX2 from C. glutamicum was regulated by nucleotides with half-maximal inhibition observed with millimolar concentrations of NTPs, dNTPs, and ADP. Similarly, exopolyphosphatase from the sponge Tethya lyncurium is inhibited by ATP and ADP (28), and SurE from E. coli is inhibited by NTPs, dNTPs, and ADP (35). The intracellular PolyP level is controlled by the “energy charge” of the C. glutamicum cell because on the one hand, PolyP hydrolysis by PPX2 is inhibited by NTPs and ADP, and on the other hand, PolyP synthesis by the PolyP kinase PPK2B is inhibited by nucleoside monophosphates (Ki values for AMP, GMP, and IMP between 3.9 and 7.9 mM) (27). This allosteric regulation is important to ensure that C. glutamicum accumulates only PolyP when the supplies of both phosphorus and energy are abundant. The genetic program of C. glutamicum to cope with phosphate limitation (13, 56) involves the induction of at least 25 genes, with the two-component regulatory system PhoS-PhoR serving a role in their transcriptional regulation (19, 46). These phosphate starvation-inducible genes code for the high-affinity phosphate uptake system PstSCAB, the glycerol-3-phosphate uptake system UgpAEBC, the glycerophosphoryl diester phosphodiesterase GlpQ1, the 5-nucleotidase UshA (40), the putative nuclease NucH, and other systems which are involved in either mobilization of phosphate from nontransportable, extracellular phosphorous compounds or in the uptake of transportable phosphorous compounds (13, 56). However, since evidence for phosphate-dependent transcriptional control of the exopolyphosphatase genes ppx1 and ppx2 and the PolyP kinase genes ppk2A and ppk2B was not obtained (13), allosteric control of the PolyP kinases and exopolyphosphatases appears to be the major type of regulation of PolyP levels in C. glutamicum.
PolyP accumulates in stationary-phase C. glutamicum cells under conditions where the carbon source is exhausted but phosphate is still abundant (18). The observation that C. glutamicum can grow for four to six generations in minimal medium without added phosphate (13) indicated that phosphorus may be mobilized from an intracellular storage. Reduction of the intracellular PolyP levels due to phosphate starvation, deletion of ppk2B (27), or overexpression of ppx2 (Fig. 1) resulted in reduced biomass formation when cells were transferred to a medium with a limited supply of phosphate (Fig. 4). The growth disadvantage observed under these conditions could be attributed to PolyP or another concomitantly reduced phosphorus storage compound. As shown here, increasing intracellular PolyP levels due to deletion of the exopolyphosphatase gene ppx2 (Fig. 1) led to growth at higher biomass concentrations after transfer to phosphate-limiting conditions (Fig. 4). Taken together, mobilization of phosphorus from the intracellular PolyP storages confers a growth advantage to C. glutamicum once the external phosphorus supply ceases.
Acknowledgments
We thank S. Willbold for help with NMR analysis, F. Wahl, Werner E. G. Müller, and Uwe Seelig for gifts of various types of sodium phosphate glasses used as standards in this study, and Hermann Sahm and Doris Rittmann for support during the initial phase of this work. We thank Peter Klauth, Katja Schmitz, and Henrike Niederholtmeyer for discussions.
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Akiyama, M., E. Crooke, and A. Kornberg. 1993. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 268:633-639. [PubMed] [Google Scholar]
- 2.Ayraud, S., B. Janvier, A. Labigne, C. Ecobichon, C. Burucoa, and J. L. Fauchere. 2005. Polyphosphate kinase: a new colonization factor of Helicobacter pylori. FEMS Microbiol. Lett. 243:45-50. [DOI] [PubMed] [Google Scholar]
- 3.Brown, M. R., and A. Kornberg. 2004. Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. USA 101:16085-16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, M. R., and A. Kornberg. 2008. The long and short of it—polyphosphate, PPK and bacterial survival. Trends Biochem. Sci. 33:284-290. [DOI] [PubMed] [Google Scholar]
- 5.Cardona, S. T., F. P. Chavez, and C. A. Jerez. 2002. The exopolyphosphatase gene from Sulfolobus solfataricus: characterization of the first gene found to be involved in polyphosphate metabolism in archaea. Appl. Environ. Microbiol. 68:4812-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz, J., E. Ingall, C. Benitez-Nelson, D. Paterson, M. D. de Jonge, I. McNulty, and J. A. Brandes. 2008. Marine polyphosphate: a key player in geologic phosphorus sequestration. Science 320:652-655. [DOI] [PubMed] [Google Scholar]
- 7.Docampo, R., W. de Souza, K. Miranda, P. Rohloff, and S. N. Moreno. 2005. Acidocalcisomes—conserved from bacteria to man. Nat. Rev. Microbiol. 3:251-261. [DOI] [PubMed] [Google Scholar]
- 8.Eggeling, L., and O. Reyes. 2005. Experiments, p. 3535-3566. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL.
- 9.Eikmanns, B. J., D. Rittmann, and H. Sahm. 1995. Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J. Bacteriol. 177:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, J., F. A. Ruiz, M. Docampo, S. Luo, J. C. Rodrigues, L. S. Motta, P. Rohloff, and R. Docampo. 2007. Overexpression of a Zn2+-sensitive soluble exopolyphosphatase from Trypanosoma cruzi depletes polyphosphate and affects osmoregulation. J. Biol. Chem. 282:32501-32510. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 12.Hooley, P., M. P. Whitehead, and M. R. Brown. 2008. Eukaryote polyphosphate kinases: is the ‘Kornberg’ complex ubiquitous? Trends Biochem. Sci. 33:577-582. [DOI] [PubMed] [Google Scholar]
- 13.Ishige, T., M. Krause, M. Bott, V. F. Wendisch, and H. Sahm. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahid, I. K., A. J. Silva, and J. A. Benitez. 2006. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl. Environ. Microbiol. 72:7043-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Kramer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Puhler, D. A. Rey, C. Ruckert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 16.Keasling, J. D., L. Bertsch, and A. Kornberg. 1993. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl. Acad. Sci. USA 90:7029-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, K. S., N. N. Rao, C. D. Fraley, and A. Kornberg. 2002. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. USA 99:7675-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klauth, P., S. R. Pallerla, D. Vidaurre, C. Ralfs, V. F. Wendisch, and S. M. Schoberth. 2006. Determination of soluble and granular inorganic polyphosphate in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 72:1099-1106. [DOI] [PubMed] [Google Scholar]
- 19.Kocan, M., S. Schaffer, T. Ishige, U. Sorger-Herrmann, V. F. Wendisch, and M. Bott. 2006. Two-component systems of Corynebacterium glutamicum: deletion analysis and involvement of the PhoS-PhoR system in the phosphate starvation response. J. Bacteriol. 188:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 21.Kristensen, O., B. Ross, and M. Gajhede. 2008. Structure of the PPX/GPPA phosphatase from Aquifex aeolicus in complex with the alarmone ppGpp. J. Mol. Biol. 375:1469-1476. [DOI] [PubMed] [Google Scholar]
- 22.Kulaev, I. S., V. M. Vagabov, and T. V. Kulakovskaya. 2004. The biochemistry of inorganic polyphosphates, 2nd ed. John Wiley & Sons, Chichester, United Kingdom.
- 23.Kuroda, A., K. Nomura, R. Ohtomo, J. Kato, T. Ikeda, N. Takiguchi, H. Ohtake, and A. Kornberg. 2001. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293:705-708. [DOI] [PubMed] [Google Scholar]
- 24.Lambert, C., D. Weuster-Botz, R. Weichenhain, E. W. Kreutz, A. A. de Graaf, and S. M. Schoberth. 2002. Monitoring of inorganic polyphosphate dynamics in Corynebacterium glutamicum using a novel oxygen sparger for real time 31P in vivo NMR. Acta Biotechnol. 22:245-260. [Google Scholar]
- 25.Lichko, L. P., T. V. Kulakovskaya, and I. S. Kulaev. 2006. Inorganic polyphosphates and exopolyphosphatases in different cell compartments of Saccharomyces cerevisiae. Biochemistry (Moscow) 71:1171-1175. [DOI] [PubMed] [Google Scholar]
- 26.Lichko, L. P., T. V. Kulakovskaya, E. V. Kulakovskaya, and I. S. Kulaev. 2008. Inactivation of PPX1 and PPN1 genes encoding exopolyphosphatases of Saccharomyces cerevisiae does not prevent utilization of polyphosphates as phosphate reserve. Biochemistry (Moscow) 73:985-989. [DOI] [PubMed] [Google Scholar]
- 27.Lindner, S. N., D. Vidaurre, S. Willbold, S. M. Schoberth, and V. F. Wendisch. 2007. NCgl2620 encodes a class II polyphosphate kinase in Corynebacterium glutamicum. Appl. Environ. Microbiol. 73:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenz, B., R. Batel, N. Bachinski, W. E. Muller, and H. C. Schroder. 1995. Purification and characterization of two exopolyphosphatases from the marine sponge Tethya lyncurium. Biochim. Biophys. Acta 1245:17-28. [DOI] [PubMed] [Google Scholar]
- 29.Lorenz, B., W. E. Muller, I. S. Kulaev, and H. C. Schroder. 1994. Purification and characterization of an exopolyphosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 269:22198-22204. [PubMed] [Google Scholar]
- 30.Mori, H., A. Iida, T. Fujio, and S. Teshiba. 1997. A novel process of inosine 5′-monophosphate production using overexpressed guanosine/inosine kinase. Appl. Microbiol. Biotechnol. 48:693-698. [DOI] [PubMed] [Google Scholar]
- 31.Mullan, A., J. P. Quinn, and J. W. McGrath. 2002. A nonradioactive method for the assay of polyphosphate kinase activity and its application in the study of polyphosphate metabolism in Burkholderia cepacia. Anal. Biochem. 308:294-299. [DOI] [PubMed] [Google Scholar]
- 32.Nishio, Y., Y. Nakamura, Y. Usuda, S. Sugimoto, K. Matsui, Y. Kawarabayasi, H. Kikuchi, T. Gojobori, and K. Ikeo. 2004. Evolutionary process of amino acid biosynthesis in corynebacterium at the whole genome level. Mol. Biol. Evol. 21:1683-1691. [DOI] [PubMed] [Google Scholar]
- 33.Pallerla, S. R., S. Knebel, T. Polen, P. Klauth, J. Hollender, V. F. Wendisch, and S. M. Schoberth. 2005. Formation of volutin granules in Corynebacterium glutamicum. FEMS Microbiol. Lett. 243:133-140. [DOI] [PubMed] [Google Scholar]
- 34.Peters-Wendisch, P. G., B. Schiel, V. F. Wendisch, E. Katsoulidis, B. Mockel, H. Sahm, and B. J. Eikmanns. 2001. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3:295-300. [PubMed] [Google Scholar]
- 35.Proudfoot, M., E. Kuznetsova, G. Brown, N. N. Rao, M. Kitagawa, H. Mori, A. Savchenko, and A. F. Yakunin. 2004. General enzymatic screens identify three new nucleotidases in Escherichia coli. Biochemical characterization of SurE, YfbR, and YjjG. J. Biol. Chem. 279:54687-54694. [DOI] [PubMed] [Google Scholar]
- 36.Ramos, A., S. A. Adham, and J. A. Gil. 2003. Cloning and expression of the inorganic pyrophosphatase gene from the amino acid producer Brevibacterium lactofermentum ATCC 13869. FEMS Microbiol. Lett. 225:85-92. [DOI] [PubMed] [Google Scholar]
- 37.Rangarajan, E. S., G. Nadeau, Y. Li, J. Wagner, M. N. Hung, J. D. Schrag, M. Cygler, and A. Matte. 2006. The structure of the exopolyphosphatase (PPX) from Escherichia coli O157:H7 suggests a binding mode for long polyphosphate chains. J. Mol. Biol. 359:1249-1260. [DOI] [PubMed] [Google Scholar]
- 38.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Remonsellez, F., A. Orell, and C. A. Jerez. 2006. Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: possible role of polyphosphate metabolism. Microbiology 152:59-66. [DOI] [PubMed] [Google Scholar]
- 40.Rittmann, D., U. Sorger-Herrmann, and V. F. Wendisch. 2005. Phosphate starvation-inducible gene ushA encodes a 5′ nucleotidase required for growth of Corynebacterium glutamicum on media with nucleotides as the phosphorus source. Appl. Environ. Microbiol. 71:4339-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodrigues, C. O., F. A. Ruiz, M. Vieira, J. E. Hill, and R. Docampo. 2002. An acidocalcisomal exopolyphosphatase from Leishmania major with high affinity for short chain polyphosphate. J. Biol. Chem. 277:50899-50906. [DOI] [PubMed] [Google Scholar]
- 42.Sable, S. B., R. Kumar, M. Kalra, I. Verma, G. K. Khuller, K. Dobos, and J. T. Belisle. 2005. Peripheral blood and pleural fluid mononuclear cell responses to low-molecular-mass secretory polypeptides of Mycobacterium tuberculosis in human models of immunity to tuberculosis. Infect. Immun. 73:3547-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaaf, S., and M. Bott. 2007. Target genes and DNA-binding sites of the response regulator PhoR from Corynebacterium glutamicum. J. Bacteriol. 189:5002-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 48.Schröder, H. C., and W. E. G. Müller. 1999. Inorganic polyphosphates: biochemistry, biology, biotechnology. Progress in molecular and subcellular biology, vol. 23. Springer, Berlin, Germany. [DOI] [PubMed]
- 49.Sethuraman, A., N. N. Rao, and A. Kornberg. 2001. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:8542-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu, H., and T. Hirasawa. 2007. Production of glutamate and glutamate-related amino acids: molecular mechanism analysis and metabolic engineering, p. 1-38. In V. F. Wendisch (ed.), Amino acid biosynthesis—pathways, regulation and metabolic engineering. Springer, Heidelberg, Germany. doi: 10.1007/7171_2006_064. [DOI]
- 51.Srivastava, V., C. Rouanet, R. Srivastava, B. Ramalingam, C. Locht, and B. S. Srivastava. 2007. Macrophage-specific Mycobacterium tuberculosis genes: identification by green fluorescent protein and kanamycin resistance selection. Microbiology 153:659-666. [DOI] [PubMed] [Google Scholar]
- 52.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 53.Tammenkoski, M., K. Koivula, E. Cusanelli, M. Zollo, C. Steegborn, A. A. Baykov, and R. Lahti. 2008. Human metastasis regulator protein H-prune is a short-chain exopolyphosphatase. Biochemistry 47:9707-9713. [DOI] [PubMed] [Google Scholar]
- 54.von Graevenitz, A., and K. Bernard. 2001. The genus Corynebacterium—medical. In M. Dworkin et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer Verlag, New York, NY.
- 55.Wehmeier, L., A. Schafer, A. Burkovski, R. Kramer, U. Mechold, H. Malke, A. Puhler, and J. Kalinowski. 1998. The role of the Corynebacterium glutamicum rel gene in (p)ppGpp metabolism. Microbiology 144:1853-1862. [DOI] [PubMed] [Google Scholar]
- 56.Wendisch, V. F., and M. Bott. 2005. Phosphorus metabolism of Corynebacterium glutamicum, p. 377-396. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL.
- 57.Wendisch, V. F., A. A. de Graaf, H. Sahm, and B. J. Eikmanns. 2000. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 182:3088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wittmann, C., and J. Becker. 2007. The l-lysine story: from metabolic pathways to industrial production, p. 39-70. In V. F. Wendisch (ed.), Amino acid biosynthesis—pathways, regulation and metabolic engineering. Springer, Heidelberg, Germany.
- 59.Wurst, H., and A. Kornberg. 1994. A soluble exopolyphosphatase of Saccharomyces cerevisiae. Purification and characterization. J. Biol. Chem. 269:10996-11001. [PubMed] [Google Scholar]
- 60.Wurst, H., T. Shiba, and A. Kornberg. 1995. The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J. Bacteriol. 177:898-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zago, A., S. Chugani, and A. M. Chakrabarty. 1999. Cloning and characterization of polyphosphate kinase and exopolyphosphatase genes from Pseudomonas aeruginosa 8830. Appl. Environ. Microbiol. 65:2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]