Abstract
Paenibacillus larvae is the etiological agent of American foulbrood (AFB) in honeybees. Recently, different genotypes of P. larvae (ERIC I to ERIC IV) were defined, and it was shown that these genotypes differ inter alia in their virulence on the larval level. On the colony level, bees mitigate AFB through the hygienic behavior of nurse bees. Therefore, we investigated how the hygienic behavior shapes P. larvae virulence on the colony level. Our results indicate that P. larvae virulence on the larval level and that on the colony level are negatively correlated.
American foulbrood (AFB) is among the economically most important honeybee diseases. The etiological agent of AFB is the gram-positive, spore-forming bacterium Paenibacillus larvae (9). The extremely tenacious spores are the infectious form of this organism. These spores drive disease transmission within colonies (11), as well as between colonies as soon as they end up in the honey stores of an infected colony (12).
The species P. larvae can be subdivided into four different genotypes designated ERIC I to ERIC IV based on results from repetitive-element PCR (20) using enterobacterial repetitive intergenic consensus (ERIC) primers (9, 10), with P. larvae ERIC I and ERIC II being the two practically most important genotypes (1, 2, 9, 10, 13, 16). The four genotypes were shown previously to differ in phenotype, including virulence on the larval level (8, 9). While larvae infected with genotypes ERIC II to ERIC IV were killed within only 6 to 7 days, it took P. larvae ERIC I around 12 to 14 days to kill all infected individuals. Therefore, genotype ERIC I was considered to be less virulent and the other three genotypes were considered to be highly virulent (7-9) on the larval level.
P. larvae is an obligately killing pathogen which must kill its host to be transmitted. The virulence of such an obligate killer is thought to be determined primarily by two factors, (i) the probability of infecting a host and (ii) the time to host death (6). The problem of ensuring a high enough probability of infecting the next host is solved for P. larvae by (i) the tenacious exospores, which remain infectious for over half a century (17) and, therefore, can wait for decades for the next host to pass by, and (ii) a high pathogen reproduction rate (23) and, thus, the production of an extremely high number of spores within each infected larva.
For evaluating the second factor determining P. larvae virulence, the time to host death, it is important to consider the two levels of honeybee hosts, the level of the individual larva dying from AFB and the level of the colony succumbing to AFB.
The virulence of P. larvae genotypes on the larval level has been analyzed recently (8, 9). We have now determined the colony-level virulence for the two most common and practically important (10, 16) genotypes of P. larvae, ERIC I and ERIC II, significantly differing in virulence on the larval level (8). We will discuss how the time to larval death relates to the time to colony death and how the hygienic response shapes P. larvae virulence.
Bacterial isolates and preparation of defined spore suspensions.
The P. larvae type strain ATCC 9545 (genotype ERIC I) was obtained from the American Type Culture Collection (ATCC) through U. Rdest (Biocenter Würzburg). P. larvae strain 04-309 (genotype ERIC II) represents a German field isolate of P. larvae isolated from a honey sample originating from an AFB-positive hive. Both strains, ATCC 9545 and 04-309, have been genotyped (10), and their degrees of virulence on the larval level (8), as well as early steps in pathogenesis (23), have also been characterized. P. larvae was cultivated on Columbia sheep blood agar plates or slant agar as described previously (8, 10). The preparation and storage of spore suspensions containing a defined concentration of CFU for use in infection assays were performed as described previously (8). All chemicals and media for culturing bacteria and preparing spore suspensions were obtained from Oxoid, Germany.
Bee and larval material for exposure bioassays in minicolonies.
To minimize the effect of the genetic background of the bees, pools of young honeybees from 10 bee colonies maintained in the apiary at the Institute for Bee Research in Hohen Neuendorf, Germany, were equally divided into 10 groups, each containing enough bees to found a minicolony. The minicolonies raised their own queens, which were then allowed to mate naturally, resulting in “queenright” colonies. The minicolonies were kept in the institute's bee yard until they were taken into the flight room for the infection experiments. Experiments were not performed until after the first young bees had emerged from the newly laid eggs.
Exposure bioassays in minicolonies.
For exposure bioassays, brood combs containing larvae at life stage 1 (L1; 10 to 12 hours after egg hatching) were taken out of the hive and a total of around 80 to 100 L1 larvae per experiment were individually infected in their brood cells (4) with 3 μl of a P. larvae spore suspension containing 8 CFU/μl for ATCC 9545 or 4 CFU/μl for 04-309. The concentrations of the spore suspensions necessary to yield the 100% lethal dose of each strain in the larval food had been extrapolated from the results of our previous studies (8) and had been verified again for this study in laboratory infection assays (data not shown). Infection was carried out on one side of the brood comb, while the other side of the comb was mock infected as a control by the application of 3 μl of water to the mouthparts of the larvae. The positions of the control and the infected brood cells were recorded on a see-through plastic sheet put onto the comb. The fates of the control and the infected larvae were monitored daily, and the following data were recorded for each cell: (i) the presence and development of the original larva, (ii) the absence of the larva, indicative of removal due to the hygienic behavior of the nurse bees, and (iii) the occupation of the cell by a newly laid egg. At day 13 postinfection, the experiments were terminated by carefully opening each capped brood cell which had not been cleaned out but still contained the original, manipulated larva and evaluating the content of the cell, i.e., the developmental stage and health status of the exposed larva (dark-eyed pupa or ropy mass) (see Fig. 2). The number of successfully infected larvae was calculated as the sum of the larvae removed in the course of the experiment and the diseased larvae/ropy masses found in the brood cells at the end of the experiment. According to this analysis, the spore concentrations used resulted in the successful infection of 93 or 84% of individuals with ATCC 9545 or 04-309, respectively. The experiments were repeated at least three times. For ATCC 9545 and 04-309, a total of 329 and 322 larvae, respectively, were exposed to infectious spores. For each experiment, new colonies as well as new hive material and flight cages were obtained to avoid cross contamination. The experiments were performed in a flight room under suitable safety precautions.
FIG. 2.
Brood comb regions at the end of the experiments. Larvae were individually infected with spores of ATCC 9545 (ERIC I) or 04-309 (ERIC II) or mock infected with water as a control as described in the text. Thirteen days postinfection, the experiment was terminated and the developmental stages and health statuses of those larvae which had not been removed in the course of the experiment were evaluated. Red arrows point to healthy pupae in the ATCC 9545-infected group, and white arrows point to ropy masses in the 04-309-infected group.
Removal rates for ERIC I and ERIC II.
In our previous studies, we determined the virulence of different genotypes of P. larvae on the larval level by using laboratory exposure bioassays (8, 9). These assays were perfect to study the interaction between P. larvae and isolated bee larvae, but they did not represent the situation the pathogen is facing in a colony where nurse bees, cleaning out diseased larvae as part of the social immune response, are present. To evaluate the success and, hence, the virulence of different P. larvae genotypes in such an environment, we performed colony exposure bioassays. We individually infected larvae in queenright minicolonies with strains ATCC 9545 and 04-309 as representatives of P. larvae genotypes ERIC I and ERIC II, respectively, and determined the time course of the removal of infected larvae by nurse bees. The mean cumulative proportion of infected larvae removed per day for each isolate was calculated and plotted against time. Infected larvae were defined as those larvae which were either removed or remained as ropy masses under the cell caps. Data for larvae that remained uninfected (i.e., healthy, dark-eyed pupae) at the end of the experiment were excluded from this calculation. For larvae infected with ATCC 9545, the time course of removal revealed a biphasic curve progression in which a first exponential phase of removal between days 1 and 5 postinfection and a second phase of removal between days 8 and 10 postinfection were separated by a phase of nearly no removal activity between days 5 and 8 postinfection (Fig. 1A). In contrast, the cumulative removal of larvae infected with 04-309 followed a classical sigmoid curve, with the majority of the infected larvae (around 86%) removed by day 5 postinfection (Fig. 1B). The mean removal rates ± standard deviations for ATCC 9545 (ERIC I) and 04-309 (ERIC II) at day 13 postinfection were 63.6% ± 4.4% and 90.0% ± 1.4%, respectively (Fig. 1). The hygienic behavior toward control larvae was more or less restricted to the first 5 days after manipulation and always resulted in the removal of less than 30% of the larvae (Fig. 1).
FIG. 1.
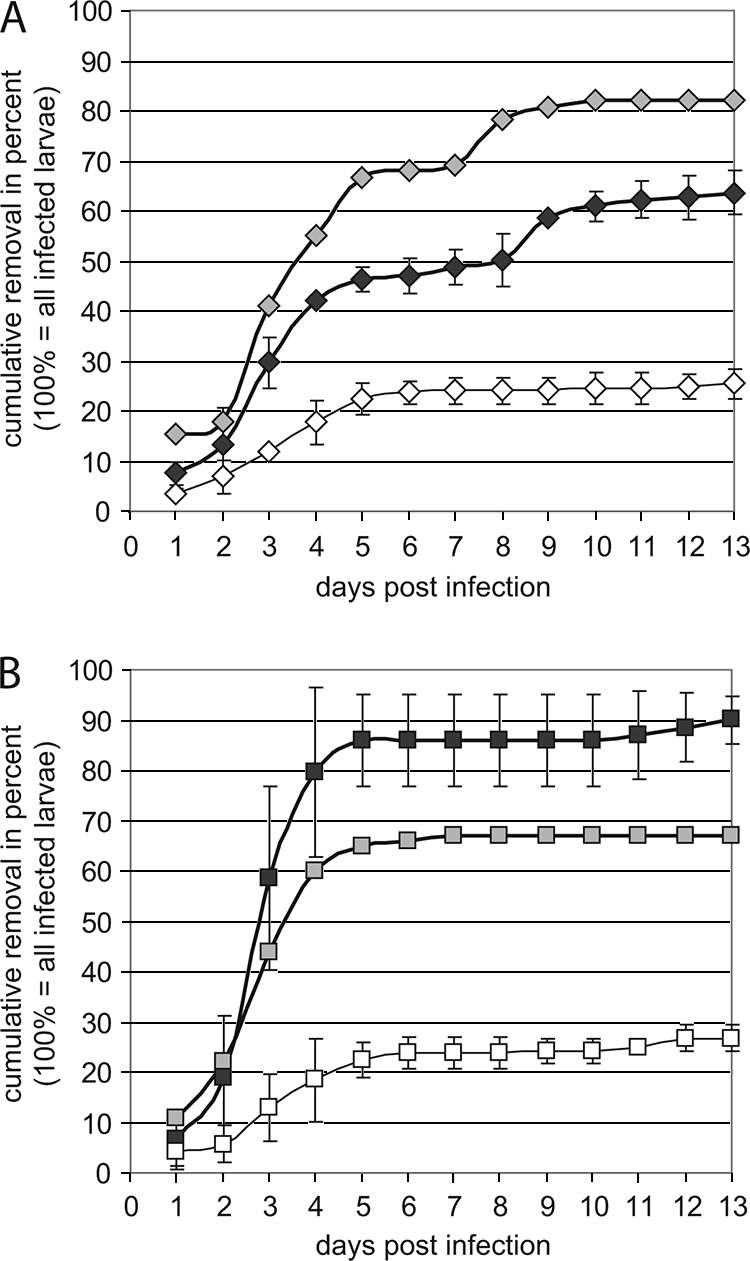
Time course of removal. The mean cumulative removal ± the standard deviation per day postinfection was calculated as described in the text and expressed as a percentage of the infected hosts (i.e., all larvae that were removed by hygienic nurse bees or were found to be AFB-diseased under the cell cap at the end of the experiments). Results for larvae infected with ATCC 9545 (ERIC I; black filled diamonds) and 04-309 (ERIC II; black filled squares) are shown in panels A and B, respectively. The outlier curves (gray filled diamonds for ATCC 9545 in panel A and gray filled squares for 04-309 in panel B) represent results from a single experiment in which the nurse bees reacted toward infected larvae in a manner contrary to those in the other experiments. Curves for the cumulative removal of mock-infected controls (open diamonds and squares) are also shown.
Production of transmission stages for ERIC I and ERIC II.
Since only those infected larvae that are not removed by nurse bees but instead remain in their brood cells and allow the pathogen to be converted into spores drive disease transmission within the colony, the proportion of larvae not cleaned out is an important measure for the prospective transmission success of P. larvae. A comparison of the rates of removal of larvae infected with either ATCC 9545 (ERIC I) or 04-309 (ERIC II) revealed that normally only around 10% of the larvae infected with 04-309 (ERIC II) had not been cleaned out in our experiments and, therefore, had the potential to serve as spore sources for the transmission of P. larvae. A four-times-greater proportion of larvae (around 40%) infected with ATCC 9545 (ERIC I) remained in their brood cells and developed into ropy masses, meaning that around four times more larvae in the ATCC 9545-infected colonies than in the 04-309-infected colonies could have allowed the pathogen to be converted into spores (Table 1). No ropy mass was ever detected in the control groups. In Fig. 2, representative areas of brood combs from one experiment are shown. In the control group, all cells contained healthy pupae in the dark-eyed stage. In the infected groups, only those cells which had not been cleaned out in the course of the experiment and which still contained the original infected larvae had been carefully opened to evaluate the developmental stages and health statuses of the engorged larvae or pupae. In the ATCC 9545-infected group, the majority of the cells shown contained ropy masses and only a few healthy pupae had survived, whereas in the 04-309-infected group, the majority of the cells within the area shown had been cleaned out and, therefore, remained unopened for evaluation. Only two cells within this area contained ropy masses.
TABLE 1.
Relationship between the time to host death on the larval level and that on the colony level and the degrees of virulence of P. larvae on the larval and colony levelsa
| Genotype | Time (days) to host death (larval death)b | Degree of virulence of P. larvae on larval level | Proportion of larvae dying after cell capping | Rate of removal of infected larvae | Level of development of ropy mass and spore production | Rate of spreading within the colony | Time to host death (colony collapse) | Degree of virulence of P. larvae on colony level |
|---|---|---|---|---|---|---|---|---|
| ERIC I | ∼12 | Low | Large | Low | High | High | Short | High |
| ERIC II | ∼7 | High | Small | High | Low | Low | Long | Low |
We also want to point out that the results of one run of experiments preceded by adverse weather conditions (a cold and rainy summer period) surprisingly differed from those of the other trials in that the hygienic bees responded contrarily to the P. larvae-infected larvae. In this single set of experiments, 82% of the larvae infected with ATCC 9545 were removed and only 67% of the larvae infected with 04-309 were cleaned out (Fig. 1). The occurrence of these outliers, with results displayed separately in the graph, may explain why sometimes colonies infected with P. larvae ERIC I survive for several seasons or why colonies infected with P. larvae ERIC II finally collapse.
Recently, the question of P. larvae virulence on the larval level has been addressed, and it was demonstrated in laboratory exposure bioassays that P. larvae ERIC II is more virulent than genotype ERIC I, since 90 to 95% of all ERIC II-infected larvae were killed before the onset of metamorphosis and 40% of the larvae infected by ERIC I died as engorged larvae after the onset of metamorphosis (8, 9). Since larval brood cells are open until the beginning of metamorphosis and larvae are regularly attended by nurse bees, it was hypothesized that in the context of a colony, the vast majority of the larvae killed by P. larvae ERIC II would be recognized as diseased and removed by hygienic nurse bees. In contrast, around a third of the larvae infected by representatives of ERIC I would die in capped cells, leaving hygienic nurse bees only a small chance of cleaning out these infected larvae in the context of the colony (3, 7, 8).
These hypotheses had so far been deduced only from the results of laboratory infection assays naturally excluding nurse bees (8, 9). We here provide experimental proof that differences in the time course of disease progression on the larval level (the times to 50 and 100% lethality) indeed influence disease progression on the colony level by influencing the efficiency with which nurse bees can remove infected larvae and reduce spore production. In our experiments, the removal behavior of the nurse bees and the rate of removal of infected larvae depended on the P. larvae strain used for infection. Larvae infected with 04-309, a representative of genotype ERIC II, which kills rapidly (on the larval level), were removed more efficiently than larvae infected with ATCC 9545, a representative of genotype ERIC I, which kills more slowly (on the larval level). These data fit well to what was predicted from the results of laboratory infection assays performed with larvae (3, 7-9). Therefore, the time course of disease progression in infected larvae indeed had an impact on the efficiency of the hygienic response in the colony.
In honeybees, hygienic behavior means the detection and removal of diseased, parasitized, or otherwise nonvital brood from the brood nest by nurse bees. This behavior helps to remove brood pathogens from the colony, and therefore, it is considered a part of the immune response of honeybees (5, 14, 15, 18, 19, 21, 22). In the case of AFB, the removal of diseased larvae by nurse bees prior to the conversion of the larval biomass into infectious spores efficiently disturbs spore production within the colony, leading to impaired disease transmission and disease development within the colony. The more AFB-infected larvae become moribund or even die before cell capping, the more larvae will be removed by nurse bees as part of the immune defense and the less ropy mass and fewer spore-containing foulbrood scales will be produced (Table 1). A low level of spore production will result in slow spreading within the colony, which in turn will result in a slow collapse of the colony, as opposed to high-level spore production resulting in fast within-colony spreading and in a rather fast colony collapse (Table 1). Given this line of thinking, the time to larval death and the virulence on the larval level are negatively correlated to the time to colony death and the virulence on the colony level. Therefore, we conclude that the faster a P. larvae isolate is killing infected larvae, the less virulent it will be on the colony level. This relationship may be the reason why the genotypes ERIC III and ERIC IV could not establish themselves in the bee population but obviously went extinct: these extremely fast-killing strains (9) may have caused the premature death of the infected larvae, allowing a too-efficient social immune response and thereby impairing disease transmission. Hence, these two genotypes may represent an evolutionary blind alley for P. larvae, where the evolution of P. larvae virulence on the larval level came to its limitation on the colony level.
In conclusion, we herein provide experimental evidence that the faster a P. larvae strain or genotype is killing infected larvae, the better these larvae will be removed from the colony by nurse bees, the fewer spores will be produced and remain within the colony, and the more slowly the disease will progress in the colony, resulting in a negative correlation between the larva-level virulence and the colony-level virulence of P. larvae.
Acknowledgments
We thank Dominique Yue for technical assistance with colony infection assays.
This work was supported by grants from the Ministries for Agriculture from Brandenburg and Sachsen-Anhalt, Germany, and through the German Research Foundation (DFG, Graduiertenkolleg 1121).
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Alippi, A. M., F. J. Reynaldi, A. C. Lopez, M. R. De Giusti, and O. M. Aguilar. 2004. Molecular epidemiology of Paenibacillus larvae larvae and incidence of American foulbrood in Argentinean honeys from Buenos Aires province. J. Apic. Res. 43:135-143. [Google Scholar]
- 2.Antunez, K., C. Piccini, S. Castro-Sowinski, A. S. Rosado, L. Seldin, and P. Zunino. 2007. Phenotypic and genotypic characterization of Paenibacillus larvae isolates. Vet. Microbiol. 124:178-183. [DOI] [PubMed] [Google Scholar]
- 3.Ashiralieva, A., and E. Genersch. 2006. Reclassification, genotypes, and virulence of Paenibacillus larvae, the etiological agent of American foulbrood in honeybees—a review. Apidologie 37:411-420. [Google Scholar]
- 4.Brodsgaard, C. J., H. Hansen, and W. Ritter. 2000. Progress of Paenibacillus larvae larvae infection in individually inoculated honey bee larvae reared singly in vitro, in micro colonies, or in full-size colonies. J. Apic. Res. 39:19-27. [Google Scholar]
- 5.Chen, Y.-W., C.-H. Wang, J. An, and K.-K. Ho. 2000. Susceptibility of the Asian honey bee, Apis cerana, to American foulbrood, Paenibacillus larvae larvae. J. Apic. Res. 39:169-175. [Google Scholar]
- 6.Ebert, D., and W. W. Weisser. 1997. Obligate killing for obligate killers: the evolution of life histories and virulence of semelparous parasites. Proc. R. Soc. Lond. Ser. B 264:965-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genersch, E. 2007. Paenibacillus larvae and American foulbrood in honeybees. Berl. Münch. Tierärztl. Wochenschr. 120:26-33. [PubMed] [Google Scholar]
- 8.Genersch, E., A. Ashiralieva, and I. Fries. 2005. Strain- and genotype-specific differences in virulence of Paenibacillus larvae subsp. larvae, a bacterial pathogen causing American foulbrood disease in honeybees. Appl. Environ. Microbiol. 71:7551-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genersch, E., E. Forsgren, J. Pentikäinen, A. Ashiralieva, S. Rauch, J. Kilwinski, and I. Fries. 2006. Reclassification of Paenibacillus larvae subsp. pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without subspecies differentiation. Int. J. Syst. Evol. Microbiol. 56:501-511. [DOI] [PubMed] [Google Scholar]
- 10.Genersch, E., and C. Otten. 2003. The use of repetitive element PCR fingerprinting (rep-PCR) for genetic subtyping of German field isolates of Paenibacillus larvae subsp. larvae. Apidologie 34:195-206. [Google Scholar]
- 11.Lindström, A., S. Korpela, and I. Fries. 2008. The distribution of Paenibacillus larvae spores in adult bees and honey and larval mortality, following the addition of American foulbrood diseased brood or spore-contaminated honey in honey bee (Apis mellifera) colonies. J. Invertebr. Pathol. 99:82-86. [DOI] [PubMed] [Google Scholar]
- 12.Lindström, A., S. Korpela, and I. Fries. 2008. Horizontal transmission of Paenibacillus larvae spores between honey bee (Apis mellifera) colonies through robbing. Apidologie 39:1-8. [Google Scholar]
- 13.Loncaric, I., I. Derakhshifar, J. T. Oberlerchner, H. Köglberger, and R. Moosbeckhofer. 2009. Genetic diversity among isolates of Paenibacillus larvae from Austria. J. Invertebr. Pathol. 100:44-46. [DOI] [PubMed] [Google Scholar]
- 14.Park, O. W. 1937. Testing for resistance to American foulbrood in honeybees. J. Econ. Entomol. 30:504-512. [Google Scholar]
- 15.Park, O. W., F. C. Pellet, and F. B. Paddock. 1937. Disease resistance and American foulbrood. Am. Bee J. 77:20-25. [Google Scholar]
- 16.Peters, M., J. Kilwinski, A. Beringhoff, D. Reckling, and E. Genersch. 2006. American foulbrood of the honey bee: occurrence and distribution of different genotypes of Paenibacillus larvae in the administrative district of Arnsberg (North Rhine-Westphalia). J. Vet. Med. B 53:100-104. [DOI] [PubMed] [Google Scholar]
- 17.Shimanuki, H., and D. A. Knox. 1994. Susceptibility of Bacillus larvae to terramycin. Am. Bee J. 134:125-126. [Google Scholar]
- 18.Spivak, M. S., and M. Gilliam. 1998. Hygienic behaviour of honey bees and its application for control of brood diseases and varroa. Part II. Studies on the hygienic behaviour since the Rothenbuhler era. Bee World 79:169-186. [Google Scholar]
- 19.Spivak, M. S., and G. S. Reuter. 2001. Resistance to American foulbrood disease by honey bee colonies Apis mellifera bred for hygienic behavior. Apidologie 32:555-565. [Google Scholar]
- 20.Versalovic, J., M. Schneider, F. J. de Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 21.Wilson-Rich, N., M. S. Spivak, N. H. Fefferman, and P. T. Starks. 2009. Genetic, individual, and group facilitation of disease resistance in insect societies. Annu. Rev. Entomol. 54:405-423. [DOI] [PubMed] [Google Scholar]
- 22.Woodrow, A. W., and E. C. Holst. 1942. The mechanism of colony resistance to American foulbrood. J. Econ. Entomol. 35:327-330. [Google Scholar]
- 23.Yue, D., M. Nordhoff, L. H. Wieler, and E. Genersch. 2008. Fluorescence in situ-hybridization (FISH) analysis of the interactions between honeybee larvae and Paenibacillus larvae, the causative agent of American foulbrood of honeybees (Apis mellifera). Environ. Microbiol. 10:1612-1620. [DOI] [PubMed] [Google Scholar]



