Abstract
Studies that assess intraspecific genetic variation in ciliates are few and quite recent. Consequently, knowledge of the subject and understanding of the processes that underlie it are limited. We sought to assess the degree of intraspecific genetic variation in Carchesium polypinum (Ciliophora: Peritrichia), a cosmopolitan, freshwater ciliate. We isolated colonies of C. polypinum from locations in the Grand River basin in Southwestern Ontario, Canada. We then used the nuclear markers—ITS1, ITS2, and the hypervariable regions of the large subunit rRNA—and an 819-bp fragment of the mitochondrial cytochrome c oxidase I gene (cox-1) to investigate the intraspecific genetic variation of C. polypinum and the degree of resolution of the above-mentioned markers at the population level. We also sought to determine whether the organism demonstrated any population structure that mapped onto the geography of the region. Our study shows that there is a high degree of genetic diversity at the isolate level, revealed by the mitochondrial markers but not the nuclear markers. Furthermore, our results indicate that C. polypinum is likely not a single morphospecies as previously thought.
Studies of intraspecific genetic variation in macroorganisms have provided insights into microevolutionary processes and speciation in these organisms. However, our understanding of the intraspecific genetic diversity of microbial eukaryotes and its temporal and spatial distribution is neither deep nor broad due to the limited number of studies that deal with these topics (2, 5, 7, 31).
Many of the studies assessing within-species genetic variation and its distribution have used random amplified polymorphic DNA (RAPD) fingerprinting. Typically, the amount of genetic variation is very low within a species (35, 36, 56), with some exceptions (20, 57). However, there are several problems arising from the use of RAPDs as the sole source of data, including reproducibility and scoring concerns (54).
Sequences of the internal transcribed spacers (ITS), namely, ITS1 and ITS2, of the rRNA region have also been used to study the genetic diversity and population structure of protists (2, 6, 44). In several instances, the ITS sequences of populations of ciliates from across the globe have been nearly identical, indicating very low genetic variation (4, 13, 59, 62). On the other hand, the hypervariable regions of the large subunit (LSU) rRNA have revealed moderate genetic diversity in ciliates at the population level (15, 51).
Mitochondrial DNA (mtDNA) has been employed only sparingly in population studies of protists, even though it is routinely used in similar studies of metazoans (3, 7, 8). Of the mitochondrial genes, the apocytochrome b gene (cob) and the cytochrome c oxidase I (cox-1) gene (cox-1) have been used for population-level analyses of ciliates. Estimation of intraspecific nucleotide diversity of the cob gene from three species of the prostome ciliate Coleps showed only minor differences (5). On the other hand, the cox-1 gene revealed genetic diversity that nuclear markers failed to detect in members of the genera Paramecium and Tetrahymena. However, the implications and the extent of this diversity were not examined in detail, since the data sets used in these studies were small (4, 42). Finally, cox-1 uncovered a modest degree of genetic diversity in members of the Paramecium aurelia sibling species complex (11).
To broaden our understanding of population-level processes in ciliates, we sought to assess the level of intraspecific genetic diversity within the morphologically cosmopolitan, freshwater peritrich ciliate Carchesium polypinum. A Canadian river system, the Grand River drainage basin, was intensively sampled. We developed primers for the cox-1 gene of C. polypinum to assess the degree of genetic variation of this organism. Furthermore, we compared the resolving power of this gene at the population level to that of the ITS regions and the LSU rRNA hypervariable region. Our study demonstrates the efficacy of cox-1 in identifying genetic diversity in C. polypinum and implies that it can be applied in other ciliate species as well.
MATERIALS AND METHODS
Sample collection, culturing, and DNA extraction.
Colonies of C. polypinum were collected from localities throughout the Grand River basin in Southwestern Ontario. This river basin consists of the main river (Grand River) and four tributaries, the Conestogo and Nith rivers on the west and Eramosa and Speed rivers on the east. Exact collection localities are indicated in Table S1 in the supplemental material. Plastic petri dishes were left in the river for 1 week and were subsequently retrieved and transported to the laboratory. Individual peritrich colonies were scraped from the bottom of the dish, washed several times in Canadian Springs (Grenville, QC, Canada) mineral water, and placed in a clean glass petri dish with mineral water, barley grains, and 20 μl of Cerophyl (Cerophyl Laboratories Inc., Kansas city, MI), which had been inoculated with Enterobacter aerogenes at least 24 h previously. The colonies were confirmed as C. polypinum with live microscopy and silver staining.
After a few days of growth and replication, colonies were picked and DNA was isolated from a minimum of 30 cells using the MasterPure DNA purification kit (Epicentre, Madison, WI).
Amplification and sequencing. (i) ITS and LSU rRNA regions.
The 3′ end of the small subunit rRNA, the full ITS region containing the 5.8S rRNA gene, and part of the LSU rRNA containing the hypervariable region were amplified using the 300 forward (5′-AGGGTTCGATTCCGGAG-3′) and reverse C primers (5′-TGGTCCGTGTTTCAAGACG-3′) (30). Each PCR contained 4 μl of DNA, 1 μM of each primer, 2 mM of MgCl2, 1 μM of each deoxynucleoside triphosphate, 1× PCR buffer, and 2.5 U of Diamond Taq DNA polymerase (Medicorp, Montreal, QC, Canada) for a total volume of 25 μl. The PCR amplification was performed in a Perkin-Elmer GeneAmp 2400 thermal cycler (PE Applied Biosystems, Mississauga, ON, Canada) with the amplification conditions for this fragment as follows: initial denaturation at 94°C for 4 min, 35 amplification cycles (94°C for 60 s, 55°C for 120 s, and 72°C for 150 s), and a final extension step at 72°C for 10 min.
(ii) cox-1 gene region.
Initially, the cox-1 sequences of Tetrahymena thermophila and Paramecium tetraurelia were aligned, and degenerate primers were designed based on highly conserved protein domains. The resulting PCR product was sequenced, and a nested forward primer (coxFper [5′-GTTGGAAGTAAAGATGTTGC-3′]) was designed, while the reverse primer (339R [5′-ATAGGATCACCTCCGTAAGC-3′]) was maintained. The resulting fragment of the cox-1 gene was 819 bp long. The PCR mix was identical to the one used for amplification of the ITS region. The PCR amplification was performed in a Perkin-Elmer GeneAmp 2400 thermal cycler (PE Applied Biosystems, Mississauga, ON, Canada) with the amplification conditions for this fragment as follows: initial denaturation at 94°C for 4 min, 35 amplification cycles (5 cycles at 94°C for 30 s, 45°C for 60 s, and 72°C for 105 s and 30 cycles at 94°C for 30 s, 55°C for 60 s, and 72°C for 105 s), and a final extension step at 72°C for 10 min.
(iii) Sequencing.
The resulting amplicons for the rRNA region and cox-1 were purified with the Qiagen MinElute gel extraction kit (Qiagen, Mississauga, ON, Canada). The amplification primer reverse C and the internal forward primer 1055F (5′-GGTGGTGCATGGCCG-3′) were used for sequencing the rRNA region in both directions with a 3730 DNA analyzer (Applied Biosystems Inc., Foster City, CA), an ABI Prism BigDye Terminator (version 3.1), and a cycle sequencing ready reaction kit. coxFper and 339R were used for sequencing of the cox-1 gene fragment.
Phylogenetic analyses.
The sequence fragments were imported into the Sequencher software program, version 4.0.5 (Gene Codes Corp.) and assembled into contigs. Subsequently, the sequences of the ITS regions were uploaded to our DCSE (dedicated comparative sequence editor [14]) database and aligned against existing peritrich sequences. The alignment was further refined by eye.
The sequences of cox-1 were aligned using the MEGA software program, version 4.0, which uses the CLUSTALW algorithm (34).
The 3′ end of the small subunit rRNA, 5.8S rRNA, the two ITS regions, and the hypervariable region of the LSU rRNA of 55 colonies of C. polypinum were sequenced for this study, but only the last three regions were used for analysis. Three separate files were constructed for phylogenetic analysis of the nuclear sequences: the ITS1 region, the ITS2 region, and the hypervariable region of the LSU rRNA. The files consisted of 128, 183, and 407 nucleotides, respectively. The 5.8S gene was omitted from the analysis due to its high degree of conservation. An 819-bp fragment of cox-1 of 42 colonies of C. polypinum was also sequenced, along with one of Epistylis plicatilis (the organism was generously provided by Chris Lobban, University of Guam [39]) and one of an Epistylis sp., as well as a Vorticella sp.
The DNADIST software program in PHYLIP, version 3.65b (J. Felsenstein, Department of Genetics, University of Washington, Seattle) was used to calculate genetic distances with the Kimura two-parameter model. Subsequently, a neighbor-joining (NJ) tree was inferred. Using the SEQBOOT software program, the data were bootstrap resampled 1,000 times, and the CONSENSE program was subsequently used to construct a consensus tree. For the Bayesian-inference (BI) and maximum-likelihood (ML) analyses, missing nucleotides were treated as missing and the gaps as a fifth character state. To determine the model of DNA substitution that best fit the data for the ML and BI calculations, the Modeltest software program (50) was used. The most suitable models for the ITS1, ITS2, and LSU rRNA data sets were Felsenstein 81 (F81), Jukes-Cantor, and the F81 model with gamma distribution and an estimate of invariable sites (F81+I+Γ), respectively, while for the cox-1 data set, the general time reversible model with gamma distribution (GTR+Γ) was used. The models were applied in the MrBayes software program, version 3.1.1 (28, 53), and the corresponding phylogenetic trees were inferred. In all cases, the hierarchical likelihood ratio test criterion of Modeltest was used. However, the data sets were also analyzed employing the Akaike information criterion proposed models (F81+I, GTR+Γ, and the Tamura Nei model with gamma distribution and an estimate of invariable sites, respectively), and the tree topologies were identical. The models were also applied in the PHYML software program for ML analysis (23, 24), and the data were bootstrap resampled 500 times.
Genetic structure analysis.
The genetic structure of C. polypinum was investigated using the software program ARLEQUIN 3.0 (S. Schneider, D. Roessli, and L. Excoffier, Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland). An analysis of molecular variance (AMOVA) was employed to investigate the genetic structure within and between populations using the cox-1 gene sequences. The sequences were grouped first by tributary and then by clade as defined in the phylogenetic analysis of the cox-1 data set.
Nucleotide sequence accession numbers.
All sequences are available through GenBank. For the cox-1 data set, the accession numbers are FJ810309 to FJ810353. For the nuclear marker data set, the corresponding numbers are FJ810354 to FJ810408.
RESULTS
Sequence analysis of nuclear data sets.
The beginning and the end of ITS1, 5.8S rRNA, and ITS2 were determined by taking into consideration the master alignment available in our laboratory, which contains data from species across the different classes and subclasses of ciliates. The lengths of ITS1, 5.8S rRNA, and ITS2 for C. polypinum were 112, 142, and 162 nucleotides, respectively. A detailed analysis of the data set is available from the author upon request.
Sequence analysis of the cox-1 data set.
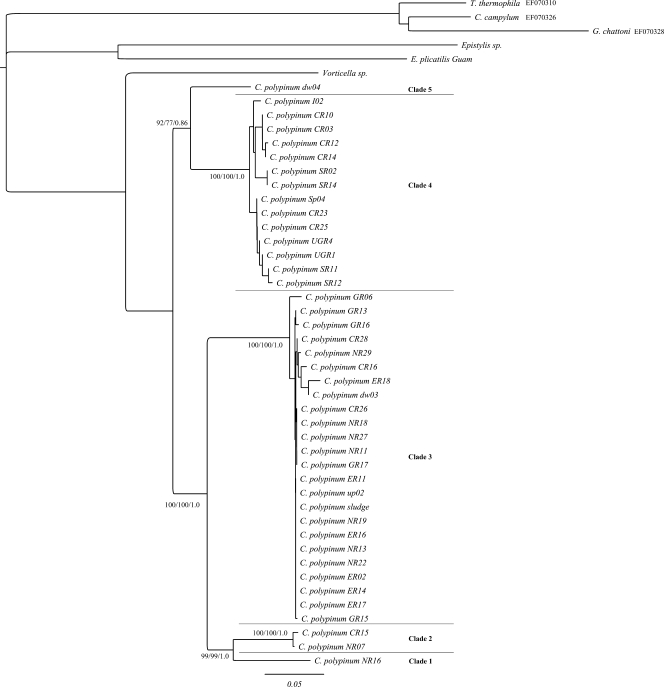
Five distinct amino acid sequences were identified for mitochondrial cox-1. These correspond to the clades described below (see “Phylogenetic analyses of the cox-1 data set”). The intraclade nucleotide sequence divergences within all the clades that contained more than one individual were <1.0%. The interclade genetic distances as calculated from the nucleotide sequences ranged from 11 to 18% (Table 1).
TABLE 1.
Mean percent nucleotide sequence divergence of an 819-bp fragment of the cox-1 gene between the five clades of the peritrich ciliate C. polypinuma
| Grouping | % Nucleotide sequence divergence
|
||||
|---|---|---|---|---|---|
| Clade 1 | Clade 2 | Clade 3 | Clade 4 | Clade 5 | |
| Clade 1 | 12 | 16 | 18 | 18 | |
| Clade 2 | 15 | 18 | 16 | ||
| Clade 3 | 18 | 11 | |||
| Clade 4 | 17 | ||||
| Clade 5 | |||||
See Fig. 4.
There was a total of 243 polymorphisms noted: 67% were at the third nucleotide position, 10% at the second, and 23% at the first position. Of all the substitutions, 18% were nonsynonymous, resulting in an amino acid change, while the rest were synonymous.
Phylogenetic analyses of the nuclear data sets.
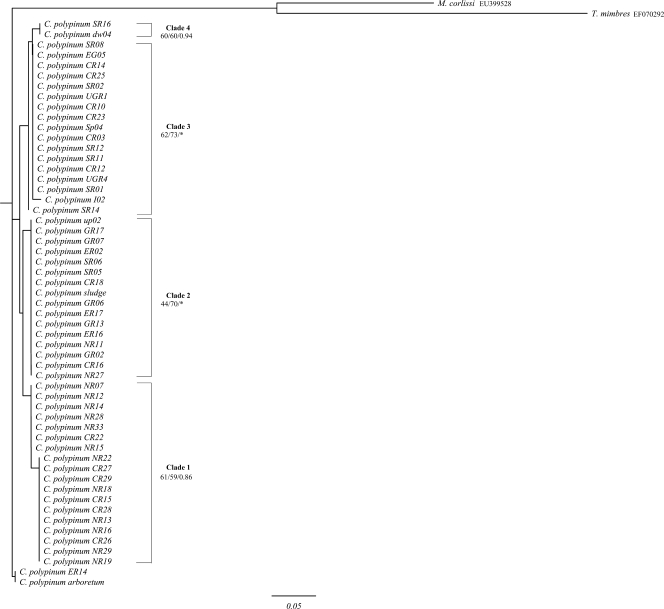
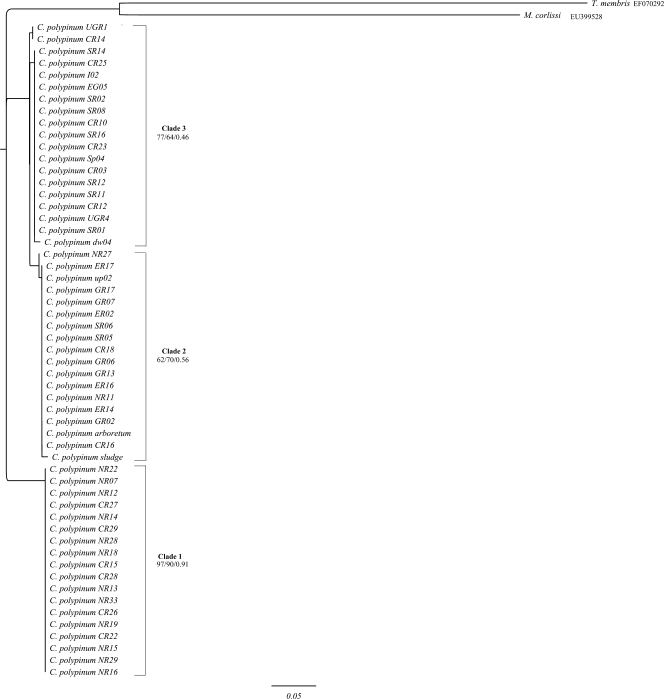
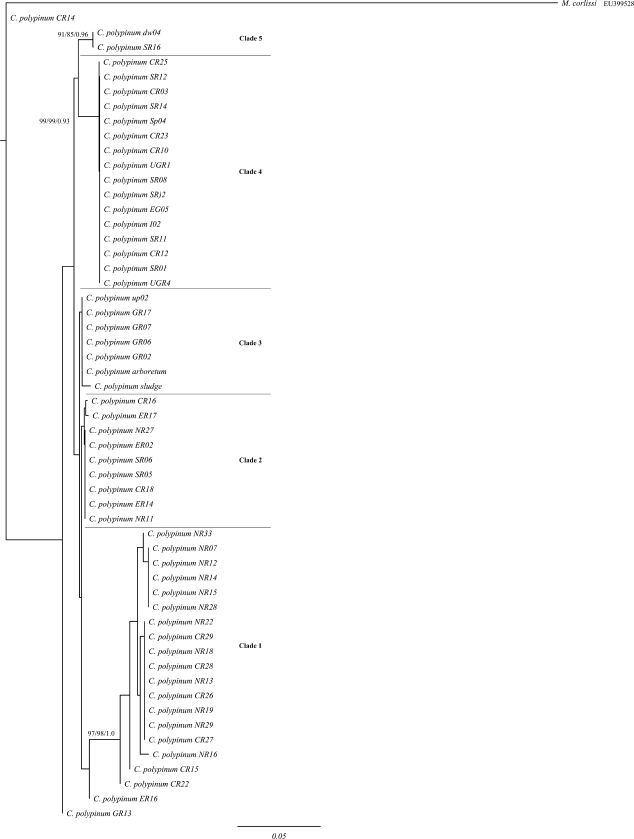
The tree topologies for ITS1, ITS2, and the 5′ end of the LSU rRNA were similar, with some notable differences (see below) (Fig. 1, 2, and 3). In addition, for the individual markers, the topologies of NJ, ML, and BI analyses were nearly identical. Based on these trees, no pattern of geographic distribution was apparent: individual genotypes collected from the five rivers were distributed throughout the tree rather than forming distinct monophyletic clades.
FIG. 1.
ML tree of the ITS1 region of environmental isolates of the peritrich ciliate Carchesium polypinum computed with PHYML, based on the F81 model, determined by Modeltest. The first and second values at the nodes represent bootstrap values for NJ and ML analyses, respectively, while the third value represents the posterior probability of the BI analysis. The scale bar represents 5 changes per 100 positions. Asterisks denote nodes that were not recovered in BI analysis. The sequences of Meseres corlissi and Tetrahymena mimbres were used to root the tree. Only bootstrap values above 40 are shown.
FIG. 2.
ML tree of the ITS2 region of environmental isolates of the peritrich ciliate Carchesium polypinum computed with PHYML, based on the Jukes-Cantor model, determined by Modeltest. The first and second values at the nodes represent bootstrap values for NJ and ML analyses, respectively, while the third value represents the posterior probability of the BI analysis. The scale bar represents 5 changes per 100 positions. The sequences of Meseres corlissi and Tetrahymena mimbres were used to root the tree. Only bootstrap values above 40 are shown.
FIG. 3.
ML tree of the 5′ end of the hypervariable region of the large subunit rRNA of environmental isolates of the peritrich ciliate Carchesium polypinum computed with PHYML, based on the F81+I+Γ model, determined by Modeltest. The first and second values at the nodes represent bootstrap values for NJ and ML analyses, respectively, while the third value represents the posterior probability of the BI analysis. The scale bar represents 5 changes per 100 positions. The sequence of Meseres corlissi was used to root the tree. Only bootstrap values above 80 are shown.
In the ITS1 tree (Fig. 1), four clades were immediately obvious. All clades were weakly to moderately supported. Clade 2 and clade 3 were not completely recovered when BI analysis was employed.
In the ITS2 tree (Fig. 2), there are three clades. All three clades were recovered with all the methods used. The support values for the clades of the ITS2 tree were higher than those of the ITS1 tree but still weak to moderate. The exception was clade 1, which was now strongly supported.
For the LSU rRNA tree (Fig. 3), five clades were recovered. Clades 2 and 3 were not completely recovered when BI analysis was employed and were very weakly supported when NJ and ML were employed (bootstrap values below 60%; not shown). The other three clades, however, were strongly supported.
Phylogenetic analyses of the cox-1 data set.
For the cox-1 tree, the tree topologies were identical for NJ, ML, and BI analyses (Fig. 4). For this data set, five clades were identified. Clades 3 and 4 were identified with very strong support in all three analyses, as were three putative clades (clades 1, 2, and 5). All clades were deeply diverging. Clades 1 and 2 consisted of individuals that belonged to clade 1 of the nuclear data sets. The rest of the individuals in Clade 1 of the nuclear data sets and all the individuals from clade 2 formed the strongly supported cox-1 clade 3. Clades 4 and 5 of the cox-1 data set were identical to clades 3 and 4 of both the ITS1 and LSU data sets.
FIG. 4.
ML tree of an 819-bp fragment of the cytochrome c oxidase I gene of environmental isolates of the peritrich ciliate Carchesium polypinum computed with PHYML, based on the GTR+Γ model, determined by Modeltest. The first and second values at the nodes represent bootstrap values for NJ and ML analyses, respectively, while the third value represents the posterior probability of the BI analysis. The scale bar represents 5 changes per 100 positions. The sequences of Tetrahymena thermophila, Colpidium campylum, and Glaucoma chattoni were used to root the tree.
In similarity to the nuclear trees, the clades did not correspond to the collection localities; rather, individual genotypes collected from various localities were distributed throughout the clades.
Population structure analysis.
Analysis of the cox-1 data set revealed 26 haplotypes. The sequences of C. polypinum were partitioned in two ways; initially a population was defined and partitioned as the individuals collected from each tributary. The AMOVA analysis based on the cox-1 data set indicated that 71.2% of the genetic variation observed was within populations and 28.9% between populations (Table 2). The pairwise FST values (statistic comparing genetic variations within and between populations) were for the most part not significant (data not shown). Similarly, when the sequences were grouped by clade, as defined in the phylogenetic analysis of the cox-1 data set, 71.15% of the variation was within populations and 28.85% among populations. However, the pairwise FST values were high and statistically significant, indicating a deep divergence in clades 2, 3, and 4 (Table 3). When the analysis was performed separately for the densely sampled clades (clades 3 and 4), the corresponding FST values were not significant (data not shown).
TABLE 2.
AMOVA analysis of an 819-bp fragment of the cox-1 gene of the peritrich ciliate C. polypinuma
| Comparison | Analysis of variation
|
||
|---|---|---|---|
| Degrees of freedom | Sum of squares | % of variation | |
| Among populations | 4 | 452.3 | 28.9 |
| Within populations | 36 | 947.2 | 71.2 |
Percentages of genetic variation within and among populations collected from different localities, as defined by the tributaries in the Grand River basin, southwestern Ontario, Canada (see Table S1 in the supplemental material).
TABLE 3.
Pairwise FST values based on AMOVA analysis and uncorrected P values using an 819-bp fragment of the cox-1 gene of five clades of the peritrich ciliate Carchesium polypinuma
| Grouping |
FST or P value for cox-1 gene fragment
|
||||
|---|---|---|---|---|---|
| Clade 1 | Clade 2 | Clade 3 | Clade 4 | Clade 5 | |
| Clade 1 | 0.377 | 0.046 | 0.085 | 0.999 | |
| Clade 2 | 0.988 | 0.005 | 0.008 | 0.999 | |
| Clade 3 | 0.980* | 0.978* | 0.000 | 0.999 | |
| Clade 4 | 0.930 | 0.934* | 0.961* | 0.999 | |
| Clade 5 | 1.000 | 0.991 | 0.981 | 0.879 | |
See Fig. 4. Numbers in the lower diagonal are pairwise FST values, while those in the upper diagonal are the uncorrected P values. Asterisks denote values of statistical significance (P ≤ 0.05).
DISCUSSION
Marker resolution.
When assessing intraspecific genetic variation, it is important to establish the appropriate markers for the organisms under investigation. In metazoan populations, such investigations involve the use of mtDNA markers, with the cox-1 gene being one of the most popular due to its higher rate of evolution (25, 29). Alternatively, ITS2 of the rRNA region is commonly used for plants (43). Traditionally, for ciliates, studies of intraspecific genetic variation are based on RAPDs (20, 35). Over the past few years, the focus has shifted from RAPDs to the use of nuclear and mitochondrial sequences (4, 5, 15, 44). Despite this, there are very few studies of intraspecific variation of ciliates, and so it is still unclear which marker is the most suitable for studying their genetic diversity. For ciliates, ITS1, ITS2, and the hypervariable regions of the LSU rRNA have been used most commonly. However, there has never been an attempt to compare all three nuclear markers on one ciliate species at the sequence level. In this study, we used all three nuclear markers in addition to one mitochondrial marker in order to compare and contrast the resolution and suitability of these molecules at the intraspecific level. We picked the cox-1 gene as the mitochondrial marker of choice for C. polypinum. The use of cox-1 in analysis of intraspecific genetic variation in ciliates is extremely limited (4). This is likely due to the fact that the only available sequences of cox-1 in ciliates are those from species of the genera Paramecium and Tetrahymena. To date, only six complete ciliate mtDNA genomes are available: for Tetrahymena thermophila, Tetrahymena pyriformis, Tetrahymena paravorax, Tetrahymena malaccensis, Tetrahymena pigmentosa, and P. aurelia (9, 10, 46, 52).
Tetrahymena and Paramecium belong to the class Oligohymenophorea but different subclasses—the subclasses Peniculia and Hymenostomatia, respectively (41). Despite this, the percent divergence of cox-1 is so high that it was not possible to align the sequences with the default settings of the Sequencher alignment software program (E. Gentekaki, personal observation). Thus, it has been extremely difficult to design primers for the cox-1 gene of other groups of ciliates. After considerable experimentation, we developed a set of primers that amplifies an 819-bp fragment of cox-1 in C. polypinum.
In our study, the best resolution was obtained with the cox-1 gene, as assessed by the resulting clades being deeply divergent and robustly and strongly supported. The nuclear gene that matched cox-1 the most closely in terms of separation of clades, evolutionary distances, robustness, and support was that of the 5′ end of the LSU rRNA. This is likely because the regions chosen for the analysis are among the most highly variable in the LSU rRNA and therefore are suitable for analysis below the morphospecies level (1). The analysis of the ITS1 and ITS2 regions gave somewhat similar topologies, but the resulting clades were not strongly supported. Our study is in agreement with the work of Barth et al. (4), who found that the cox-1 gene provided better separation of Paramecium species than the corresponding ITS markers. Since ITS regions contain signals used for processing of the rRNA transcript (27, 38), these spacers are constrained to evolve more slowly. Consequently, they did not have adequate time to accumulate polymorphisms that would be useful for an intraspecies-level analysis. Alternatively, the high degree of variation and fast evolutionary rates in the mtDNA genome versus those of the nuclear genome are well documented for metazoans, and this seems to also be the case for protists (22). Unlike the case for metazoans, the mutation rate of mtDNA in protists is unknown. It has been speculated that it is 10% per million years (5), which would make mtDNA genes good candidates for assessing intraspecific variation.
Intraspecific genetic variation within Carchesium polypinum.
Studies of intraspecific genetic diversity of ciliates have yielded varying results. On the one hand, low genetic diversity was revealed for T. thermophila (32), Stentor coeruleus (35), and Euplotes daidaleos (36). On the other hand, high genetic diversity was evident in others, including Paramecium multimicronucleatum and Paramecium caudatum (4), Halteria grandinella (31), Cyclidium glaucoma (15), Coleps spetai (5), and Coleps hirtus hirtus (5). It is worth noting, however, that all the studies that used mitochondrial markers revealed a high intraspecific genetic diversity (4, 5). In agreement with results of these studies, ours also indicated a high genetic diversity of C. polypinum populations, particularly in the case of cox-1. Moreover, the degree of variation revealed when analyzing the cox-1 data set indicated that the majority of the variation was at the isolate level rather than among populations. A similar degree of genetic variation within populations of C. polypinum was also found when intersimple sequence analysis was employed (63). The authors of that report suggested that the high genetic diversity of C. polypinum at the isolate level was due to high gene flow and the ability of telotrochs, the dispersal stage, to range over large distances, resulting in a large panmictic population.
Phylogeography.
It has been a longstanding debate whether or not protists form uniform global populations or if, at least in the case of some, they form distinct endemic populations (16, 17, 18, 19, 45, 60). So far the debate remains inconclusive due to the very small number of studies that deal with the spatial and temporal distribution of protists (7). For instance, no geographic isolation of populations was observed for the heterotrich Stentor or the hypotrich Euplotes (35, 37). Alternatively, when RAPDs were assessed along with morphological features for the hypotrich Gonostomum affine, a rudimentary population structure was revealed (20). Use of the ITS1 region, a nuclear marker, has yielded conflicting results. On the one hand, Miao et al. (44) demonstrated distinct population structures in populations of C. polypinum in China, represented by two distinct clades that mapped onto the geography of the region. Similarly, H. grandinella showed variation among geographic isolates (31). Conversely, there was no geographic structure for populations of Meseres corlissi, Isotricha prostoma, and P. caudatum, despite the fact that some individuals from all of these species were from across the globe (4, 59, 62). In agreement with results in the latter studies, we did not detect any pattern that maps to the geography of the Grand River basin when we used any of the four markers. In the case of the mitochondrial marker, we obtained deeply divergent clades, but those were not geographically partitioned. The absence of population structure could be due to a high effective population size and a high gene flow or a combination of both (3). It has been argued that effective population size is inversely proportional to body size: as body size decreases, the effective population size increases (40). Effective population size estimates of ciliates are extremely limited and are restricted to members of the genera Paramecium and Tetrahymena (32, 55). These analyses have indicated that the effective population of T. thermophila is small but that of Paramecium is large. However, recent studies of the protein evolution of ciliates suggest an elevated rate of substitutions that might make analysis of their effective population sizes difficult to determine (64). At present, we do not know the effective population size of C. polypinum.
Morphospecies and cryptic species.
Over the past couple of decades, the advent of molecular techniques has uncovered a large degree of genetic diversity. Several species that were once considered cosmopolitan are now considered cryptic; in fact, discovery of cryptic species has grown exponentially (21, 26, 33, 49, 61). The most common current method of identifying cryptic species is based on the amount of genetic divergence as defined by genetic distances. Analyses of our mitochondrial data set show that at least three genetically distinct and deeply divergent groups of C. polypinum occur in sympatry in the Grand River basin. The minimum genetic distance between the groups was 11%, while the maximum was 18%. Given this amount of genetic divergence, C. polypinum is very likely a cryptic species complex compared to genetic divergences of this gene for known biological species of Tetrahymena and Paramecium.
Currently it is speculated that the concept of morphospecies might be too conservative for assessing protist diversity (58) or that their morphological and molecular evolution might be decoupled (19). Previous research has shown that the amount of genetic divergence differs between groups. For instance, a 10% divergence in the cox-1 gene of Tetrahymena would be enough to designate a new species, while this value is higher for Paramecium (4, 12). Since there are very few studies of such nature with ciliates, we cannot be sure what percentage of variation constitutes a new species or a cryptic species of C. polypinum.
If the uncovered diversity is indeed an indication of cryptic species of C. polypinum, then how did seemingly sympatric species occur in a continuous system like a river, where there are no barriers to disrupt gene flow long enough for isolation to occur? Lately there have been speculations that sympatric speciation might be more common than initially thought (48). However, studies that make a strong case for this are very few (47). Alternatively, the observed genetic diversity could be due to the transfer of a few founder cells from other bodies of water. While we currently have no way to determine which of the two processes accounts for the observed variation, it seems that there is a lot more underlying genetic diversity in protists than initially thought.
Supplementary Material
Acknowledgments
This research was partly funded by the Natural Sciences and Engineering Research Council of Canada (NSERC). The research was also supported through funding to the Canadian Barcode of Life Network from Genome Canada through the Genomics Institute, awarded to D.H.L. and other sponsors (listed at http://www.BOLNET.ca).
We thank Chris Lobban for providing us with the E. plicatilis isolate. We also thank Strüder-Kypke for critical review of the manuscript. We are grateful to Guelph wastewater treatment plant personnel for their help in obtaining the sludge C. polypinum isolate. We also thank Angela Holliss and Elizabeth Holmes for their invaluable help in sequencing all of our samples. Finally, we thank two anonymous reviewers for their constructive comments.
Footnotes
Published ahead of print on 20 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ali, A. B., J. Wuyts, R. De Wachter, A. Meyer, and Y. Van de Peer. 1999. Construction of a variability map for eukaryotic large subunit ribosomal RNA. Nucleic Acids Res. 27:2825-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amato, A., W. H. C. F. Kooistra, J. H. L. Ghiron, D. G. Mann, T. Pröschold, and M. Montresor. 2007. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist 158:193-207. [DOI] [PubMed] [Google Scholar]
- 3.Avise, C. J. 2000. Phylogeography: the history and formation of species, 1st ed. Harvard University Press, Harvard, MA.
- 4.Barth, D., S. Krenek, S. I. Fokin, and T. U. Berendonk. 2006. Intraspecific genetic variation in Paramecium revealed by mitochondrial cytochrome c oxidase I sequences. J. Eukaryot. Microbiol. 53:20-25. [DOI] [PubMed] [Google Scholar]
- 5.Barth, D., K. Tischer, H. Berger, M. Schlegel, and T. U. Berendonk. 2008. High mitochondrial haplotype diversity of Coleps sp. (Ciliophora: Prostomatida). Environ. Microbiol. 10:626-634. [DOI] [PubMed] [Google Scholar]
- 6.Bass, D., T. A. Richards, L. Matthai, V. Marsh, V., and T. Cavalier-Smith. 2007. DNA evidence for global dispersal and probable endemicity of protozoa. BMC Evol. Biol. 7:162-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beheregaray, L. B. 2008. Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Mol. Ecol. 17:3754-3774. [DOI] [PubMed] [Google Scholar]
- 8.Blair, D., T. Agatsuma, T. Watanobe, M. Okamoto, and A. Ito. 1997. Geographical genetic structure within the human lung fluke, Paragonimus westermani, detected from DNA sequences. Parasitology 115:411-417. [DOI] [PubMed] [Google Scholar]
- 9.Brunk, F. C., L. C. Lee, A. B. Tran, and J. Li. 2003. Complete sequence of the mitochondrial genome of Tetrahymena thermophila and comparative methods for identifying highly divergent genes. Nucleic Acids Res. 31:1673-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burger, G., Y. Zhu, T. G. Littlejohn, S. J. Greenwood, M. N. Schnare, B. F. Lang, and M. W. Gray. 2000. Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA. J. Mol. Biol. 297:365-380. [DOI] [PubMed] [Google Scholar]
- 11.Catania, F., F. Wurmser, A. A. Potekhin, E. Przyboś, and M. Lynch. 2009. Genetic diversity in the Paramecium aurelia species complex. Mol. Biol. Evol. 26:421-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chantangsi, C., D. H. Lynn, M. T. Brandl, J. C. Cole, N. Hetrick, and P. Ikonomi. 2007. Barcoding ciliates: a comprehensive study of 75 isolates of the genus Tetrahymena. Int. J. Syst. Evol. Microbiol. 57:2412-2425. [DOI] [PubMed] [Google Scholar]
- 13.Coleman, A. W. 2005. Paramecium aurelia revisited. J. Eukaryot. Microbiol. 52:68-77. [DOI] [PubMed] [Google Scholar]
- 14.De Rijk, P., and R. De Wachter. 1993. DCSE, an interactive tool for sequence alignment and secondary structure research. Comput. Appl. Biol. Sci. 9:735-740. [DOI] [PubMed] [Google Scholar]
- 15.Finlay, J. B., G. F. Esteban, S. Brown, T. Fenchel, and K. Hoef-Emden. 2006. Multiple cosmopolitan ecotypes within a microbial eukaryote morphospecies. Protist 157:377-390. [DOI] [PubMed] [Google Scholar]
- 16.Finlay, J. B., G. F. Esteban, and T. Fenchel. 1998. Protozoan diversity, converging estimates of the global number of free-living ciliate species. Protist 149:29-37. [DOI] [PubMed] [Google Scholar]
- 17.Finlay, J. B., and T. Fenchel. 1999. Divergent perspectives on protist species richness. Protist News 150:229-233. [DOI] [PubMed] [Google Scholar]
- 18.Foissner, W. 2006. Biogeography and dispersal of micro-organisms, a review emphasizing protists. Acta Protozool. 45:111-136. [Google Scholar]
- 19.Foissner, W., A. Chao, and L. A. Katz. 2008. Diversity and geographic distribution of ciliates (Protista: Ciliophora). Biodivers. Conserv. 17:345-363. [Google Scholar]
- 20.Foissner, W., T. Stoeck, H. Schmidt, and H. Berger. 2001. Biogeographical differences in a common soil ciliate, Gonostomum affine (Stein), as revealed by morphological and RAPD-fingerprint analysis. Acta Protozool. 40:83-97. [Google Scholar]
- 21.Gomez, A., M. Serra, G. R. Carvalho, and D. H. Lunt. 2002. Speciation in ancient cryptic species complexes: evidence from the molecular phylogeny of Brachionus plicatilis (Rotifera). Evolution 56:1431-1444. [DOI] [PubMed] [Google Scholar]
- 22.Gray, M. W., B. F. Lang, and G. Burger. 2004. Mitochondria of protists. Annu. Rev. Genet. 38:477-524. [DOI] [PubMed] [Google Scholar]
- 23.Guindon, S., and O. Gascuel. 2003. PhyML, a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 24.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33(Web server issue):W557-W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hajibabaei, M., G. A. C. Singer, P. D. N. Hebert, and D. A. Hickey. 2007. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 23:167-172. [DOI] [PubMed] [Google Scholar]
- 26.Hebert, P. D. N., E. H. Penton, J. M. Burns, D. H. Janzen, and W. Hallwachs. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 101:14812-14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillis, D. M., and M. T. Dixon. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 66:411-453. [DOI] [PubMed] [Google Scholar]
- 28.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES, Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 29.Hwang, U. W., and W. Kim. 1999. General properties and phylogenetic utilities of nuclear ribosomal DNA and mitochondrial DNA commonly used in molecular systematics. Korean J. Parasitol. 37:215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jerome, C. A., and D. H. Lynn. 1996. Identifying and distinguishing sibling species in the Tetrahymena pyriformis complex (Ciliophora, Oligohymenophorea) using PCR/RFLP analysis of nuclear ribosomal DNA. J. Eukaryot. Microbiol. 43:492-497. [DOI] [PubMed] [Google Scholar]
- 31.Katz, L. A., G. B. McManus, L. O. Snoeyenbos-West, A. Griffin, K. Pirog, B. Costas, and W. Foissner. 2005. Reframing the “everything is everywhere” debate: evidence for high gene flow and diversity in ciliate morphospecies. Aquat. Microb. Ecol. 41:55-65. [Google Scholar]
- 32.Katz, L. A., L. O. Snoeyenbos-West, and F. P. Doerder. 2006. Unusual patterns of molecular evolution at the SerH surface antigen locus in Tetrahymena thermophila: implication for estimates of effective population size. Mol. Biol. Evol. 23:608-614. [DOI] [PubMed] [Google Scholar]
- 33.King, J. L., and R. Hanner. 1998. Cryptic species in a “living fossil” lineage: taxonomic and phylogenetic relationships within the genus Lepidurus (Crustacea: Notostraca) in North America. Mol. Phylogenet. Evol. 10:23-36. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2, molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 35.Kusch, J. 1998. Local and temporal distribution of different genotypes of pond-dwelling Stentor coeruleus. Protist 149:147-154. [DOI] [PubMed] [Google Scholar]
- 36.Kusch, J., and K. Heckmann. 1996. Population structure of Euplotes ciliates revealed by RAPD fingerprinting. Ecoscience 3:378-384. [Google Scholar]
- 37.Kusch, J., H. Welter, M. Stremmel, and H. J. Schmidt. 2000. Genetic diversity in populations of a freshwater ciliate. Hydrobiologia 431:185-192. [Google Scholar]
- 38.Leary, D. J., and J. Huang. 2001. Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 509:145-150. [DOI] [PubMed] [Google Scholar]
- 39.Lobban, C. S., and M. Schefter. 2008. Freshwater biodiversity in Guam. I. Introduction with new records of ciliates and a heliozoan. Micronesica 40:253-273. [PMC free article] [PubMed] [Google Scholar]
- 40.Lynch, M. 2006. The origins of eukaryotic gene structure. Mol. Biol. Evol. 23:450-468. [DOI] [PubMed] [Google Scholar]
- 41.Lynn, D. H. 2008. The ciliated protozoa. Characterization, classification and guide to the literature, 3rd ed. Springer, New York, NY.
- 42.Lynn, D. H., and M. C. Strüder-Kypke. 2006. Species of Tetrahymena identical by small subunit rRNA gene sequences are discriminated by mitochondrial cytochrome c oxidase I gene sequences. J. Eukaryot. Microbiol. 53:385-387. [DOI] [PubMed] [Google Scholar]
- 43.Mai, J. C., and A. W. Coleman. 1997. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J. Mol. Evol. 44:258-271. [DOI] [PubMed] [Google Scholar]
- 44.Miao, W., Y. Yu, Y. H. Shen, and X. Zhang. 2004. Intraspecific phylogeography of Carchesium polypinum (Peritrichia, Ciliophora) from China, inferred from 18S-ITS1-5.8S ribosomal DNA. Sci. China 47:11-17. [DOI] [PubMed] [Google Scholar]
- 45.Mitchell, E. A. D., and R. Meisterfeld. 2005. Taxonomic confusion blurs the debate on cosmopolitanism versus local endemism of free-living protists. Protist 156:263-267. [DOI] [PubMed] [Google Scholar]
- 46.Moradian, M. M., D. Beglaryan, J. M. Skozylas, and V. Kerikorian. 2007. Complete mitochondrial genome sequence of three Tetrahymena species reveals mutation hot spots and accelerated nonsynonymous substitutions in Ymf genes. PLoS ONE 2:e650. doi: 10.1371/journal.pone.0000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niemiller, M. L., B. M. Fitzpatrick, and B. T. Miller. 2008. Recent divergence with gene flow in Tennessee cave salamanders (Plethodontidae: Gyrinophilus) inferred from gene genealogies. Mol. Ecol. 17:2258-2275. [DOI] [PubMed] [Google Scholar]
- 48.Nosil, P. 2008. Speciation with gene flow could be common. Mol. Ecol. 17:2103-2106. [DOI] [PubMed] [Google Scholar]
- 49.Pfenninger, M., and K. Schwenk. 2007. Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evol. Biol. 7:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Posada, D., and K. A. Crandall. 1998. MODELTEST, testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 51.Preparata, R. M., E. B. Meyer, F. P. Preparata, E. M. Simon, C. R. Vossbrinck, and D. L. Nanney. 1989. Ciliate evolution: the ribosomal phylogenies of the tetrahymenine ciliates. J. Mol. Evol. 28:427-441. [DOI] [PubMed] [Google Scholar]
- 52.Pritchard, A. E., J. J. Seilhamer, R. Mahalingam, L. C. Sable, E. S. Venuti, and J. D. Cummings. 1990. Nucleotide sequence of the mitochondrial genome of Paramecium. Nucleic Acids Res. 18:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes3, Bayesian inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 54.Skroch, P., and J. Nienhuis. 1995. Impact of scoring error and reproducibility of RAPD data on RABD based estimates of genetic distance. Theor. Appl. Genet. 91:1086-1091. [DOI] [PubMed] [Google Scholar]
- 55.Snoke, M. S., T. U. Berendonk, and D. Barth. 2006. Large global effective population sizes in Paramecium. Mol. Biol. Evol. 23:2474-2479. [DOI] [PubMed] [Google Scholar]
- 56.Stoeck, T., and H. J. Schmidt. 1998. Fast and accurate identification of European species of the Paramecium aurelia complex by RAPD-fingerprinting. Microb. Ecol. 35:311-317. [DOI] [PubMed] [Google Scholar]
- 57.Stoeck, T., E. Przybos, J. Kusch, and H. J. Schmidt. 2000. Intra-specific differentiation and level of inbreeding of different sibling species of the Paramecium aurelia complex. Acta Protozool. 39:15-22. [Google Scholar]
- 58.Weisse, T. 2008. Distribution and diversity of aquatic protists: an evolutionary and ecological perspective. Biodivers. Conserv. 17:243-259. [Google Scholar]
- 59.Weisse, T., M. C. Strüder-kypke, H. Berger, and W. Foissner. 2008. Genetic, morphological, and ecological diversity of spatially separated clones of Meseres corlissi Petz & Foissner, 1992 (Ciliophora, Spirotrichea). J. Eukaryot. Microbiol. 55:257-270. [DOI] [PubMed] [Google Scholar]
- 60.Wilkinson, D. M. 2001. What is the upper size limit for cosmopolitan distribution in free-living microorganisms? J. Biogeogr. 28:285-291. [Google Scholar]
- 61.Witt, S. D. J., and P. D. N. Hebert. 2000. Cryptic species diversity and evolution in the amphipod genus Hyalella within central glaciated North America: a molecular phylogenetic approach. Can. J. Fish. Aquat. Sci. 57:687-698. [Google Scholar]
- 62.Wright, A. D. G. 1999. Analysis of intraspecific sequence variation among eight isolates of the rumen symbiont, Isotricha prostoma (Ciliophora) from two continents. J. Eukaryot. Microbiol. 46:445-446. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, W. J., J. Yang, Y. H. Yu, S. W. Shu, and Y. F. Shen. 2006. Population genetic structure of Carchesium polypinum (Ciliophora: Peritrichia) in four Chinese lakes inferred from ISSR fingerprinting: high diversity but low differentiation. J. Eukaryot. Microbiol. 53:358-363. [DOI] [PubMed] [Google Scholar]
- 64.Zufall, R. A., C. McGrath, S. V. Muse, and L. A. Katz. 2006. Genome architecture drives protein evolution in ciliates. Mol. Biol. Evol. 23:1681-1687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.