Abstract
Lactobacillus helveticus strains possess an efficient proteolytic system that releases peptides which are essential for lactobacillus growth in various fermented dairy products and also affect textural properties or biological activities. Cell envelope proteinases (CEPs) are bacterial enzymes that hydrolyze milk proteins. In the case of L. helveticus, two CEPs with low percentages of amino acid identity have been described, i.e., PrtH and PrtH2. However, the distribution of the genes that encode CEPs still remains unclear, rendering it difficult to further control the formation of particular peptides. This study evaluated the diversity of genes that encode CEPs in a collection of strains of L. helveticus isolated from various biotopes, both in terms of the presence or absence of these genes and in terms of nucleotide sequence, and studied their transcription in dairy matrices. After defining three sets of primers for both the prtH and prtH2 genes, we studied the distribution of the genes by using PCR and Southern blotting experiments. The prtH2 gene was ubiquitous in the 29 strains of L. helveticus studied, whereas only 18 of them also exhibited the prtH gene. Sequencing of a 350-bp internal fragment of these genes revealed the existence of intraspecific diversity. Finally, expression of these two CEP-encoding genes was followed during the growth in dairy matrices of two strains, ITG LH77 and CNRZ32, which possess one and two CEP-encoding genes, respectively. Both genes were shown to be expressed by L. helveticus at each stage of growth in milk and at different stages of mini-Swiss-type cheese making and ripening.
Lactobacillus helveticus belongs to the group of lactic acid bacteria (LAB) and is widely used as a starter in the manufacture of dairy products such as hard cheeses like Emmental, Grana Padano, or Provolone (9, 11) and in many fermented milks (14, 15, 17, 28, 37, 38) because of its ability to produce lactic acid by catabolizing lactose.
L. helveticus strains are among the most nutritionally fastidious LAB as they show multiple amino acid auxotrophies. In order to overcome nutritional requirements when grown in milk, L. helveticus possesses a strong proteolytic system capable of producing short peptides and liberating amino acids from the casein matrix. This proteolytic system is composed of (i) cell envelope proteinases (CEPs) that hydrolyze caseins into oligopeptides, (ii) transport systems that allow uptake of oligopeptides, and (iii) various intracellular peptidases with differing and partly overlapping specificities, leading to a pool of free amino acids (13, 26).
Many LAB typically possess only one CEP-encoding genes (7, 12, 23, 32), but the presence of two or more different genes that encode CEPs has been reported in L. bulgaricus and L. helveticus (22, 31, 34). Among LAB, the proteolytic system of L. helveticus is considered to be one of the most efficient in cheese proteolysis (25), as well as in generating various bioactive peptides from caseins that are antihypertensive, are antimicrobial, reduce the risk of colon cancer, or are immunomodulators in fermented milk (15, 17, 19, 28, 37, 38) and in cheese (2). Several studies have biochemically characterized the CEPs in different strains of L. helveticus such as CP53, Zuc2, or L89 (16, 20, 27), but little information is available at the genetic level despite the widespread use of L. helveticus in dairy products. The prtH gene of L. helveticus CNRZ32 was identified and showed significant similarity to known lactococcal prtP genes (22). Moreover, the L. helveticus CNRZ32 strain possesses at least two distinct CEPs, as a prtH deletion mutant was indistinguishable from the wild type in growth rate and acid production in milk (22). The existence of a second CEP-encoding gene, named prtH2, in strain CNRZ32 has been reported, as well as the presence of two other putative genes that encode CEPs, named prtH3 and prtH5, which are significantly induced during growth in milk versus MRS medium (Difco, Detroit, MI) (31). Only one complete genome of L. helveticus is currently available, that of strain DPC4571 (3). At least one gene that encodes a serine proteinase, lhv_1641, was found but was annotated as a pseudogene (3). Finally, analysis of the substrate specificities of proteinases for caseins in eight L. helveticus strains has confirmed the existence of CEP biodiversity in this species (18).
All of these data suggest that CEP-encoding genes could be variable among L. helveticus strains, and this biodiversity could influence some parameters of technological interest such as the growth rate in milk, acid production, or peptide liberation.
The aims of this study were to evaluate the diversity of genes that encode CEPs in a collection of strains of L. helveticus isolated from various biotopes and to study their transcription in dairy matrices.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Twenty-nine strains of L. helveticus were chosen according to their technological significance in dairy products, as well as their geographic origins (Table 1). L. helveticus strains were propagated three times without shaking at 43°C in MRS medium (Difco, Detroit, MI) from a cryoball stored at −80°C. For growth in milk, 10 ml of ultra-high-temperature (UHT) milk was inoculated with an overnight MRS culture at different concentrations (1%, 0.5%, 0.1%, 0.05%, 0.01%, 0.005%, and 0.001%). After 12 h of culture at 43°C without shaking, the first nonclotted dilution was used to inoculate 10 ml of UHT milk at 5%, which was then incubated at 43°C without shaking until the appropriate pH was reached.
TABLE 1.
L. helveticus strains used in this study
| Strain | Biotope | Origin | Reference or source |
|---|---|---|---|
| CNRZ32a | Artisanal starter used in Comté | France | INRA of Jouy-en-Josas |
| CNRZ32JS | Artisanal starter used in Comté | France | J. Steele, University of Wisconsin, Madison |
| CNRZ32A | Mutant derived from CNRZ32JS | United States | J. Steele |
| CNRZ32B | Mutant derived from CNRZ32JS | United States | J. Steele |
| CNRZ32C | Mutant derived from CNRZ32JS | United States | J. Steele |
| CNRZ67 | Artisanal lactic starter of Gruyère de Comté | France | |
| CNRZ240 | Artisanal starter of Gruyère de Comté | France | |
| CNRZ241 | Artisanal starter used in Comté | France | |
| CNRZ303 | Artisanal starter used in Comté | France | 6 |
| CNRZ328 | Artisanal starter of Emmental | Finland | |
| CNRZ414 | Cow milk Koumis | Russia | |
| CNRZ450 | Commercial starter of Emmental | France | |
| CNRZ498 | Yogurt | France | |
| CNRZ762 | Artisanal starter of Gruyère de Comté | France | |
| CNRZ891 | Commercial starter | France | |
| CNRZ1080 | NDc | China | |
| CNRZ1095 | ND | Finland | |
| CNRZ1111 | Commercial starter | France | |
| CNRZ1317 | Commercial starter used in Tallegio | Italy | |
| ISLC5b | Artisanal lactic starter of Grana Padano | Italy | 36 |
| CP615 | ND | Japan | |
| CIP 103146Td | Artisanal starter used in Emmental | France | |
| ROSELL 5088 | ND | Canada | |
| ROSELL 5089 | ND | Canada | |
| DPC4571 | Swiss cheese whey | Ireland | T. Beresford, Teagasc |
| ITG LH1 | Whey | France | 5 |
| ITG LH2 | Aminopeptidase-deficient mutant strain derived from ITG LH1 | France | 1 |
| ITG LH3 | Aminopeptidase-deficient mutant strain derived from ITG LH1 | France | 1 |
| ITG LH77 | Artisanal starter | France | 5 |
CNRZ, Centre National de Recherche Zootechnique collection, INRA Jouy-en-Josas, France. The strains in bold were studied by RT-PCR and Southern hybridization.
ISLC, Istituto Sperimentale Latiero-Caesario, Italy.
ND, not determined.
CIP, Collection of Institut Pasteur, France.
Small-scale Swiss-type cheese making.
Two small-scale Swiss-type cheeses (800 g) were made according to a standardized Emmental cheese-making process (24) from thermized skim milk and from an overnight culture in UHT milk of L. helveticus strain ITG LH77 or CNRZ32. A subculture was made at 44°C until 1.1% acidity was achieved in Phagex medium (Phagex LB; Laboratoires Standa, Caen, France) reconstituted at 13.5% (wt/vol) and heat treated at 85°C for 30 min. Three milliliters of this culture was added to the cheese milk (∼105 CFU/ml). A freeze-dried starter culture of Streptococcus thermophilus (PAL-ITG ST20-87; Laboratoires Standa) for vat inoculation was also prepared in 13.5% (wt/vol) reconstituted Marstar 412A (Danisco, Dangé Saint Romain, France) until 0.95% acidity was achieved. Twelve milliliters of this culture (∼106 CFU/ml) was added to 1 liter of the cheese milk.
Briefly, the milk was ripened with starters (S. thermophilus and either strain ITG LH77 or CNRZ32 of L. helveticus) at 32°C for 60 min and coagulated by the addition of calf rennet. The gel was cut into curd grains with a mean diameter of 4 mm. After 15 min of fore working, the whey-curd mixture was cooked at 53.5°C for 33 min. The stir-out duration was 45 min. The curd was then molded in 12-cm-diameter molds. The pressing step (6 h) and the acidification step (14 h) were conducted in thermostat-controlled ovens at 48 and 36°C, respectively, in order to reproduce the cooling rate of the center of a full-size Emmental wheel. The cheeses were then cooled and salted for 5 h in NaCl-saturated brine at 7°C. The cheeses were wrapped under vacuum in BK1L ripening bags (F-28; Cryovac, Epernon, France) and ripened at 12°C for 12 days.
Primer design and PCR experiments.
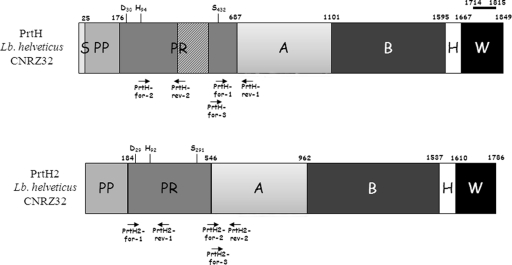
Previous results have shown that L. helveticus CNRZ32 has at least two genes that encode CEPs (22, 31). We decided to align the two genes to highlight specific regions. At the nucleotide level, no significant identity was observed between the two genes (accession no. AF133727 and DQ826130). When the amino acid sequences (accession no. AAD50643 and ABI13574) were aligned, only 22% identity was observed. A homology search for both proteins was carried out with the BLAST program (http://www.ncbi.nlm.nih.gov/blast/; February 2008). PrtH of L. helveticus CNRZ32 was homologous to the cell wall-associated serine proteinase of L. johnsonii NCC533 (60% identity, 74% similarity), to the PII-type proteinase precursor of Lactococcus lactis subsp. cremoris (51% identity, 65% similarity), and to the cell wall-associated serine proteinase of L. casei ATCC 334 and BL23 (50% identity, 64% similarity), whereas PrtH2 of L. helveticus CNRZ32 was homologous to PrtP of L. acidophilus NCFM (60% identity, 74% similarity), to PrtR of L. rhamnosus BGT10 (40% identity, 53% similarity), and to PrtR of L. casei BL23 (35% identity, 51% similarity).
Specific primers for the prtH gene were chosen by aligning its sequence with the gene that encodes the cell wall-associated serine proteinase of L. johnsonii NCC533 and with prtH2 of CNRZ32. Alignments were achieved with the BioEdit software. Primers were chosen both in regions conserved between L. helveticus CNRZ32 and L. johnsonii NCC533 genes to increase the hybridization probability in all of the strains to be tested and in regions not conserved between prtH and prtH2 to avoid cross-hybridization. The same methodology was used to choose prtH2-specific primers by alignment with the gene that encodes the L. acidophilus NCFM PrtP proteinase. One of the primer pairs chosen corresponded to primers used previously to amplify the prtH gene (22).
The sequences and localizations of the different primers used are shown in Table 2 and Fig. 1. Each PCR was carried out with a Veriti 96-well thermal cycler (Applied Biosystems, Foster City, CA) in a final volume of 20 μl with 1.25 U of Taq DNA polymerase (Invitrogen, Cergy-Pontoise, France), 20 ng of genomic DNA, each deoxynucleoside triphosphate (Invitrogen, Cergy-Pontoise, France) at 200 μmol liter−1, and each primer (Sigma-Aldrich, Lyon, France) at 25 μmol liter−1. After a first denaturation step of 4 min at 95°C, the mixture was subjected to 30 cycles of three steps of 30 s (95°C, 58°C, and 72°C). A final extension at 72°C for 10 min was performed. PCR products were checked on a 2% (wt/vol) agarose gel (Interchim, Montluçon, France) in 1× Tris-borate-EDTA (TBE 1x; Sigma-Aldrich, Lyon, France) at 100 V for 45 min in an i-Mupid system (Eurogentec, Angers, France). The ready-to-use O'RangeRuler 100-bp DNA ladder (Fermentas, St. Rémy Lès Chevreuse, France) was used as a molecular weight marker.
TABLE 2.
Sequences and localizations of primers used in this work
| Primer | Sequence (5′-3′) | Target sequence |
|---|---|---|
| PrtH-for-1 | GGTACTTCAATGGCTTCTCC | 1816-1835a |
| PrtH-rev-1 | GATGCGCCATCAATCTTCTT | 2282-2301a |
| PrtH-for-3 | CTCAGCACCAGGTGGACATA | 1755-1774a |
| PrtH-for-2 | CGATGATAATCCTAGCGAGC | 786-805a,c |
| PrtH-rev-2 | TGGCAGAACCTGTGCCTA | 1405-1422a,c |
| PrtH2-for-1 | CCAGCTAATAATCAAGACCA | 547-566b |
| PrtH2-rev-1 | AACAGCATATTGAACTGCTC | 1083-1102b |
| PrtH2-for-2 | GAAGACAAGGTGCTGGTCAA | 1592-1611b |
| PrtH2-rev-2 | TAGCATTTTGGTCAAAGACA | 2044-2063b |
| PrtH2-for-3 | GTTGGTGCCGCAACTAAATC | 1618-1637b |
FIG. 1.
Schematic representation of the predicted domains of PrtH (22) and PrtH2 of L. helveticus CNRZ32. Each number refers to the last amino acid of a domain. The catalytic triad and the amino acid number corresponding in the catalytic domain are indicated. The localization of the primers used in this work is shown by arrows. S, signal sequence; PP, preprodomain; PR, catalytic proteinase domain; A, A domain; B, B domain; H, helical domain; W, cell wall domain. The hatched part of the PrtH catalytic domain corresponds to insertion domain I. The C-terminal 101-aa region of PrtH showing homology with S-layer proteins is overlined.
RNA and DNA extraction.
Total RNA extraction was carried out either from an overnight UHT milk culture (10 ml at 43°C) or from 10 g of mini-Emmental cheeses in order to have more than 108 CFU ml−1. RNAs were obtained as described by Ulvé et al. (35), with the RNeasy Mini Kit (Qiagen, Courtaboeuf, France) after adding 2 U of RNase-free DNase I (Ambion, Courtaboeuf, France) and incubation for 30 min at 37°C. DNA was extracted from 10 ml of L. helveticus strains cultivated for 16 h in MRS medium as previously described (8). DNA and RNA concentrations were estimated at a wavelength of 260 nm with a Nanodrop ND-1000 spectrophotometer (Labtech, Palaiseau, France).
Reverse transcription (RT)-PCR experiments.
Total RNAs obtained from strains grown on MRS broth, milk, or cheeses were converted into cDNA with the QScript cDNA synthesis kit (Quanta Biosciences, Gaithersburg, MD). PCR was achieved on cDNA with primers PrtH-for-1/PrtH-rev-1 and PrtH2-for-3/PrH2-rev-2 as described above for genomic DNA. For each experiment, a PCR negative control was achieved with extracted RNA to verify the total absence of DNA before conversion into cDNA.
Southern blotting.
Internal fragments of the prtH and prtH2 genes were used as probes. These fragments were amplified from the genomic DNA of strain CNRZ32 with primers PrtH-for-1/PrtH-rev-1, PrtH-for-2/PrtH-rev-2, PrtH2-for-1/PrtH2-rev-1, and PrtH2-for-3/PrtH2-rev-2 (Table 2 and Fig. 1) and labeled with the DIG DNA labeling and detection kit (Roche Molecular Biochemicals, Mannheim, Germany). The probes were hybridized against EcoRV (New England BioLabs, Ozyme, Saint-Quentin, France) restriction digestion of the genomic DNAs of 16 L. helveticus strains separated on a 0.8% (wt/vol) agarose gel (Interchim, Montluçon, France) in TBE 1x (Sigma-Aldrich, Lyon, France) at 40 V for 16 h after transfer onto a Hybond-N membrane (GE Healthcare, Orsay, France). The ready-to-use O'GeneRuler 1-kb DNA ladder (Fermentas, Saint-Rémy Lès Chevreuse, France) was used as a molecular weight marker. The Southern blotting, prehybridization, and hybridization conditions used were described previously (33). Hybridization was carried out at 65°C for high-stringency conditions and at 42°C in the presence of 10% formamide for low-stringency conditions. Detection of specific hybrids was performed with the DIG DNA labeling and detection kit under the conditions recommended by the supplier (Roche Molecular Biochemicals, Mannheim, Germany).
DNA sequencing.
DNA sequencing was performed on a double-stranded template from a PCR product by the Sanger method by AGOWA (Berlin, Germany). Homology searching was done with the BLAST program (http://www.ncbi.nlm.nih.gov/blast/), and sequence comparisons and alignments were performed with the BioEdit software and the ClustalW algorithm.
RESULTS
Organization of PrtH and PrtH2.
PrtH of L. helveticus CNRZ32 is a 1,849-amino-acid (aa) protein with a deduced molecular mass of 204 kDa (Fig. 1) (22). It is synthesized as a preproenzyme and becomes mature after removal of the N-terminal 176 aa containing the 25-aa signal sequence and the 151-aa prodomain. It belongs to the subtilase family characterized by the catalytic triad Asp30-His94-Ser432 (Fig. 1) (30). It contains A, B, H, and W domains homologous to those of other subtilisin-like serine proteinases and an insertion domain in the catalytic domain. It lacks the LPxTG anchor motif but has a C-terminal 101-aa region (from aa 1714 to aa 1815) homologous to that of S-layer proteins. When compared with PrtH and other members of the subtilisin-like serine proteinase family, PrtH2 exhibited a putative prosequence of 184 aa (Fig. 1) with a putative cleavage site most probably located between Ala184 and Asn185, as found in PrtR of L. rhamnosus BGT10 (21). PrtH2 could be synthesized as a proenzyme leading to a mature proteinase of 1,602 residues. As in PrtH, the N-terminal region of the mature PrtH2 proteinase showed the characteristics of subtilisin-like serine proteinases with the catalytic triad Asp29, His92, and Ser291 where residue 1 defines the first residue of the mature protein (Fig. 1). The oxyanion hole residue well conserved among CEPs was present in the catalytic domain of PrtH2, at position Asn189. Unlike PrtH of L. helveticus CNRZ32, PrtB of L. bulgaricus NCDO1489, or PrtP of L. paracasei 151, PrtH2 did not contain the I domain inserted in the catalytic domain (Fig. 1). The putative A domain, located just after the catalytic domain, could contain 416 aa. The nine well-conserved segments identified in the A domain of other characterized CEPs and of PrtR of L. rhamnosus BGT10 were also present in PrtH2, i.e., 643-LVEGFLRF-652, 661-VPYLSYYGDM-672, 733-AFSPNGD-741, 770-VRVLADS-778, 781-KSYHS-787, 806-WDGKLYDA-815, 823-DGNYTYRF-832, 848-PVIIDTTAPVL-860, and 941-FSDVADN-949 (21, 29). In the B domain, sequences were poorly conserved and the domain limits for PrtH2 became unclear. Compared with PrtH, the B domain could spread from aa 962 to aa 1536 and the H domain could spread from aa 1537 to aa 1609 (Fig. 1). As for PrtH, no LPxTG anchor motif can be identified in the C terminus of PrtH2. However, as found for PrtH, homology with S-layer proteins was found, suggesting a similar attachment mechanism.
Detection of the prtH and prtH2 genes by PCR.
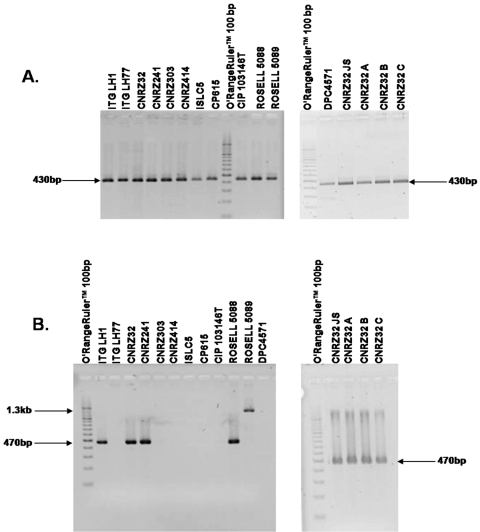
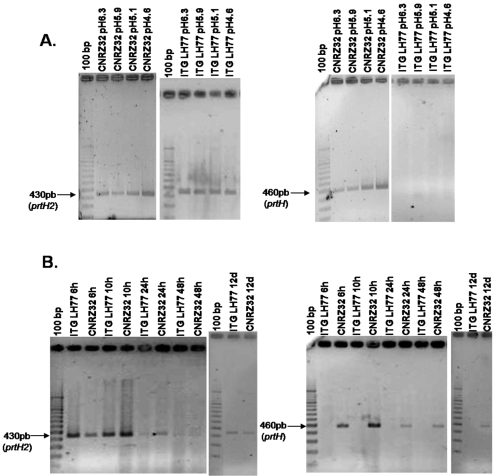
To determine the presence or absence of the prtH and prtH2 genes among various strains of L. helveticus, six pairs of primers (PrtH-for-1/PrtH-rev-1, PrtH-for-2/PrtH-rev-2, PrtH-for-3/PrtH-rev-1, PrtH2-for-1/PrtH2-rev-1, PrtH2-for-2/PrtH2-rev-2, and PrtH2-for-3/PrtH2-rev-2; Table 2 and Fig. 1) were used (see Materials and Methods). In all of the cases, either one product of the expected size or no product was obtained, confirming the specificity of the primers used. As illustrated in Fig. 2A for the PrtH2-for-3/PrtH2-rev-2 primers, the 16 L. helveticus strains tested (in bold in Table 1) exhibited a band of about 430 bp likely corresponding to the prtH2 gene whereas only 8 of the 16 strains also exhibited a band with the expected size (470 bp) corresponding to the prtH gene (Fig. 2B). For the ROSELL 5089 strain, a band of about 1.3 kb was obtained that could correspond to the prtH gene but was different from the other strains or could be a nonspecific product (see below).
FIG. 2.
Results of amplification of the prtH2 (A) and prtH (B) genes of 16 strains of L. helveticus. PCR products were obtained with the PrtH2-for-3/PrtH2-rev-2 (A) and PrtH-for-1/PrtH-rev-1 (B) primers and separated on a 2% (wt/vol) agarose gel in TBE 1x at 100 V for 45 min. The O'RangeRuler 100-bp DNA ladder was used as a molecular weight marker.
These results suggest that prtH2, not prtH, is the ubiquitous CEP-encoding gene in L. helveticus. These results were further confirmed by using primers PrtH-for-1/PrtH-rev-1 and PrtH2-for-3/PrH2-rev-2 for 13 supplementary strains of L. helveticus (reported with normal typing in Table 1). All of the strains exhibited a band corresponding to the prtH2 gene, and nine of them also exhibited a band corresponding to the prtH gene, i.e., ITGLH2, ITGLH3, CNRZ67, CNRZ450, CNRZ498, CNRZ891, CNRZ1080, CNRZ1111, and CNRZ1317.
Detection of the prtH and prtH2 genes by Southern blotting.
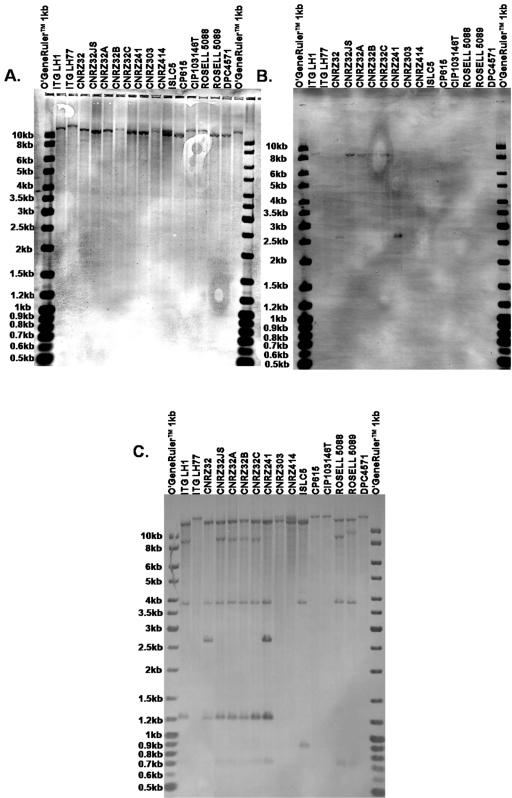
To confirm our PCR results, we performed Southern blotting analyses. Two probes corresponding to internal fragments of the prtH and prtH2 genes were obtained from the genomic DNA of strain CNRZ32 with primers PrtH-for-2/PrtH-rev-2 and PrtH2-for-3/PrtH2-rev-2 (Table 2 and Fig. 1) and hybridized against EcoRV restriction digests of genomic DNAs of 16 of the 29 L. helveticus strains (in bold in Table 1) under high-stringency conditions (65°C). With the PrtH2-for-3/PrtH2-rev-2 PCR product as a probe, one band with a molecular size of greater than 10 kb was detected in all of the strains (Fig. 3A) and there was a slight variation in the molecular mass of the band in ITG LH77, CP615, CIP 103146T, and DPC4571. This result showed that all of the L. helveticus strains tested showed sequences homologous to that of the prtH2 gene. With the PrtH-for-2/PrtH-rev-2 PCR product as a probe, a band of about 9 kb was detected in ITG LH1, CNRZ32JS, and the mutants CNRZ32A, CNRZ32B, and CNRZ32C while a band of 2.7 kb was present in CNRZ32 and CNRZ241 (Fig. 3B). These bands probably correspond to the prtH gene and highlight a restriction polymorphism in the latter two strains compared to the other five. Moreover, they were present only in strains for which an amplicon had been obtained for the prtH gene. No band corresponding to the prtH gene was detected in ROSELL 5088 and ROSELL 5089, despite the fact that they have been detected by PCR.
FIG. 3.
Southern hybridization of the EcoRV-digested genomic DNAs of 16 L. helveticus strains with probes corresponding to internal fragments of prtH2 and prtH obtained from the genomic DNA of the CNRZ32 strain with the PrtH2-for-3/PrtH2-rev-2 (A) and PrtH-for-2/PrtH-rev-2 (B) primers under high-stringency conditions (65°C). (C) Southern hybridization of the EcoRV-digested genomic DNAs of the 16 L. helveticus strains with probes corresponding to internal fragments of prtH2 and prtH obtained from the genomic DNA of the CNRZ32 strain with the PrtH2-for-1/PrtH2-rev-1, PrtH2-for-3/PrtH2-rev-2, PrtH-for-1/PrtH-rev-1, and PrtH-for-2/PrtH-rev-2 primers under low-stringency conditions (42°C). A labeled O'GeneRuler 1-kb DNA ladder was used to estimate molecular weights.
To increase the probability of detection of prtH in strains ROSELL 5088 and ROSELL 5089 and to highlight the potential presence of additional genes that encode CEPs of the subtilisin family, we used low-stringency conditions (42°C) and mixed four probes corresponding to internal fragments of the prtH and prtH2 genes and obtained from the genomic DNA of strain CNRZ32 with primers PrtH-for-1/PrtH-rev-1, PrtH-for-2/PrtH-rev-2, PrtH2-for-1/PrtH2-rev-1, and PrtH2-for-3/PrtH2-rev-2 (Table 2 and Fig. 1). These probes were hybridized against the EcoRV restriction enzyme-digested genomic DNAs of the 16 L. helveticus strains. As shown in Fig. 3C, a band with a molecular size of greater than 10 kb was present in all of the restriction patterns and probably corresponded to the prtH2 gene, as detected in Fig. 3A. Moreover, the virtual EcoRV digestion of the DPC4571 genome with RestrictionMapper version 3 (www.restrictionmapper.org) predicted a 16-kb fragment containing lhv_1641, the expected prtH2 gene in strain DPC4571. The band of about 9 kb detected in Fig. 3B and corresponding to the prtH gene was present in the ITG LH1, CNRZ32JS, CNRZ32A, CNRZ32B, and CNRZ32C digestion patterns, whereas the band of about 2.7 kb, due to a polymorphism of EcoRV restriction around this gene, was detected for CNRZ32 and CNRZ241. Another band of 1.2 kb was always associated with and probably corresponded to the EcoRV fragments recognized by the PrtH-for-1/PrtH-rev-1 probe. The 9-kb band was also present in ROSELL 5088, while ROSELL 5089 showed a band of about 9.8 kb. This coincides with the PCR results in which this strain showed an amplicon 800 bp longer than that of other strains that possess the prtH gene. However, in both strains, no band at 1.2 kb was detected, suggesting that the PrtH-for-1/PrtH-rev-1 probe did not hybridize, unlike the PrtH-for-2/PrtH-rev-2 probe.
Finally, two other bands of about 3.8 and 0.75 kb (except for ISCL5 [0.9 kb]) were detected in the ITG LH1, CNRZ32, CNRZ32JS, CNRZ32A, CNRZ32B, CNRZ32C, CNRZ241, ISLC5, ROSELL 5088, and ROSELL 5089 digestion patterns. This suggests that another sequence in the genomes of these strains possessed homology with the prtH or prtH2 genes which can be detected only under low-stringency conditions.
Southern blotting experiments confirmed that all of the strains possess the prtH2 gene, while strains ITG LH1, CNRZ32, CNRZ32JS, CNRZ32A, CNRZ32B, CNRZ32C, CNRZ241, ROSELL 5088, and ROSELL 5089 also had the prtH gene. However, the latter two strains seemed to have a more divergent prtH allele, as it was detected only with one of two probes and only under low-stringency conditions. Finally, another band with homology to prtH or prtH2 was present in 10/16 strains.
PCR product sequencing.
In order to ensure that the PCR products obtained for all of the strains actually corresponded to the prtH and prtH2 genes and to observe a possible biodiversity in the sequences of these genes, the nine PCR products obtained with the PrtH-for-1/PrtH-rev-1 primers and the 16 products obtained with the PrtH2-for-3/PrtH2-rev-2 primers were sequenced.
Analyses of the prtH2 sequences of the 16 strains revealed that strains CIP 103146T, CP615, DPC4571, ITG LH77, ROSELL 5088, and ROSELL 5089 led to PCR products with 100% identity (99% for strains ROSELL 5088 and ROSELL 5089) with a region of the DPC4571 genome (accession no. NC_010080; from nucleotide [nt] 1617908 to nt 1617607) that corresponds to lhv_1641. This gene is annotated as a pseudogene with a predicted function of proteinase PrtP (3). The 1% divergence between the sequences of ROSELL 5088 and ROSELL 5089 corresponds to a G-to-A substitution at position 1617790 of the DPC4571 genome (NC_010080). This substitution could provoke a Glu-to-Lys modification at the amino acid level.
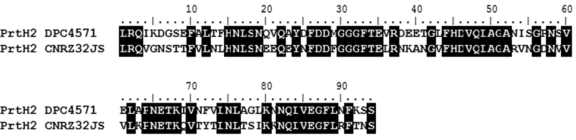
Strains ITG LH1, CNRZ32, CNRZ32JS, CNRZ32A, CNRZ32B, CNRZ32C, CNRZ241, CNRZ303, CNRZ414, and ISLC5 led to PCR products showing 100% identity with the published sequence of the prtH2 gene of CNRZ32 (accession no. DQ826130). A comparison of the prtH2 sequences of these two groups of strains reveals 60% identity at both the nucleotide and amino acids levels. Figure 4 shows an alignment of the partial PrtH2 protein sequences deduced from the nucleotide sequences of two representative strains, DPC4571 and CNRZ32JS. Some regions are well conserved, including one described as the first region involved in the recognition of the substrate in the A domain of subtilisin-like serine proteinases (29).
FIG. 4.
Alignment of the deduced amino acid sequences of part of PrtH2 from the sequenced region of the prtH2 genes of DPC4571 and CNRZ32JS. The alignment was made with the BioEdit software. Black boxes indicate residues that are similar between the two sequences, and white boxes indicate residues that differ. A well-conserved region in the A domain is overlined (29).
Thus, all of the strains tested possess a prtH2 gene, as shown by PCR and Southern blotting experiments, but sequencing of an internal fragment allowed us to distinguish two groups with only 60% identity in the sequenced part. The emerging question is if both forms detected correspond to two alleles of prtH2 or if the group including ROSELL 5088, ROSELL 5089, CIP 103146T, CP615, DPC4571, and ITG LH77 does not, in fact, have the prtH2 gene first described in CNRZ32 but another homologue such as, for example, prtH3 or prtH5 (22, 31).
The prtH gene sequences obtained for ITG LH1, CNRZ32, CNRZ32JS, CNRZ32A, CNRZ32C, and CNRZ241 showed 99% identity with the published sequence of prtH of CNRZ32 (accession no. AF133727). One GC-to-CG inversion at positions 1935 and 1936 compared to the published sequence was detected that caused a Pro-to-Ala modification at position 646 of the PrtH protein of CNRZ32 (accession no. AAD50543). The sequence of the mutant CNRZ32B showed two substitutions compared to the strains listed above, a T-to-C substitution at position 1887 of the prtH sequence (accession no. AF133727) and a G-to-A modification at position 1917. The first would have no impact at the protein level (Asn at position 629 is unchanged), while the second would change Met to Ile at position 639 of the sequence with accession no. AAD50543.
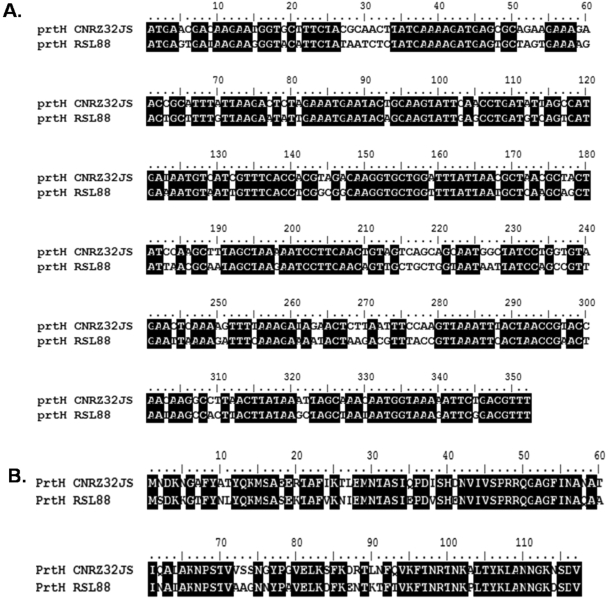
The ROSELL 5088 strain differed from the others because the sequenced region showed only 72% identity at the nucleotide level (Fig. 5A) and 74% identity at the amino acid level (Fig. 5B) with the sequences of the six strains listed above. For the ROSELL 5089 strain, whose PCR product was about 800 bp longer than those of the other strains, the region sequenced with the PrtH-for-1 primer showed 100% identity with that of prtH of the ROSELL 5088 strain in the 5′ part but not in the 3′ part (Fig. 6A) and the region sequenced with the PrtH-rev-1 primer showed 100% identity with that of prtH of the ROSELL 5088 strain in the 3′ part but not in the 5′ part (Fig. 6B). A homology search carried out with the BLAST program revealed that the 3′ part of “prtH for RSL89” and the 5′ part of “prtH rev RSL89” showed 99% identity with the mobile genetic element ISL2 (accession no. X77332), showing that this element was inserted into the prtH gene of the ROSELL 5089 strain. As this element consists of 858 bp, it explains why the PCR product from ROSELL 5089 was about 800 bp longer than those of the other strains. When this element was deleted from the sequence of ROSELL 5089 in silico, 99% identity with the sequence of the gene of ROSELL 5088 was observed, the difference consisting of an insertion of 3 nt (ATT), the mark of ISL2 insertion (39).
FIG. 5.
Alignment of the sequenced region of the prtH genes (A) and deduced amino acid sequences (B) of CNRZ32JS and ROSELL 5088 (RSL88). The alignment was made with the BioEdit software. Black boxes indicate residues that are similar between the two sequences, and white boxes indicate residues that differ.
FIG. 6.
Sequence alignment obtained for ROSELL 5088 and ROSELL 5089 with the PrtH-for-1 primer (A; prtH for RSL88 versus prtH for RSL89) and the PrtH-rev-1 primer (B; prtH rev RSL88 versus prtH rev RSL89). The alignment was made with the BioEdit software. Black boxes indicate residues that are similar between the two sequences, and white boxes indicate residues that differ.
In conclusion, sequence analyses confirmed the results of our PCR and Southern blotting experiments, as ITG LH1, CNRZ32, CNRZ32JS, CNRZ32A, CNRZ32B, CNRZ32C, CNRZ241, ROSELL 5088, and ROSELL 5089 had two different genes showing homology with both the prtH and prtH2 genes whereas the seven remaining strains possessed only one gene showing the highest homology with the prtH2 gene. Interestingly, the ROSELL strains showed a prtH allele different from those of the others and the ROSELL 5089 strain could not express an active PrtH protein because of the presence of the mobile genetic element ISL2 inserted in its prtH gene. Sequencing also revealed different alleles. For prtH2, 6 strains showed an allele similar to that of DPC4571 and the other 10 strains showed an allele similar to that of CNRZ32 (31). The CEP-encoding gene of DPC4571 was annotated as a pseudogene (3). Alignment of lhv_1641 (from nt 1614455 to nt 1619425) and prtH2 of CNRZ32 (accession no. DQ826130) revealed 56% nucleotide identity and 59% amino acid identity. However, the CEP of DPC4571 could be truncated (3), as a STOP codon was present in the region corresponding to the B domain, compared with PrtH2 of CNRZ32 (ABI13574), probably leading to the synthesis of a 1,286-aa protein lacking the H and W domains. As strains CP615, CIP 103146T, ITG LH77, ROSELL 5088, and ROSELL 5089 showed a sequenced product similar to that of DPC4571, their gene could also contain a STOP codon and therefore produce a nonfunctional proteinase.
Detection of prtH and prtH2 transcripts in dairy matrices.
The presence of two genes that encode CEPs in L. helveticus raised the question of their functionality. In order to determine if both genes are transcribed, the total RNAs of the 16 L. helveticus strains were extracted from UHT milk cultures at pH 5.6, i.e., in the exponential phase of growth. The total RNAs were converted into cDNA, and PCRs were achieved with primers PrtH-for-1/PrtH-rev-1 for prtH detection and PrtH2-for-3/PrtH2-rev-2 for prtH2 detection. A transcript was detected in all of the strains with the PrtH2-for-3/PrtH2-rev-2 primers, whereas a prtH transcript was observed in the nine strains with the prtH gene in their genomes (data not shown). For the ROSELL 5089 strain, as observed with genomic DNA, a band of 1.3 kb was observed (data not shown).
So, in all of the strains studied, the presence of a CEP-encoding gene in the L. helveticus genome leads to the detection of a transcript. The emerging question was if both genes are or are not transcribed at the same stages of growth. To verify this, we extracted the total RNAs of two strains, CNRZ32 and ITG LH77, during growth in MRS broth and in UHT milk. In MRS broth, both genes were expressed at different growth stages (1, 4, 6, 8, 10, and 12 h of culture; data not shown). In UHT milk, total RNAs were extracted during the lag phase (i.e., at pH 6.3), during the early and late exponential growth phases (i.e., at pHs 5.9 and 5.1), and during the stationary phase (i.e., at pH 4.6). RNA was converted into cDNA, and PCRs were achieved as described above. With the PrtH2-for-3/PrtH2-rev-2 primers, a transcript of about 430 bp was observed at each point for the ITG LH77 strain (Fig. 7A, left), indicating that the corresponding gene was transcribed at all stages of growth in UHT milk. No prtH transcript was observed, as expected since this strain was devoid of this gene (Fig. 7A, right). For the CNRZ32 strain, both transcripts were detected at all stages of growth (Fig. 7A).
FIG. 7.
Results of amplification of prtH2 (left) and prtH (right) with cDNAs obtained from total RNA extraction at different stages of growth of L. helveticus strains CNRZ32 and ITG LH77 in UHT milk (A) or mini-Swiss-type cheese (B). PCR products were obtained with the PrtH2-for-3/PrtH2-rev-2 (on the left) and PrtH-for-1/PrtH-rev-1 (on the right) primers and separated on a 2% (wt/vol) agarose gel in TBE 1x at 100 V for 45 min. The O'RangeRuler 100-bp DNA ladder (lane 100 bp) was used as a molecular weight marker.
Finally, in order to know if these genes are actually transcribed during cheese making, total RNAs were extracted during the manufacture of two mini-Emmental cheeses containing as starters the same strains used in milk experiments (CNRZ32 and ITG LH77). Ten-gram samples of cheese were collected after 6 h (i.e., during molding), 10 h (i.e., when the pH reached 5.3), 24 h, 48 h (i.e., after salting and the beginning of ripening), and 12 days (i.e., during ripening in a cold room), and total RNAs were extracted. With PrtH2-for-3/PrtH2-rev-2, a transcript was detected for both strains at each of the five time points tested (Fig. 7B, left). No prtH transcript was detected in cheese containing the ITG LH77 strain, whereas it was detected at each time in CNRZ32 (Fig. 7B, right).
DISCUSSION
The intraspecific diversity of CEP-encoding genes in L. helveticus was highlighted and further explored in this work by screening a collection of 29 strains from various origins. Two CEPs were previously described in L. helveticus, PrtH2 and PrtH, both belonging to the subtilisin-like serine proteinase family, as suggested by conserved domains, but exhibiting only 22% amino acid sequence identity. prtH2 was shown here to be a ubiquitous gene in L. helveticus (100% of the strains tested), whereas the presence of prtH was shown to be strain dependent (in 18 of the 29 strains). Previously published Southern blotting analysis revealed that only two of eight strains of L. helveticus tested showed a band corresponding to the prtH gene (22), in accordance with our observations. All of the strains of L. helveticus thus possessed at least one gene that encodes a CEP, with the highest homology with prtH2. For some strains, Southern blotting experiments revealed the presence of two or even more CEP-encoding genes, a result which was not widely obtained before, even if the existence of prtH3 and prtH5, two potential genes induced during growth in milk, has been suggested (31). L. helveticus is described as one of the most proteolytic species of LAB, and the presence of several genes constituting its proteolytic system could explain its efficiency.
The transcriptome assays described here for 16 strains of L. helveticus grown in milk revealed that the prtH2 and prtH genes, when present, systematically led to the corresponding transcripts in the exponential growth phase. In a more detailed kinetic analysis of two strains grown in MRS broth, in milk, and in mini-Swiss-type cheeses, prtH and prtH2 were shown to be transcribed at all growth phases. The absence of repression by free peptides or final amino acids (like branched-chain amino acids) is different from what was observed previously in L. lactis, where CEP expression is under the control of the repressor CodY. One explanation can be that prtH and prtH2 are not under the control of CodY in L. helveticus as we did not find any homologues of codY in the whole genome of L. helveticus strain DPC 4571 (4). Quantitative RT-PCR experiments could highlight different levels of expression of the genes according to the growth phase or environmental conditions. Microarray analyses revealed that transcription of prtH and prtH2 was induced, respectively, 5.81- and 4.34-fold in milk compared to that in MRS medium (31). It has also been proposed that both CEPs of L. helveticus show a specific medium-dependent regulation (10) whose mechanisms remain to be elucidated.
prtH and prtH2 both encode CEPs of the subtilisin family with little identity, resembling, respectively, the proteinase of L. johnsonii and that of L. acidophilus. This raises the question of what advantage is conferred by the occurrence of two different CEPs. They could have different substrate and cleavage specificities, or they could be redundant. One hypothesis would be that strains containing only one CEP-encoding gene had lost additional redundant genes such as, for example, prtH. The other hypothesis is that CEPs have distinct and complementary properties and that some strains have acquired an additional CEP-encoding gene that was maintained because it provides an adaptive advantage, in particular regarding milk protein hydrolysis. The facts that a prtH deletion mutant of the CNRZ32 strain was indistinguishable from the wild type in growth rate and showed a casein hydrolysis pattern different from that of the wild type (22) support the second hypothesis. Moreover, previous studies have shown that a large phenotypic variability exists among L. helveticus strains regarding proteolytic activity, with varied casein breakdown abilities (9). These phenotypic differences could be explained by the number of genes that encode CEPs or by their sequence variability, as detected in this work. Indeed, the partial sequencing of PCR products obtained for 16 strains revealed the existence of different alleles for both the prtH2 and prtH genes. For prtH2, six strains showed an allele similar to that of sequenced strain DPC 4571 and the other 10 strains showed an allele similar to that of CNRZ32 (31). These two alleles showed only 60% identity in the sequenced region. For prtH, seven strains showed an allele similar to that of CNRZ32 (22) while the two ROSELL strains showed a specific allele. Interestingly, the PrtH proteinase of ROSELL 5089 might not be functional because of the presence in the gene of the mobile genetic element ISL2. The complete genome sequence of DPC 4571 reveals the presence of 213 IS elements of 21 different types and in particular the presence of the mobile genetic element ISL2 18 times in the genome (3). This is consistent with what had been reported previously (39); the ISL2 element, which is specific to the L. helveticus species of LAB, is present 4 to 21 times, depending on the strain. It is therefore not surprising to find it inserted in CEP-encoding genes in some strains, as is the case for ROSELL 5089.
As proteolysis is important in the manufacture of cheese, by influencing texture and flavor, as well as the level of some bioactive peptides, improvement of the selection of L. helveticus starter strains would be highly valuable. Further work on phenotypic characteristics, and in particular the peptides released from caseins, by the 16 strains well characterized in this work would help in clarifying the relationship between genomic CEP content and strain activity on milk caseins.
Acknowledgments
This work was supported by research grants from the INRA and the Brittany Region.
We are grateful to James Steele for kindly providing strains CNRZ32JS, CNRZ32A, CNRZ32B, and CNRZ32C and to Tom Beresford for strain DPC4571. We thank Florence Valence-Bertel and Marie-Noëlle Madec for the choice of strains, Romain Richoux and Lydie Aubert from ITFF (Institut Technique Français du Fromage) for mini-Swiss-type cheese manufacture, and Danièle Atlan for fruitful discussion of the manuscript.
Footnotes
Published ahead of print on 13 March 2009.
REFERENCES
- 1.Blanc, B., P. Laloi, D. Atlan, C. Gilbert, and R. Portalier. 1993. Two cell-wall-associated aminopeptidases from Lactobacillus helveticus and the purification and characterization of APII from strain ITGL1. J. Gen. Microbiol. 139:1441-1448. [DOI] [PubMed] [Google Scholar]
- 2.Bütikofer, U., J. Meyer, R. Sieber, B. Walther, and D. Wechsler. 2008. Occurrence of the angiotensin-converting enzyme inhibiting tripeptides Val-Pro-Pro and Ile-Pro-Pro in different cheese varieties of Swiss origin. J. Dairy Sci. 91:29-38. [DOI] [PubMed] [Google Scholar]
- 3.Callanan, M., P. Kaleta, J. O'Callaghan, O. O'Sullivan, K. Jordan, O. McAuliffe, A. Sangrador-Vegas, L. Slattery, G. F. Fitzgerald, T. Beresford, and R. P. Ross. 2008. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J. Bacteriol. 190:727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callanan, M. J., R. P. Ross, and T. P. Beresford. 2007. Insertion sequence elements as mediators of strain diversity in Lactobacillus helveticus. Int. J. Food Microbiol. 120:120-123. [DOI] [PubMed] [Google Scholar]
- 5.Deutsch, S. M., D. Mollé, V. Gagnaire, M. Piot, D. Atlan, and S. Lortal. 2000. Hydrolysis of sequenced β-casein peptides by peptidases from thermophilic lactic acid bacteria highlights the intrinsic resistance of phosphopeptides. Appl. Environ. Microbiol. 66:5360-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deutsch, S. M., A. Neveu, S. Guezenec, P. Ritzenthaler, and S. Lortal. 2003. Early lysis of Lactobacillus helveticus CNRZ 303 in Swiss cheese is not prophage-related. Int. J. Food Microbiol. 81:147-157. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Espla, M. D., P. Garault, V. Monnet, and F. Rul. 2000. Streptococcus thermophilus cell wall-anchored proteinase: release, purification and biochemical and genetic characterization. Appl. Environ. Microbiol. 66:4772-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer, G., B. Decaris, and P. Leblond. 1997. Occurrence of deletions, associated with genetic instability in Streptomyces ambofaciens, is independent of the linearity of the chromosomal DNA. J. Bacteriol. 179:4553-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortina, M. G., G. Nicastro, D. Carminati, E. Neviani, and P. L. Manachini. 1998. Lactobacillus helveticus heterogeneity in natural cheese starters: the diversity in phenotypic characteristics. J. Appl. Microbiol. 84:72-80. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, C., B. Blanc, J. Frot-Coutaz, R. Portalier, and D. Atlan. 1997. Comparison of cell surface proteinase activities within the Lactobacillus genus. J. Dairy Res. 64:561-571. [Google Scholar]
- 11.Klaenhammer, T., E. Altermann, F. Arigoni, A. Bolotin, F. Breidt, J. Broadbent, R. Cano, S. Chaillou, J. Deutscher, M. Gasson, M. van de Guchte, J. Guzzo, A. Hartke, T. Hawkins, P. Hols, R. Hutkins, M. Kleerebezem, J. Kok, O. Kuipers, M. Lubbers, E. Maguin, L. McKay, D. Mills, A. Nauta, R. Overbeek, H. Pel, D. Pridmore, M. Saier, D. van Sinderen, A. Sorokin, J. Steele, D. O'Sullivan, W. de Vos, B. Weimer, M. Zagorec, and R. Siezen. 2002. Discovering lactic acid bacteria by genomics. Antonie van Leeuwenhoek 82:29-58. [DOI] [PubMed] [Google Scholar]
- 12.Kok, J., and W. M. De Vos. 1994. The proteolytic system of lactic acid bacteria, p. 169-210. In M. J. Gasson and W. M. De Vos (ed.), Genetics and biotechnology of lactic acid bacteria. Blackie Academic & Professional, Glasgow, United Kingdom.
- 13.Kunji, E. R. S., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie van Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 14.LeBlanc, J. G., C. Matar, J. C. Valdez, J. Leblanc, and G. Perdigon. 2002. Immunomodulating effects of peptidic fractions issued from milk fermented with Lactobacillus helveticus. J. Dairy Sci. 85:2733-2742. [DOI] [PubMed] [Google Scholar]
- 15.Maeno, M., N. Yamamoto, and T. Takano. 1996. Identification of an antihypertensive peptide from casein hydrolysate produced by a proteinase from Lactobacillus helveticus CP790. J. Dairy Sci. 79:1316-1321. [DOI] [PubMed] [Google Scholar]
- 16.Martin-Hernandez, M. C., A. C. Alting, and F. A. Exterkate. 1994. Purification and characterization of the mature, membrane-associated cell-envelope proteinase of Lactobacillus helveticus L89. Appl. Microbiol. Biotechnol. 40:828-834. [Google Scholar]
- 17.Matar, C., S. S. Nadathur, A. T. Bakalinsky, and J. Goulet. 1997. Antimutagenic effects of milk fermented by Lactobacillus helveticus L89 and a protease-deficient derivative. J. Dairy Sci. 80:1965-1970. [DOI] [PubMed] [Google Scholar]
- 18.Oberg, C. J., J. R. Broadbent, M. Strickland, and D. J. McMahon. 2002. Diversity in specificity of the extracellular proteinases in Lactobacillus helveticus and Lactobacillus delbrueckii subsp. bulgaricus. Lett. Appl. Microbiol. 34:455-460. [DOI] [PubMed] [Google Scholar]
- 19.Ong, L., and N. P. Shah. 2008. Influence of probiotic Lactobacillus acidophilus and L. helveticus on proteolysis, organic acid profiles, and ACE-inhibitory activity of cheddar cheeses ripened at 4, 8, and 12°C. J. Food Sci. 73:M111-M120. [DOI] [PubMed] [Google Scholar]
- 20.Ono, H., N. Yamamoto, M. Maeno, T. Takano, and H. Momose. 1997. Purification and characterization of a cell-wall associated proteinase of Lactobacillus helveticus CP53. Milchwissenschaft 52:373-377. [Google Scholar]
- 21.Pastar, I., I. Tonic, N. Golic, M. Kojic, R. van Kranenburg, M. Kleerebezem, L. Topisirovic, and G. Jovanovic. 2003. Identification and genetic characterization of a novel proteinase, PrtR, from the human isolate Lactobacillus rhamnosus BGT10. Appl. Environ. Microbiol. 69:5802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pederson, J. A., G. J. Mileski, B. C. Weimer, and J. L. Steele. 1999. Genetic characterization of a cell envelope-associated proteinase from Lactobacillus helveticus CNRZ32. J. Bacteriol. 181:4592-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard, G. G., and T. Coolbear. 1993. The physiology and biochemistry of the proteolytic system in lactic acid bacteria. FEMS Microbiol. Rev. 12:179-206. [DOI] [PubMed] [Google Scholar]
- 24.Richoux, R., L. Aubert, G. Roset, and J.-R. Kerjean. 2008. Impact of the proteolysis due to lactobacilli on the stretchability of Swiss-type cheese. Dairy Sci. Technol. 89:31-41. [Google Scholar]
- 25.Sasaki, M., B. W. Bosman, and P. S. T. Tan. 1995. Comparison of proteolytic activities in various lactobacilli. J. Dairy Res. 62:601-610. [DOI] [PubMed] [Google Scholar]
- 26.Savijoki, K., H. Ingmer, and P. Varmanen. 2006. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 71:394-406. [DOI] [PubMed] [Google Scholar]
- 27.Scolari, G., M. Vescovo, C. Zacconi, and F. Vescovi. 2006. Extraction and partial characterization of proteolytic activities from the cell surface of Lactobacillus helveticus Zuc2. J. Dairy Sci. 89:3800-3809. [DOI] [PubMed] [Google Scholar]
- 28.Seppo, L., T. Jauhiainen, T. Poussa, and R. Korpela. 2003. A fermented milk high in bioactive peptides has a blood pressure-lowering effect in hypertensive subjects. Am. J. Clin. Nutr. 77:326-330. [DOI] [PubMed] [Google Scholar]
- 29.Siezen, R. J. 1999. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie van Leeuwenhoek 76:139-155. [PubMed] [Google Scholar]
- 30.Siezen, R. J., and J. A. Leunissen. 1997. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6:501-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smeianov, V. V., P. Wechter, J. R. Broadbent, J. E. Hughes, B. T. Rodriguez, T. K. Christensen, Y. Ardo, and J. L. Steele. 2007. Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl. Environ. Microbiol. 73:2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smid, E. J., B. Poolman, and W. N. Konings. 1991. Casein utilization by lactococci. Appl. Environ. Microbiol. 57:2447-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 34.Stefanitsi, D., G. Sakellaris, and J.-R. Garel. 1995. The presence of two proteinases associated with the cell wall of Lactobacillus bulgaricus. FEMS Microbiol. Lett. 128:53-58. [Google Scholar]
- 35.Ulvé, V. M., C. Monnet, F. Valence, J. Fauquant, H. Falentin, and S. Lortal. 2008. RNA extraction from cheese for analysis of in situ gene expression of Lactococcus lactis. J. Appl. Microbiol. 105:1327-1333. [DOI] [PubMed] [Google Scholar]
- 36.Valence, F., and S. Lortal. 1995. Zymogram and preliminary characterization of Lactobacillus helveticus autolysins. Appl. Environ. Microbiol. 61:3391-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vinderola, G., C. Matar, and G. Perdigon. 2007. Milk fermented by Lactobacillus helveticus R389 and its non-bacterial fraction confer enhanced protection against Salmonella enteritidis serovar Typhimurium infection in mice. Immunobiology 212:107-118. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto, N., A. Akino, and T. Takano. 1994. Antihypertensive effect of the peptides derived from casein by an extracellular proteinase from Lactobacillus helveticus CP 790. J. Dairy Sci. 77:917-922. [DOI] [PubMed] [Google Scholar]
- 39.Zwahlen, M. C., and B. Mollet. 1994. ISL2, a new mobile genetic element in Lactobacillus helveticus. Mol. Gen. Genet. 245:334-338. [DOI] [PubMed] [Google Scholar]