Abstract
Campylobacter infections have been reported at prevalences ranging from 2 to 50% in a range of wild bird species, although there have been few studies that have investigated the molecular epidemiology of Campylobacter spp. Consequently, whether wild birds are a source of infection in humans or domestic livestock or are mainly recipients of domestic animal strains and whether separate cycles of infection occur remain unknown. To address these questions, serial cross-sectional surveys of wild bird populations in northern England were carried out over a 2-year period. Fecal samples were collected from 2,084 wild bird individuals and screened for the presence of Campylobacter spp. A total of 56 isolates were recovered from 29 birds sampled at 15 of 167 diverse locales. Campylobacter jejuni, Campylobacter lari, and Campylobacter coli were detected by PCR, and the prevalences of different Campylobacter spp. in different avian families ranged from 0% to 33%. Characterization of 36 C. jejuni isolates by multilocus sequence typing revealed that wild birds carry both livestock-associated and unique strains of C. jejuni. However, the apparent absence of unique wild bird strains of C. jejuni in livestock suggests that the direction of infection is predominantly from livestock to wild birds. C. lari was detected mainly in wild birds sampled in an estuarine or coastal habitat. Fifteen C. lari isolates were analyzed by macrorestriction pulsed-field gel electrophoresis, which revealed genetically diverse populations of C. lari in Eurasian oystercatchers (Haematopus ostralegus) and clonal populations in magpies (Pica pica).
Infection with Campylobacter spp. continues to be the leading cause of human infectious intestinal disease in the United Kingdom and has a significant economic impact (39). Consequently, there is a continuing effort to identify effective control methods. The majority of human infections (∼90%) are caused by Campylobacter jejuni subsp. jejuni (46). Other Campylobacter species, including Campylobacter coli and Campylobacter lari, can also cause enteritis in humans, but their prevalence is lower. Most C. jejuni infections are believed to result from consumption of contaminated food, including poultry meat (27, 40), red meat (52), and milk (13), which is thought to be contaminated primarily by feces. It is well established that most livestock species, including poultry, ruminants, and pigs, carry C. jejuni asymptomatically (27), making control at the farm level difficult. However, the epidemiology of C. jejuni cannot be explained solely by food-borne exposure; C. jejuni has also been isolated from a range of environmental samples, including samples of soil, water, sand, and the feces of a number of wildlife species, including wild birds (1-3). However, the role that non-food-borne exposure plays in the epidemiology of C. jejuni is currently not well defined.
High prevalences of Campylobacter species infections have been found in a wide range of wild bird species, although there is great variation between taxa (2, 4, 7, 16, 35, 47, 48). Given their ability to fly long distances and their ubiquity, wild birds have the potential to play an important role in the epidemiology and evolution of Campylobacter spp. However, whether wild birds are a source of infection for humans or domestic livestock or are mainly recipients of domestic animal strains or, indeed, whether separate cycles of infection occur remain unknown. These questions remain unanswered in part because investigations of the epidemiology of Campylobacter spp. have been complicated by their high inter- and intraspecies genetic diversity (6).
The methods that have been routinely used to characterize Campylobacter isolates are restricted due to genomic instability in Campylobacter populations (10, 38, 45). Multilocus sequence typing (MLST) is a method that has the advantage of being objective since it is sequence based, which allows comparison of isolates from different laboratories and accurate determination of relationships between isolates from diverse sources (11). MLST studies of C. jejuni in farm animals and the environment, including wildlife, suggest that some strains may be associated with particular host groups (6, 10, 15, 30). However, in the same studies other strains were found to occur in several host species or habitats. Few studies have investigated the molecular epidemiology of Campylobacter infection in wild bird populations using MLST, and because only a relatively small number of isolates from wild birds have been characterized by MLST, conclusions have not been drawn yet about how wild bird isolates fit into the overall phylogenetic scheme or whether wild birds act as reservoirs, amplifiers, or merely indicators of infection of domestic animals with zoonotic genotypes.
In the current study a large cross-sectional survey of wild bird populations in northern England was undertaken to investigate the epidemiology of Campylobacter infection. Previous studies that have focused on the epidemiology of Campylobacter spp. solely in wild birds have investigated either a narrow range of taxonomic groups (2, 5, 17, 23, 29, 33, 43, 50) or wild birds from a limited range of habitats (18, 25, 48). Studies that have investigated a broad range of wild bird species have used Campylobacter characterization techniques that do not allow conclusions about possible host associations to be drawn or comparison of the genetic diversity of isolates between studies (21, 25, 34, 47, 53). Therefore, the aims of this study were (i) to determine the host range and prevalence of Campylobacter spp. in a wild bird population and (ii) through molecular characterization of isolates to determine whether wild birds were a likely source of infection in humans or domestic livestock and whether separate cycles of infection with host-adapted strains of Campylobacter spp. were maintained in the wild bird population.
MATERIALS AND METHODS
Study design.
Serial cross-sectional surveys of wild bird populations throughout northern England were performed between July 2004 and October 2006. Samples were collected from a broad range of wild bird species, including representatives of the majority of wild bird families that are present in the United Kingdom for part of the year (migratory species) or the entire year (resident species). Wild bird individuals were the basic sampling unit.
Sample sources.
Samples were collected from both live and dead wild birds. All live wild birds sampled in this study were trapped in collaboration with licensed British Trust for Ornithology bird ringers (36), and birds were caught by a variety of methods, including mist netting, cannon netting, ground traps, wildfowl round-ups, sampling nestlings in the nest, and environmental sampling.
Dead wildfowl and corvids were provided by licensed shooters, and dead garden birds were collected as part of the Garden Bird Health Initiative, a study investigating causes of garden bird mortality (8).
Sample collection.
Once captured, each live bird was placed in a paper bag (R. S. Mulvihill, and R. C. Leberman, Powdermill Banding Station protocol, 2006 [http://www.westol.com/∼banding/PowdermillBandingProtocol_Jan2006b.pdf]), in which it usually defecated, allowing a fecal sample to be collected. A new paper bag was used for each bird. Fecal samples were collected from the bags with sterile cotton-tipped swabs, placed in bacterial transport medium (Medical Wire and Equipment Ltd., Bath, United Kingdom), and transported to the laboratory in a cool box. If the bird did not defecate in the bag or the bird was too large to place in a bag, a cloacal swab sample was obtained. Occasionally, fecal samples were collected from the environment with sterile swabs. Fecal samples were also collected from the lower intestine of dead birds during postmortem examination.
Data collection.
Data for each bird and the site at which the bird had been caught or found were recorded in the field. The date, location, and type of habitat where the bird had been trapped or found dead were recorded along with the capture method, British Trust for Ornithology ring number, species, age, and sex (36). Depending on the species or time of year, not all of these data could be obtained.
Bacterial culture.
Campylobacter spp. were isolated from fecal samples using Campylobacter enrichment broth (LabM, Bury, United Kingdom) containing 10% lysed horse blood and were cultured on modified charcoal-cefoperazone-deoxycholate agar (LabM). Up to four colonies showing typical morphological characteristics of Campylobacter spp. (gray or white, small, round [diameter, 1 to 3 mm] colonies that are flat often with a raised center on modified charcoal-cefoperazone-deoxycholate agar) were subcultured onto Columbia agar (LabM) supplemented with 5% defibrinated horse blood. Isolates that grew under microaerobic conditions and did not grow under aerobic conditions were identified as presumptive Campylobacter isolates and were frozen in Microbank tubes (Pro-Lab Diagnostics, Neston, United Kingdom) at −80°C until they were required.
Molecular assignment to Campylobacter species by PCR.
DNA extracts were prepared for each isolate by boiling one bead from each Microbank tube in 0.5 ml of sterile distilled water for 20 min. Cell lysates were kept at 4°C for no longer than 14 days.
Isolates were confirmed to be C. jejuni, C. coli, or C. lari using a previously described multiplex PCR that targeted the hipO and 23S rRNA genes of C. jejuni and the glyA gene of both C. coli and C. lari (49).
Three further single-PCR assays were used to amplify the ceuE gene of both C. jejuni and C. coli (19) and the 16S rRNA gene of C. lari (28) to confirm the results generated by the multiplex PCR assay described above (49). A duplex PCR was used to detect the 16S rRNA gene of Campylobacter fetus (both C. fetus subsp. fetus and C. fetus subsp. venerealis) and Campylobacter hyointestinalis (28).
MLST of C. jejuni isolates.
Chromosomal DNA was extracted from freshly grown C. jejuni using a NucleoSpin tissue DNA extraction kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions. Segments of seven housekeeping genes, aspA (aspartase), glnA (glutamine synthetase), gltA (citrate synthase), glyA (serine hydroxymethyl transferase), pgm (phosphoglucomutase), tkt (transketolase), and uncA (ATP synthase alpha subunit), were amplified by PCR and sequenced using protocols, primers, and reaction conditions described previously (11). Sequencing reactions were carried out using an ABI Prism 3130x genetic analyzer (Applied Biosystems, California).
The STADEN software package (44) was used to assemble sequences from the chromatograms generated by the ABI Prism 3130x genetic analyzer (Applied Biosystems), and allele numbers were assigned by comparing sequences with sequences in the public MLST profile database (http://pubmlst.org/campylobacter). Sequence types (STs) and clonal complexes were also assigned by comparison with the STs and clonal complexes in the MLST database. Novel sequences and STs were submitted to the database, and new allele and ST numbers were assigned.
MLST was not attempted with the C. coli isolates as three of the four PCR-positive C. coli isolates could not be revived from storage at −80°C.
Genetic analysis of C. jejuni isolates.
The genotypic relatedness of C. jejuni isolates was investigated by construction of an unweighted pair group method with arithmetic mean (UPGMA) dendrogram based on a distance matrix of pairwise differences in allelic profiles using the START (Sequence Type Assignment Recombination Tests) software package (22).
Relationships between isolates and identification of founder strains were explored using the burst algorithm (based on related STs) (14, 22) in the START software package. Isolates were grouped together if they shared five, six, or seven alleles. Allelic profiles of isolates from this study were compared with the allelic profiles of all C. jejuni isolates obtained from wild bird sources in the public MLST database (http://pubmlst.org/campylobacter). The population structure of the isolates collected as part of our study was then compared with the population structure of the larger unstructured data set using the eBURST program to obtain a “population snapshot” of the data set (14).
Linkage analysis using the standardized index of association (ISA) (20) was carried out using the LIAN (linkage analysis) program in the START package (22).
PFGE of C. lari isolates.
Pulsed-field gel electrophoresis (PFGE) of all C. lari isolates was performed using a previously described protocol (37). The BioNumerics software (version 4.0; Applied Maths BVBA, Sint-Martens-Latem, Belgium) was used for image analysis. Levels of similarity between pulsed-field banding patterns were determined using the Dice similarity coefficient method with a 2% tolerance window, and a UPGMA dendrogram was constructed.
Urease production assay with C. lari isolates.
To detect any urease-positive thermophilic strains of C. lari, all C. lari isolates were grown on urea agar containing 5% (vol/vol) urea (LabM), which was incubated for 48 h under microaerobic conditions. The urea agar contained a pH-sensitive indicator that was pink in the presence of the hydrolysis products of urea, which was indicative of urease production.
Statistical analysis.
The prevalence of each Campylobacter sp. detected and the 95% confidence intervals (95% CI) were determined using the frequency command in the statistical software package EpiInfo 2002 (CDC, Atlanta, GA).
RESULTS
Bacterial isolates.
Fecal samples were collected from 2,084 individual wild birds belonging to 99 species from 167 locations between July 2004 and October 2006. A total of 59 Campylobacter species isolates were recovered from fecal samples from 29 wild bird individuals belonging to 19 species sampled in 15 different locations, giving an overall Campylobacter species prevalence of 1.4% (95% CI, 1.0 to 2.0%) (Table 1). Three Campylobacter species were detected, C. jejuni (prevalence, 0.6% [95% CI, 0.4 to 1.1%]), C. lari (prevalence, 0.6% [95% CI, 0.4 to 1.1%]), and C. coli (prevalence, 0.1% [95% CI, 0.05 to 0.4%]).
TABLE 1.
Number of individuals of each wild bird species that were positive for Campylobacter spp. as determined by PCR
| Family | Common name | Latin name | na | No. of PCR-positive wild bird individuals (no. of isolates)
|
Location(s)b | ||
|---|---|---|---|---|---|---|---|
| C. jejuni | C. coli | C. lari | |||||
| Anatidae | Mute swan | Cygnus olor | 30 | 0 | 1 (2) | 0 | 6 |
| Eurasian wigeon | Anas penelope | 43 | 0 | 0 | 1 (1) | 12 | |
| Falconidae | Common kestrel | Falco tinnunculus | 14 | 0 | 1 (1) | 0 | 6 |
| Rallidae | Common moorhen | Gallinula chloropus | 3 | 1 (3) | 0 | 0 | 4 |
| Hematopodidae | Eurasian oystercatcher | Hematopus ostralegus | 95 | 0 | 0 | 6 (10) | 13, 14 |
| Scolopacidae | Red knot | Calidris canutus | 14 | 0 | 1 (1) | 0 | 14 |
| Ruddy turnstone | Arenaria interpres | 27 | 0 | 0 | 1 (1) | 15 | |
| Laridae | Black-headed gull | Larus ridibundus | 13 | 0 | 0 | 1 (1) | 4 |
| Columbidae | Rock dove or feral pigeon | Columba livia | 47 | 2 (7) | 0 | 0 | 3, 5 |
| Tytonidae | Barn owl | Tyto alba | 24 | 1 (1) | 0 | 0 | 8 |
| Turdidae | Common blackbird | Turdus merula | 53 | 1 (3) | 0 | 0 | 1 |
| Paridae | Great tit | Parus major | 154 | 1 (1) | 0 | 0 | 1 |
| Corvidae | Magpie | Pica pica | 71 | 2 (6) | 0 | 3 (5) | 7, 9, 12 |
| Eurasian jay | Garrulus glandarius | 3 | 1 (1) | 0 | 0 | 10 | |
| Eurasian jackdaw | Corvus monedula | 6 | 1 (3) | 0 | 0 | 6 | |
| Sturnidae | Common starling | Sturnus vulgaris | 9 | 1 (3) | 0 | 0 | 4 |
| Passeridae | House sparrow | Passer domesticus | 86 | 1 (4) | 0 | 0 | 2 |
| Fringillidae | Chaffinch | Fringilla coelebs | 84 | 1 (4) | 0 | 0 | 11 |
| European greenfinch | Carduelis chloris | 93 | 0 | 0 | 1 (1) | 3 | |
| Total | 13 (36) | 3 (4) | 13 (19) | ||||
Total number of individuals sampled.
Location(s) at which birds were sampled or were found dead. Locations 1 to 4 and 6 to 11, mixed pasture and arable farmland on the Cheshire plain in northwest England; location 5, a small town in Cheshire in northwest England; location 12, salt marsh habitat in the Mersey estuary in northwest England; locations 13 and 15, Morecambe Bay estuary in northwest England; location 14, Dee estuary in northwest England.
Thirty-six C. jejuni isolates were isolated from fecal samples from 13 individual wild birds belonging to 11 species, including the common moorhen (Gallinula chloropus), feral pigeon (Columba livia), barn owl (Tyto alba), common blackbird (Turdus merula), great tit (Parus major), magpie (Pica pica), Eurasian jay (Garrulus glandarius), Eurasian jackdaw (Corvus monedula), common starling (Sturnus vulgaris), house sparrow (Passer domesticus), and chaffinch (Fringilla coelebs).
Nineteen C. lari isolates were recovered from fecal samples from 13 wild bird individuals belonging to six species, including the Eurasian wigeon (Anas penelope), Eurasian oystercatcher (Haematopus ostralegus), ruddy turnstone (Arenaria interpres), black-headed gull (Larus ridibundus), magpie, and European greenfinch (Carduelis chloris).
Four C. coli isolates were isolated from fecal samples from three wild bird individuals belonging to three species, the mute swan (Cygnus olor), common kestrel (Falco tinnunculus), and red knot (Calidris canutus). On no occasion were individual wild birds infected with more than one species of Campylobacter. For all but one wild bird species, one species of Campylobacter was isolated from fecal samples; magpies were the only wild bird species from which both C. jejuni and C. lari were isolated (Table 1).
Diversity of MLST STs.
By using MLST, a total of 15 C. jejuni STs were identified for the 36 C. jejuni isolates examined (Table 2). The 36 C. jejuni isolates were detected in samples collected from 13 wild bird individuals belonging to 11 species. There were only two wild bird species from which C. jejuni was isolated from more than one individual (two feral pigeons and two magpies). All isolates derived from different individuals of the same species had different STs. Sixty-seven different alleles were identified for the seven loci, three of which were novel. Six of the 15 STs identified were also novel (ST3001, ST3002, ST3003, ST3274, ST3275, and ST3276) (Table 2). The most common ST and the most common clonal complex were ST45 and the ST45 complex, which were identified for 19% of the isolates from three wild bird individuals (a moorhen, a barn owl, and a magpie) from three separate locations (Table 2). Twenty-five isolates (69%) were assigned to a previously described clonal complex when their allelic profiles were compared with those in the larger public MLST database for isolates, and 11 isolates (31%) could not be assigned to a clonal complex (Table 2). Four of the six novel allele profiles identified were among the profiles that could not be assigned to a clonal complex. The number of STs in each clonal complex ranged from one (ST42, ST45, ST177, ST353, and ST1275 complexes) to three (ST179 complex) (Table 2). The ST of the clonal complex founder strain was the most predominant ST in four of the clonal complexes (ST42, ST45, ST48, and ST177 complexes). However, in three of the clonal complexes, the founder ST was not present in the isolates (ST179, ST353, and ST1275 complexes) (Table 2). For the majority (77%) of the wild bird individuals sampled, all of the isolates recovered had the same ST. However, for 23% of the individuals sampled, isolates with more than one ST were recovered from the same sample.
TABLE 2.
MLST allelic profiles and STs of 36 C. jejuni isolates detected in the fecal samples from 13 wild birdsa
| Clonal complex | ST | Allelic profile (allele no.)
|
Sourcee | Locationf | Sample collection date (day/mo/yr) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | |||||
| 48 | ST66 | 2 | 4 | 5 | 2 | 7 | 1 | 5 | Great tit 1 | 1 | 22/7/2004 |
| ST48 | 2 | 4 | 1 | 2 | 7 | 1 | 5 | House sparrow 1 | 2 | 14/1/2005 | |
| 2 | 4 | 1 | 2 | 7 | 1 | 5 | House sparrow 1 | 2 | 14/1/2005 | ||
| 2 | 4 | 1 | 2 | 7 | 1 | 5 | House sparrow 1 | 2 | 14/1/2005 | ||
| 2 | 4 | 1 | 2 | 7 | 1 | 5 | House sparrow 1 | 2 | 14/1/2005 | ||
| 179 | ST3001c | 1 | 6 | 29 | 40 | 40 | 32 | 3 | Feral pigeon 1 | 3 | 28/1/2005 |
| ST220 | 1 | 6 | 29 | 2 | 40 | 32 | 3 | Feral pigeon 1 | 3 | 28/1/2005 | |
| ST3002c | 1 | 6 | 29 | 289 | 40 | 32 | 3 | Feral pigeon 1 | 3 | 28/1/2005 | |
| Ub | ST3274c | 18 | 33 | 22 | 326d | 1 | 86 | 16 | Blackbird 1 | 1 | 3/3/2005 |
| 18 | 33 | 22 | 326d | 1 | 86 | 16 | Blackbird 1 | 1 | 3/3/2005 | ||
| 18 | 33 | 22 | 326d | 1 | 86 | 16 | Blackbird 1 | 1 | 3/3/2005 | ||
| 45 | ST45 | 4 | 7 | 10 | 4 | 1 | 7 | 1 | Moorhen 1 | 4 | 11/3/2005 |
| 4 | 7 | 10 | 4 | 1 | 7 | 1 | Moorhen 1 | 4 | 11/3/2005 | ||
| 4 | 7 | 10 | 4 | 1 | 7 | 1 | Moorhen 1 | 4 | 11/3/2005 | ||
| 4 | 7 | 10 | 4 | 1 | 7 | 1 | Barn owl 1 | 8 | 6/6/2005 | ||
| 4 | 7 | 10 | 4 | 1 | 7 | 1 | Magpie 2 | 9 | 20/6/2005 | ||
| 4 | 7 | 10 | 4 | 1 | 7 | 1 | Magpie 2 | 9 | 20/6/2005 | ||
| 4 | 7 | 10 | 4 | 1 | 7 | 1 | Magpie 2 | 9 | 20/6/2005 | ||
| 42 | ST42 | 1 | 2 | 3 | 4 | 5 | 9 | 3 | Feral pigeon 2 | 5 | 16/3/2005 |
| 1 | 2 | 3 | 4 | 5 | 9 | 3 | Feral pigeon 2 | 5 | 16/3/2005 | ||
| 1 | 2 | 3 | 4 | 5 | 9 | 3 | Feral pigeon 2 | 5 | 16/3/2005 | ||
| 1 | 2 | 3 | 4 | 5 | 9 | 3 | Feral pigeon 2 | 5 | 16/3/2005 | ||
| U | ST3003c | 64 | 98 | 20 | 100 | 134 | 94 | 16 | Jackdaw 1 | 6 | 26/4/2005 |
| 64 | 98 | 20 | 100 | 134 | 94 | 16 | Jackdaw 1 | 6 | 26/4/2005 | ||
| U | ST3275c | 64 | 288d | 71 | 189 | 134 | 94 | 60 | Jackdaw 1 | 6 | 26/4/2005 |
| 1275 | ST1231 | 27 | 2 | 22 | 104 | 43 | 86 | 31 | Magpie 1 | 7 | 9/5/2005 |
| 27 | 2 | 22 | 104 | 43 | 86 | 31 | Magpie 1 | 7 | 9/5/2006 | ||
| 27 | 2 | 22 | 104 | 43 | 86 | 31 | Magpie 1 | 7 | 9/5/2005 | ||
| U | ST2303 | 18 | 243 | 208 | 18 | 345 | 86 | 47 | Jay 1 | 10 | 1/12/2005 |
| U | ST3276c | 216d | 22 | 22 | 98 | 116 | 101 | 16 | Chaffinch 1 | 11 | 15/5/2006 |
| 216d | 22 | 22 | 98 | 116 | 101 | 16 | Chaffinch 1 | 11 | 15/5/2006 | ||
| 216d | 22 | 22 | 98 | 116 | 101 | 16 | Chaffinch 1 | 11 | 15/5/2006 | ||
| 216d | 22 | 22 | 98 | 116 | 101 | 16 | Chaffinch 1 | 11 | 15/5/2006 | ||
| 353 | ST5 | 7 | 2 | 5 | 2 | 10 | 3 | 6 | Starling 1 | 4 | 19/10/2006 |
| 7 | 2 | 5 | 2 | 10 | 3 | 6 | Starling 1 | 4 | 19/10/2006 | ||
| 177 | ST177 | 17 | 2 | 8 | 5 | 8 | 2 | 4 | Starling 1 | 4 | 19/10/2006 |
Allele numbers were assigned by comparing sequences with sequences in the public MLST profile database (http://pubmlst.org/campylobacter). STs and clonal complexes were also assigned by comparison with STs and clonal complexes in the MLST database. Novel sequences and STs were submitted to the database, and new allele or ST numbers were assigned.
U, not assigned to a clonal complex.
Novel ST in this study.
Novel allele in this study.
The number indicates the individual from which the isolate was derived.
Location at which the bird was sampled or found dead. Locations 1 to 4 and 6 to 11, mixed pasture and arable farmland on the Cheshire plain in northwest England; location 5, a small town in Cheshire in northwest England; location 12, salt marsh habitat in the Mersey estuary in northwest England; locations 13 and 15, Morecambe Bay estuary in northwest England; location 14, Dee estuary in northwest England.
Relatedness and population structure of C. jejuni isolates.
Using the criterion that members of a group should share five loci with at least one other member of the group, analysis of the isolates collected during this study using the burst algorithm grouped eight isolates with five STs into two groups. Group 1 contained all isolates of ST48 and ST66, and group 2 contained isolates of ST220, ST3001, and ST3002. The founder strains could not be predicted for either of these groups due to the small number of STs for each group. The grouping of STs as determined by burst analysis was confirmed by construction of a UPGMA dendrogram based on allele profiles (Fig. 1). Isolates in group 1 (ST48 and ST66) differed from each other by one allele (gltA). ST66 was represented by one isolate from a great tit sampled on a dairy farm in Cheshire (location 1) (Table 2). ST48 was represented by four isolates from an individual house sparrow sampled on a beef farm in Cheshire (location 2) (Table 2). These two farms were separated by 5 km, and the samples were collected 6 months apart. Isolates in group 2 differed from each other by one allele (glyA). The three isolates in this group, one each of ST220, ST3001, and ST3002, were isolated from the same feral pigeon that was sampled at location 3. This bird was the only C. jejuni-positive individual sampled at this location. The remainder of the STs identified in this study were determined by burst analysis to be singletons. Using less stringent criteria, the UPGMA dendrogram grouped isolates with ST3275 and ST3003 together, which differ from each other by three alleles. Isolates with these STs were recovered from one jackdaw. Two isolates that had only one allele in common (ST5 and ST177 isolates) were detected in a sample collected from one starling.
FIG. 1.
UPGMA dendrogram based on a distance matrix of pairwise differences in MLST allelic profiles of C. jejuni isolates used in this study. Allele numbers were assigned by comparing sequences with sequences in the public MLST profile database (http://pubmlst.org/campylobacter). Locations 1 to 4 and 6 to 11 were mixed pasture and arable farmland on the Cheshire plain in northwest England. Location 5 was a small town in Cheshire in northwest England. Location 12 was a salt marsh habitat in the Mersey estuary in northwest England. Locations 13 and 15 were in the Morecambe Bay estuary in northwest England. Location 14 was in the Dee estuary in northwest England. CC, clonal complex.
The ISA for the 15 STs detected during this study was 0.366. The observed variance was greater than the maximum variance obtained in 1,000 trials (P = 0.000), indicating that there was significant linkage disequilibrium. This suggests there was a degree of clonality in the study population of C. jejuni isolates. The dominant clonal complex in this study was the ST45 complex. The ISA for all STs in the ST45 clonal complex in the public MLST database (373 isolates with 180 different STs) was 0.003, indicating that there was no significant linkage disequilibrium, which may suggest that recombination within this clonal complex is common.
Comparison with a wider population of C. jejuni isolates.
C. jejuni isolates from our study were compared with all C. jejuni strains isolated from wild bird sources for which there are data in the public MLST database (last database query, 14 August 2008). The larger data set included 234 isolates with 177 different STs isolated from wild birds in Europe, North America, and Australasia between 1982 and 2007. Thirteen of the 15 STs identified during our study had not been reported for wild birds in the public MLST database previously. Only isolates of ST42 and ST45 recovered from wild birds were in both our data set and the larger data set. The ISA for STs belonging to the larger data set was 0.314, indicating that there was significant linkage disequilibrium in the group of STs reported for wild bird sources in the public MLST database.
Six (40%) of the STs identified in our study have previously been associated with either sporadic cases or outbreaks of human gastroenteritis (ST5, ST42, ST45, ST48, ST66, and ST2303). Isolates of ST66 and ST2303 in the public MLST database have been reported solely from human stool samples from patients with sporadic cases of gastroenteritis. In addition to isolation from human gastroenteritis cases, isolates of ST42, ST45, and ST48 have been recovered from ruminant meat, offal, animal carriers, and environmental samples. Isolates with ST45 and ST48 have also been isolated from poultry meat and carriers. One isolate with ST5 has been recovered from a pig carrier, as well as from human gastroenteritis cases. Isolates with ST177, ST220, and ST1231 in the public MLST database have been recovered from environmental samples, including samples of environmental water and samples of sand from bathing beaches. However, isolates with other STs in the ST177, ST179, and ST1275 complexes, to which STs identified in this study belong, have previously been recovered from both wild bird sources and stool samples from humans with sporadic cases of gastroenteritis.
PFGE.
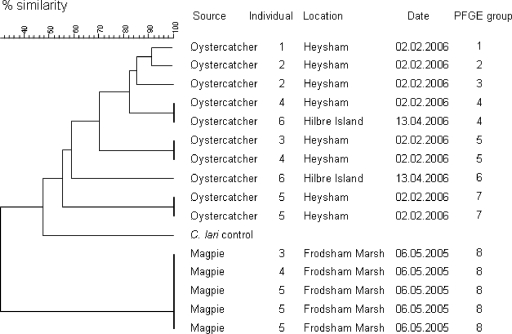
PFGE was carried out for 15 of 19 PCR-positive C. lari isolates. Four of the PCR-positive isolates could not be revived from storage at −80°C. Isolates that were subjected to PFGE were from nine wild bird individuals belonging to two species, magpies and oystercatchers. The magpie individuals were sampled on the same date in a salt marsh habitat. The samples from oystercatcher individuals were collected from two oystercatcher flocks in two estuaries separated by approximately 95 km on two dates 2 months apart. PFGE revealed eight SmaI banding patterns that were labeled PFGE groups 1 to 8 (Fig. 2). These SmaI PFGE groups were confirmed by PFGE after digestion with the KpnI restriction enzyme. The level of genetic similarity within each PFGE group was very high (>95%). One PFGE pattern was identified for C. lari isolates from all magpie individuals sampled in the same area on the same day (PFGE group 8); these isolates, including multiple isolates from one individual, showed 100% genetic similarity to each other. Seven PFGE patterns were identified for 10 isolates collected from six oystercatcher individuals (PFGE groups 1 to 7), and the overall genetic similarity between the groups was 56%. Multiple C. lari isolates were isolated from fecal samples from four oystercatcher individuals. Of these four individuals, only one oystercatcher yielded multiple isolates with the same PFGE banding pattern (PFGE group 7). Analysis of the only C. lari-positive oystercatcher from a population sampled in the Dee estuary during April 2006 (oystercatcher 6) (Fig. 2) yielded two isolates with different PFGE patterns (PFGE groups 4 and 6); one of these isolates had a banding pattern identical to that of an isolate obtained from a different oystercatcher (oystercatcher 4) sampled during February 2006 from a different population 95 km north of the Dee estuary (PFGE group 4). PFGE group 5 contained two isolates from different individuals in the same population that had identical PFGE banding patterns. Conversely, PFGE groups 2 and 3 each consist of one isolate showing 85% genetic similarity, and both isolates were isolated from a sample from one individual oystercatcher. For all C. lari isolates, the PFGE banding patterns suggested that the overall level of genetic similarity was 30%. All C. lari isolates were found to be urease negative.
FIG. 2.
Dendrogram showing the genetic similarity between C. lari isolates digested with the SmaI restriction enzyme. The level of similarity between PFGE banding patterns was computed using the Dice similarity coefficient method with a 2% tolerance window.
DISCUSSION
The overall prevalence of Campylobacter spp. for all wild birds sampled in this study was 1.4% (95% CI, 1.0 to 2.0%). However, the prevalences of different Campylobacter spp. in members of different wild bird taxonomic families were heterogeneous, ranging from 0% to 33%. Compared with the results of other studies describing Campylobacter in wild bird populations, which reported prevalences ranging from 2% to 50% (5, 21, 25, 29, 34, 47, 48), the prevalence reported in this study is relatively low. Prevalence estimates are likely to vary between studies due to the use of different sampling regimens and culture methods, which vary in sensitivity. Furthermore, there is evidence that the survival of Campylobacter spp. in fecal samples from different wild bird species is variable (48). The extent to which survival of Campylobacter spp. varies in fecal samples from most wild bird species is not known. Previous studies of Campylobacter occurrence in wild bird populations have reported differences in prevalence between different wild bird life stages, including breeding, migration, molting, and wintering, and differences depending on the feeding guild to which wild bird species belonged (21, 25, 29, 47, 48). It is clear that the ecology of Campylobacter infection in wild birds is complex, involving many intrinsic and extrinsic factors, and unless such factors are carefully considered, comparisons of prevalence values between studies may be misleading.
High prevalences of Campylobacter spp. in wild birds have often been interpreted as evidence that there is nonpathogenic coexistence of Campylobacter spp. and wild bird hosts (30, 31). The findings of this and previous studies suggest that this may be the case for some, but not all, wild bird taxa. In this and similar studies, Campylobacter spp. were not found to be equally distributed among all wild bird species investigated (21, 25, 34, 47). This might indicate that certain species of Campylobacter have coevolved with specific wild bird taxa. The occurrence of Campylobacter in wild birds is rarely associated with disease and was never associated with obvious disease in our study. Considering that campylobacters are known to survive in the environment (1, 3, 26, 32), it is more likely that wild bird taxa which share particular ecological factors (for example, habitat, diet, and/or behavioral characteristics) are exposed to Campylobacter more frequently than other wild bird taxa.
Despite the fact that there is no apparent association between specific C. jejuni strains and wild birds, there are clonal complexes that appear to be associated with environmental samples, which have also been detected in wild birds (6, 10, 30). Five of the 15 STs identified in this study (ST177, ST220, ST1231, ST3001, and ST3002) belonged to clonal complexes associated with environmental samples in previous studies (ST177 complex, ST179 complex, and ST1275 complex). These STs may be better adapted to survive in the environment, or the results may simply reflect contamination of the environment by bird feces. Until recently, it was thought that such strains may be nonpathogenic in humans, as they had never been isolated from human campylobacteriosis cases. However, recently, strains belonging to these clonal complexes have been isolated from sporadic cases of human gastroenteritis (30, 42).
Forty percent of the STs detected were unique to this study. A previous study investigating the occurrence of C. jejuni in wildlife and wild birds in a farmland habitat also found a high proportion of unique STs (15). None of the STs unique to this study have been reported for domestic animals or humans (last public MLST database query, 14 August 2008). However, livestock-associated strains were detected in wild bird samples, probably as a result of wild birds sharing a common environment or interacting with livestock. Therefore, if wild birds act as a significant source of Campylobacter strains for livestock, as has been suggested previously (4, 7), it might be expected that the novel strains detected in wild birds would also have been detected in livestock. The apparent absence of these novel strains in livestock may suggest that the predominant direction of infection is from livestock to wild birds.
Two of the novel strains, strains with ST3001 and ST3002, differed from each other and previously recognized ST220 strains by one locus (glyA). Strains with all three STs were isolated from one pigeon. These unique STs may represent recent variants of the previously described ST220, which may have arisen due to in vivo genetic recombination between strains in the host. A further two novel strains were isolated from one jackdaw (ST3003 and ST3275), and they shared three of seven alleles. It is possible that these strains are variants of a common strain, that one strain evolved from the other, or that the jackdaw acquired the two strains from one or multiple sources. In addition to coinfection with genetically similar strains, strains with two previously described unrelated STs (ST177 and ST5) were isolated from the fecal sample from one starling. It is thought that coinfection with more than one strain of C. jejuni may lead to recombination between strains and evolution of new strains within hosts (9, 38), which may explain the occurrence of unique genetically diverse STs in the wild bird populations examined.
It is possible that the unique STs identified in this study are parts of larger clonal complexes that are currently unidentified due to the small number of studies that have characterized wild bird isolates using MLST. Analysis of all C. jejuni isolates from wild birds in the public MLST database (n = 234; last database query, 14 August 2008) indicated that there is a degree of clonality in this C. jejuni population (ISA, 0.314) and that some STs are members of small complexes (STs having five or more MLST loci in common). However, the majority of STs were singletons or related to one other strain, suggesting that there is a high degree of genetic diversity among isolates from wild birds. Furthermore, 13 of the 15 STs identified in our study have not been reported previously for wild birds. This suggests that numerous strains of C. jejuni, many of which have not been identified yet, are carried by wild birds. Further studies would be useful for determining if the singleton STs are indeed singletons or parts of larger clonal complexes.
C. lari was detected in 13 of 2,084 (0.6%; 95% CI, 0.4 to 1.1%) wild birds sampled, and the occurrence in different wild bird taxa was not very heterogeneous since this species was obtained from only 6 of 99 wild bird species that were sampled. Accordingly, due to the small number of isolates examined, the results presented here should be treated as a starting point for further examination rather than a final conclusion. The prevalence of C. lari was greatest among wading birds (Haematopodidae, 7.8%; Scolopacidae and Charadriidae, 1.3%), followed by gulls (Laridae, 4.4%) and corvids (Corvidae, 3.0%). Very low prevalences were detected for ducks, geese, and swans (Anatidae, 0.4%) and finches (Fringillidae, 0.4%). These findings are largely in keeping with the findings of previous studies (47, 48). C. lari has long been associated with members of the gull family (16, 23, 24, 41) and was isolated from gulls in this study, but it was isolated less frequently from gulls than from wading birds. However, more wading birds than gulls were sampled in our study; consequently these observations should be treated as a starting point for further examination.
The majority of C. lari isolates were obtained from estuarine or estuarine marsh habitats. In these habitats, C. lari was most commonly isolated from Eurasian oystercatchers (H. ostralegus) and magpies (P. pica). Eurasian oystercatchers are gregarious birds that feed predominantly in estuaries on bivalve mollusks, which they locate by probing in estuarine mud with their bills. Studies investigating the occurrence of Campylobacter spp. in shellfish detected a high incidence of genetically diverse C. lari in mussels from The Netherlands, Germany, Denmark, England, and Ireland (12, 51). It is possible that there is a cycle between oystercatchers and their bivalve prey whereby oystercatchers acquire C. lari from eating bivalve prey and subsequently shed C. lari into the estuarine environment in which mollusks live.
PFGE analysis of the C. lari isolates from oystercatchers in this study revealed that these isolates are genetically diverse. Multiple isolates from the same fecal sample with different PFGE banding patterns were detected for three birds, showing that individual birds can harbor more than one strain of C. lari and illustrating the high level of genetic diversity even in this small number of isolates. A high prevalence of genetically hypervariable urease-positive thermophilic strains of C. lari was recently reported in a population of Swedish shore birds (48). The isolates examined in our study were urease negative; nevertheless, they showed the same high levels of genetic diversity. It has been suggested that generation of genetic diversity is likely an adaptive strategy employed by campylobacters to increase their chances of surviving environmental stresses that are likely to be encountered during host-to-host transmission (38). However, C. lari isolates with the same PFGE banding pattern were isolated from fecal samples from three oystercatcher individuals. Similarly, C. lari isolates having the same PFGE pattern were isolated from three magpies sampled in the same salt marsh habitat. This may indicate that despite high levels of genetic diversity among C. lari isolates, there are some strains that are more environmentally persistent than other strains and consequently more widely distributed in the environment. Alternatively, the birds from which these isolates were derived may have acquired them from the same source. However, due to the small number of isolates examined it is not possible to draw solid conclusions from these data; rather, hypotheses for further investigation are created.
In conclusion, MLST analysis of C. jejuni isolates revealed that wild birds carry both livestock-associated and unique strains of C. jejuni. The occurrence of livestock-associated strains in wild birds may suggest that these birds can act as a source of C. jejuni for livestock. However, the apparent absence of unique wild bird strains of C. jejuni in livestock may suggest that the direction of infection is predominantly from livestock to wild birds and less often from wild birds to livestock. Further studies of the C. jejuni occurrence in wild bird populations would be valuable for determining if unique strains identified in this and other studies belong to larger clonal complexes.
PFGE analysis revealed both genetically diverse and clonal populations of C. lari in two different wild bird species sharing the same habitat. The differences are likely to be influenced by complex interactions between the ecology of C. lari populations and differences in host life history, which warrant further investigation.
Acknowledgments
We thank the Merseyside Ringing Group for collecting field samples, Frodsham and District Wildfowlers' Club for providing wildfowl and corvid samples, and the RSPCA Stapeley Grange Wildlife Centre, the Garden Bird Health Initiative (British Veterinary Association Animal Welfare Foundation, CJ Wildbird Foods Ltd., Cranswick Pet Products, Gardman Ltd., RSPB, The Birdcare Standards Association, Universities Federation for Animal Welfare), and members of the public who submitted dead birds for postmortem examination.
This work was funded through the Defra Veterinary Teaching and Research Initiative program. L.A.H. received a scholarship from the Faculty of Veterinary Science, University of Liverpool.
Footnotes
Published ahead of print on 13 March 2009.
REFERENCES
- 1.Abulreesh, H. H., T. A. Paget, and R. Goulder. 2006. Campylobacter in waterfowl and aquatic environments: incidence and methods of detection. Environ. Sci. Technol. 40:7122-7131. [DOI] [PubMed] [Google Scholar]
- 2.Broman, T., H. Palmgren, S. Bergstrom, M. Sellin, J. Waldenstrom, M. L. Danielsson-Tham, and B. Olsen. 2002. Campylobacter jejuni in black-headed gulls (Larus ridibundus): prevalence, genotypes, and influence on C. jejuni epidemiology. J. Clin. Microbiol. 40:4594-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, P. E., O. F. Christensen, H. E. Clough, P. J. Diggle, C. A. Hart, S. Hazel, R. Kemp, A. J. Leatherbarrow, A. Moore, J. Sutherst, J. Turner, N. J. Williams, E. J. Wright, and N. P. French. 2004. Frequency and spatial distribution of environmental Campylobacter spp. Appl. Environ. Microbiol. 70:6501-6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuma, T., S. Hashimoto, and K. Okamoto. 2000. Detection of thermophilic Campylobacter from sparrows by multiplex PCR: the role of sparrows as a source of contamination of broilers with Campylobacter. J. Vet. Med. Sci. 62:1291-1295. [DOI] [PubMed] [Google Scholar]
- 5.Colles, F. M., K. E. Dingle, A. J. Cody, and M. C. Maiden. 2008. Comparison of Campylobacter populations in wild geese with those in starlings and free-range poultry on the same farm. Appl. Environ. Microbiol. 74:3583-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colles, F. M., K. Jones, R. M. Harding, and M. C. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craven, S. E., N. J. Stern, E. Line, J. S. Bailey, N. A. Cox, and P. Fedorka-Cray. 2000. Determination of the incidence of Salmonella spp., Campylobacter jejuni, and Clostridium perfringens in wild birds near broiler chicken houses by sampling intestinal droppings. Avian Dis. 44:715-720. [PubMed] [Google Scholar]
- 8.Cunningham, A. A., B. Lawson, M. Bennett, J. Chantrey, J. Kirkwood, T. Pennycott, and V. Simpson. 2005. Garden bird health. Vet. Rec. 156:656. [DOI] [PubMed] [Google Scholar]
- 9.de Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. van Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 10.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endtz, H. P., J. S. Vliegenthart, P. Vandamme, H. W. Weverink, N. P. van den Braak, H. A. Verbrugh, and A. van Belkum. 1997. Genotypic diversity of Campylobacter lari isolated from mussels and oysters in The Netherlands. Int. J. Food Microbiol. 34:79-88. [DOI] [PubMed] [Google Scholar]
- 13.Fahey, T., D. Morgan, C. Gunneburg, G. K. Adak, F. Majid, and E. Kaczmarski. 1995. An outbreak of Campylobacter jejuni enteritis associated with failed milk pasteurisation. J. Infect. 31:137-143. [DOI] [PubMed] [Google Scholar]
- 14.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.French, N., M. Barrigas, P. Brown, P. Ribiero, N. Williams, H. Leatherbarrow, R. Birtles, E. Bolton, P. Fearnhead, and A. Fox. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7:1116-1126. [DOI] [PubMed] [Google Scholar]
- 16.Fricker, C. R., R. W. Girdwood, and D. Munro. 1983. A comparison of procedures for the isolation of campylobacters from seagull faeces. J. Hyg. (London) 91:445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fricker, C. R., and N. Metcalfe. 1984. Campylobacters in wading birds (Charadrii): incidence, biotypes and isolation techniques. Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. B 179:469-475. [PubMed] [Google Scholar]
- 18.Fukuyama, M., T. Kamimura, T. Itoh, K. Saito, M. Takahashi, S. Sakai, M. Murata, K. Kohzaki, M. Hara, T. Shimizu, et al. 1986. Distribution of Campylobacter jejuni in wild birds and serogroup of isolates by slide agglutination technique. Nippon Juigaku Zasshi 48:487-493. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez, I., K. A. Grant, P. T. Richardson, S. F. Park, and M. D. Collins. 1997. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 35:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage analysis. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 21.Ito, K., Y. Kubokura, K. Kaneko, Y. Totake, and M. Ogawa. 1988. Occurrence of Campylobacter jejuni in free-living wild birds from Japan. J. Wildl. Dis. 24:467-470. [DOI] [PubMed] [Google Scholar]
- 22.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko, A., M. Matsuda, M. Miyajima, J. E. Moore, and P. G. Murphy. 1999. Urease-positive thermophilic strains of Campylobacter isolated from seagulls (Larus spp.). Lett. Appl. Microbiol. 29:7-9. [DOI] [PubMed] [Google Scholar]
- 24.Kaneuchi, C., T. Imaizumi, Y. Sugiyama, Y. Kosako, M. Seki, T. Itoh, and M. Ogata. 1987. Thermophilic campylobacters in seagulls and DNA-DNA hybridization test of isolates. Nippon Juigaku Zasshi 49:787-794. [DOI] [PubMed] [Google Scholar]
- 25.Kapperud, G., and O. Rosef. 1983. Avian wildlife reservoir of Campylobacter fetus subsp. jejuni, Yersinia spp., and Salmonella spp. in Norway. Appl. Environ. Microbiol. 45:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemp, R., A. J. Leatherbarrow, N. J. Williams, C. A. Hart, H. E. Clough, J. Turner, E. J. Wright, and N. P. French. 2005. Prevalence and genetic diversity of Campylobacter spp. in environmental water samples from a 100-square-kilometer predominantly dairy farming area. Appl. Environ. Microbiol. 71:1876-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer, J. M., J. A. Frost, F. J. Bolton, and D. R. Wareing. 2000. Campylobacter contamination of raw meat and poultry at retail sale: identification of multiple types and comparison with isolates from human infection. J. Food Prot. 63:1654-1659. [DOI] [PubMed] [Google Scholar]
- 28.Linton, D., R. J. Owen, and J. Stanley. 1996. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147:707-718. [DOI] [PubMed] [Google Scholar]
- 29.Luechtefeld, N. A., M. J. Blaser, L. B. Reller, and W. L. Wang. 1980. Isolation of Campylobacter fetus subsp. jejuni from migratory waterfowl. J. Clin. Microbiol. 12:406-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newell, D. G. 2002. The ecology of Campylobacter jejuni in avian and human hosts and in the environment. Int. J. Infect. Dis. 6:S16-S21. [DOI] [PubMed] [Google Scholar]
- 32.Obiri-Danso, K., N. Paul, and K. Jones. 2001. The effects of UVB and temperature on the survival of natural populations and pure cultures of Campylobacter jejuni, Camp. coli, Camp. lari and urease-positive thermophilic campylobacters (UPTC) in surface waters. J. Appl. Microbiol. 90:256-267. [DOI] [PubMed] [Google Scholar]
- 33.Pacha, R. E., G. W. Clark, E. A. Williams, and A. M. Carter. 1988. Migratory birds of central Washington as reservoirs of Campylobacter jejuni. Can. J. Microbiol. 34:80-82. [DOI] [PubMed] [Google Scholar]
- 34.Palmgren, H., M. Sellin, S. Bergstrom, and B. Olsen. 1997. Enteropathogenic bacteria in migrating birds arriving in Sweden. Scand. J. Infect. Dis. 29:565-568. [DOI] [PubMed] [Google Scholar]
- 35.Petersen, L., E. M. Nielsen, J. Engberg, S. L. On, and H. H. Dietz. 2001. Comparison of genotypes and serotypes of Campylobacter jejuni isolated from Danish wild mammals and birds and from broiler flocks and humans. Appl. Environ. Microbiol. 67:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redfern, C. P. E., and J. A. Clark. 2001. Ringer's manual. British Trust for Ornithology, Thetford, United Kingdom.
- 37.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ridley, A. M., M. J. Toszeghy, S. A. Cawthraw, T. M. Wassenaar, and D. G. Newell. 2008. Genetic instability is associated with changes in the colonization potential of Campylobacter jejuni in the avian intestine. J. Appl. Microbiol. 105:95-104. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, J. A., P. Cumberland, P. N. Sockett, J. Wheeler, L. C. Rodrigues, D. Sethi, and P. J. Roderick. 2003. The study of infectious intestinal disease in England: socio-economic impact. Epidemiol. Infect. 130:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shane, S. M. 1992. The significance of Campylobacter jejuni infection in poultry: a review. Avian Pathol. 21:189-213. [DOI] [PubMed] [Google Scholar]
- 41.Skirrow, M. B., and J. Benjamin. 1988. Campylobacters: cultural characteristics of intestinal campylobaters from man and animals. J. Hyg. 85:427-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sopwith, W., A. Birtles, M. Matthews, A. Fox, S. Gee, M. Painter, M. Regan, Q. Syed, and E. Bolton. 2006. Campylobacter jejuni multilocus sequence types in humans, northwest England, 2003-2004. Emerg. Infect. Dis. 12:1500-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Southern, J. P., R. M. Smith, and S. R. Palmer. 1990. Bird attack on milk bottles: possible mode of transmission of Campylobacter jejuni to man. Lancet 336:1425-1427. [DOI] [PubMed] [Google Scholar]
- 44.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 45.Steinbrueckner, B., F. Ruberg, and M. Kist. 2001. Bacterial genetic fingerprint: a reliable factor in the study of the epidemiology of human campylobacter enteritis? J. Clin. Microbiol. 39:4155-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tam, C. C., S. J. O'Brien, G. K. Adak, S. M. Meakins, and J. A. Frost. 2003. Campylobacter coli—an important foodborne pathogen. J. Infect. 47:28-32. [DOI] [PubMed] [Google Scholar]
- 47.Waldenstrom, J., T. Broman, I. Carlsson, D. Hasselquist, R. P. Achterberg, J. A. Wagenaar, and B. Olsen. 2002. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waldenstrom, J., S. L. On, R. Ottvall, D. Hasselquist, and B. Olsen. 2007. Species diversity of campylobacteria in a wild bird community in Sweden. J. Appl. Microbiol. 102:424-432. [DOI] [PubMed] [Google Scholar]
- 49.Wang, G., C. G. Clark, T. M. Taylor, C. Pucknell, C. Barton, L. Price, D. L. Woodward, and F. G. Rodgers. 2002. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 40:4744-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whelan, C. D., P. Monaghan, R. W. Girdwood, and C. R. Fricker. 1988. The significance of wild birds (Larus sp.) in the epidemiology of Campylobacter infections in humans. Epidemiol. Infect. 101:259-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, I. G., and J. E. Moore. 1996. Presence of Salmonella spp. and Campylobacter spp. in shellfish. Epidemiol. Infect. 116:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, T. L., L. Hollis, A. Cornelius, C. Nicol, R. Cook, and J. A. Hudson. 2007. Prevalence, numbers, and subtypes of Campylobacter jejuni and Campylobacter coli in uncooked retail meat samples. J. Food Prot. 70:566-573. [DOI] [PubMed] [Google Scholar]
- 53.Yogasundram, K., S. M. Shane, and K. S. Harrington. 1989. Prevalence of Campylobacter jejuni in selected domestic and wild birds in Louisiana. Avian Dis. 33:664-667. [PubMed] [Google Scholar]