Abstract
Bifidobacteria are normal inhabitants of the human gut. Some strains of this genus are considered health promoting or probiotic, being included in numerous food products. In order to exert their health benefits, these bacteria must overcome biological barriers, including bile salts, to colonize and survive in specific parts of the intestinal tract. The role of multidrug resistance (MDR) transporters in bile resistance of probiotic bacteria and the effect of bile on probiotic gene expression are not fully understood. In the present study, the effect of subinhibitory concentrations of bile on the expression levels of predicted MDR genes from three different bifidobacterial strains, belonging to Bifidobacterium longum subsp. longum, Bifidobacterium breve, and Bifidobacterium animalis subsp. lactis, was tested. In this way, two putative MDR genes whose expression was induced by bile, BL0920 from B. longum and its homolog, Bbr0838, from B. breve, were identified. The expression of the BL0920 gene in Escherichia coli was shown to confer resistance to bile, likely to be mediated by active efflux from the cells. To the best of our knowledge, this represents the first identified bifidobacterial bile efflux pump whose expression is induced by bile.
In the gut, commensal bacteria, including those that have health-promoting or probiotic activity, are challenged by the presence of several toxic compounds of intestinal origin, such as bile salts. Bile is secreted into the duodenum to an estimated concentration of 0.2 to 2% for a given bile salt (14). Bile salts are detergent-like compounds with strong antimicrobial activity (2). Therefore, intestinal microorganisms have developed strategies to tolerate physiological concentrations of bile salts during passage through, or colonization of, the gut.
Bifidobacteria are common inhabitants of the human gastrointestinal tract (GIT) (19, 39). Some strains of this genus are known to have probiotic activity and are used as functional ingredients in food products around the world (40). The health-promoting effects attributed to these microorganisms are numerous (20, 42). To exert beneficial actions, these bacteria must overcome biological barriers including acid in the stomach and bile in the intestine in order to, at least temporarily, colonize specific parts of the GIT. Thus, understanding the mechanisms of resistance of a given commensal or probiotic microorganism to toxic substances, such as bile salts, is important in the context of its physiology in the GIT. Although bifidobacterial bile tolerance mechanisms are currently poorly understood, bifidobacteria with increased resistance to bile salts have been obtained by progressive adaptation to gradually increasing concentrations of these compounds (27, 29). Through the analysis of such bile-tolerant derivatives, it was shown that the acquisition of bile resistance coincides with changes in membrane protein profiles (27) and carbohydrate metabolism (35, 38), while also conferring cross-resistance to other environmental stresses (29, 36). This indicates that the bifidobacterial response to bile entails a complex cellular activation/repression process, which impacts on general metabolic pathways (37).
Specific bile resistance mechanisms have been described in intestinal bacteria, with bile efflux and bile salt hydrolysis being the most prevalent (31). In this respect, multidrug resistance (MDR) transporters seem to play a crucial role in conferring a bile resistance phenotype. MDR proteins are present in all organisms and frequently confer resistance against several structurally unrelated toxic compounds (31). Research on these transporters has mainly been focused on their role in antibiotic resistance. However, given their ubiquitous presence this does not seem to be their primary function. In fact, recent studies support a role for various MDR transporters in allowing microorganisms to survive, establish, and persist in their (human) host (31).
It has been shown elsewhere that MDR proteins confer resistance to bile in different enteric bacteria (32). Bile was found to upregulate the expression of the multidrug efflux system cmeABC in Campylobacter jejuni (22), and inhibition of this pump was found to reduce the colonization ability of the microorganism by reducing bile resistance (21). In vitro and in vivo induction of acrAB expression by bile was observed in Vibrio cholerae (6), and also other MDR proteins appear to be induced in this microorganism (5). In the intestinal bacterium Bacteroides fragilis, the expression of different MDR pumps was also found to be upregulated by bile (34).
Several MDR systems have been identified in gram-positive bacteria (including probiotic bacteria) (8, 28), but their role in conferring resistance to intestinal toxic compounds, such as bile salts, and the effect of bile on gene expression have not received much scientific attention. Transporters able to extrude bile salts have been found in gram-positive bacteria such as Lactococcus lactis (44, 45) and Lactobacillus johnsonii (7). In Lactobacillus plantarum a membrane protein whose expression is induced by bile, both in vitro and in vivo, was previously identified (3). Proteins conferring resistance to bile, and whose expression is induced by it, have also been reported in other lactobacilli (30, 43). Just a couple of studies have been published on MDR transporters in bifidobacteria. Margolles and coworkers identified and characterized two MDR transporters from Bifidobacterium breve, BbmAB and BbmR, conferring resistance to antimicrobials (25, 26). In Bifidobacterium longum an MDR transporter, Ctr, was found to export cholate from the cell, conferring resistance to this compound when cloned in a heterologous bacterial host (33). However, there is still a knowledge gap with regard to possible effects of bile on MDR gene expression and the potential role of MDR transporters in bile resistance in bifidobacteria.
In the present study, the effect of subinhibitory concentrations of bile on the expression levels of genes encoding bifidobacterial MDR protein homologs was tested. For this purpose, known or putative MDR genes were selected from the genomes of different Bifidobacterium strains belonging to the species B. longum, B. breve, and Bifidobacterium animalis. A putative MDR-encoding gene, present as a homolog in both B. breve and B. longum and whose expression was strongly induced by bile, was identified, and the B. longum gene was then characterized.
MATERIALS AND METHODS
Strains and growth conditions.
The bifidobacterial strains Bifidobacterium longum subsp. longum NCC2705, B. breve UCC2003, and Bifidobacterium animalis subsp. lactis BB-12 (BB-12 is a registered trademark of Chr. Hansen A/S) were selected for this study. Subinhibitory concentrations of bile (oxgall; Sigma, St. Louis, MO) were determined by adding various concentrations of bile to DeMan, Rogosa, Sharpe broth (Difco, Becton, Dickinson and Company, Le Pont de Claix, France) supplemented with 0.25% cysteine-HCl (Sigma) followed by inoculation (0.1%, vol/vol) with the corresponding bacterial culture. Anaerobic incubation was carried out at 37°C in a Mac 500 chamber (Don Whitley Scientific, West Yorkshire, United Kingdom), and the optical density at 600 nm (OD600) was monitored at different time points.
MDR genes selected and primers used.
A total of 13, 16, and 4 putative MDR-encoding genes were identified on the genomes of B. longum NCC2705, B. breve UCC2003, and B. animalis subsp. lactis BB-12, respectively, and selected for further analysis. Primers targeting those genes were designed (Table 1), and their specificity was tested against the corresponding bifidobacterial strains.
TABLE 1.
Oligonucleotides used for expression studies of predicted MDR-encoding genes
| Organism | Primer name | Sequence, 5′-3′ |
|---|---|---|
| B. longum | BL0332F | TTCATCATCACCGCCATCTG |
| BL0332R | CACTTGGCGGTAGCTGGTAAG | |
| BL1102F | CCACGATGACAAGCACGAGAT | |
| BL1102R | CCGCTGAAACGGTGCTCTT | |
| BL0179F | GCGATACGGCTACCAGATGTG | |
| BL0179R | CGGAGACGAGGCGGTAGAC | |
| BL0037F | ATCGGCATGAAGTGCATTGAG | |
| BL0037R | CCAAGCTGAATCGGGTCAAGA | |
| BL0919F | GAAGAAGAGCGCGGAACATG | |
| BL0919R | GCGGGTTGCCCGCTATT | |
| BL0920F | GCCGCCTCCCTGTTCAA | |
| BL0920R | TAGGGCTGATCGTCCATGATTC | |
| BL1369F | GACGACCGCAGCGTCAA | |
| BL1369R | GCGGGTCAGGTTGTTTTCG | |
| BL0759F | GGGCCCTGACCAAAAGGA | |
| BL0759R | CGCAGGCGATGTTCACAAT | |
| BL0842F | GCGCCACTGACGACACTTG | |
| BL0842R | GGCGAGCCGACGATGAT | |
| BL1082F | CGGCCGTCATCTCCAACTT | |
| BL1082R | CCGAGCTGACCAATAAAGAACGT | |
| BL0251F | GGCTTTGGCGGGCTTATGT | |
| BL0251R | ACCGGTGCCGCGAATC | |
| BL0681F | GCGGTACCTTGTCCGACAGA | |
| BL0681R | GCCGATTTCGCCGATGA | |
| BL0162F | TGCCCGACTATATGTCAGACATCA | |
| BL0162R | CCGCGATCCAGATGTCCTT | |
| B. breve | Bbr0244F | CGCCTATACACGCGCCATT |
| Bbr0244R | TTCGCTGCGTATACCAGCAA | |
| Bbr0428F | TGCTTATGCCGATTACCACATG | |
| Bbr0428R | CGATCCTGCAGACACCTCATC | |
| Bbr0619F | CCTGCGTATGCGGTTCGT | |
| Bbr0619R | GCCGACTGGCCTTGGTATC | |
| Bbr0821F | TCCGGCCACTTCTGCTGAT | |
| Bbr0821R | CCCGTCCATTCCCCAGATAC | |
| Bbr0838F | CGCCCTGATCATCTGCATCT | |
| Bbr0838R | TTGCGTGCGGCCTTCTC | |
| Bbr0859F | TCAGGCGTTCTCGCTGTTG | |
| Bbr0859R | GCAATACGATCCACCACCAGAT | |
| Bbr0957F | TGAGACGTGACTTCGGTTCGT | |
| Bbr0957R | CGAGCGGTGCGGCTATC | |
| Bbr1169F | GCTGGGCCAGCCTATTCTATG | |
| Bbr1169R | TGGTCAGGAAGCCGGAAAG | |
| Bbr1334F | GCTCAAATGCATCGAGCAAAA | |
| Bbr1334R | CCAACTGGACCGGATCAAGA | |
| Bbr1421F | CCCCGAAGATGCTCACCAA | |
| Bbr1421R | GCTGAAGAAGTAGCCGACAAAGAT | |
| Bbr1489F | GTCACCTCGCTCTACAACATCGT | |
| Bbr1489R | GAAGATCACTGTACCGACTTGAAACA | |
| Bbr1826F | CGCGAGAACATCGCCTACTC | |
| Bbr1826R | TTGCATGCGCTGATGATCTG | |
| Bbr1881F | GCCTCCCAAGCGTCAACAG | |
| Bbr1881R | CCGATCGCATACCCGAAATT | |
| BbmA(A1)F | ACGACTGGCAGCGATTCAAC | |
| BbmA(A1)R | GACCTGCTCGACGATTGTGA | |
| BbmB(A2)F | TTCTCCGACATCTTCACATTCT | |
| BbmB(A2)R | CAGTACGAGGCTCCACAATGC | |
| BbmRF | GAAGTCGGCATGCAGGATTC | |
| BbmRR | CGGGTTGGAGGCAAAGGA | |
| B. animalis | RDBD02062F | GTTCGGGTCAACGTACATCGA |
| subsp. lactis | RDBD02062R | GCCGCCCAGACATGCAT |
| RDBD00308F | CGACACAGGGCGGAAAGAA | |
| RDBD00308R | GGAAATGCGATCGTCATTCG | |
| RDBD00009F | CTTCGGCCTGCATAATTTCG | |
| RDBD00009R | GAAAGACCCACCGAGAAGAAGTC | |
| RDBD01274F | CGCGAATACGACTTGAAGACACT | |
| RDBD01274R | TCGACGGTGCCTTTGAACAG |
Expression studies of predicted MDR-encoding genes.
Cultures of the three selected bifidobacterial strains were grown in the presence of the corresponding subinhibitory concentration of bile until they reached an OD600 of 1.8 ± 0.2. Culture medium without bile was used as a control. Incubation times needed to obtain the desired ODs ranged between 7 and 9 h depending on the strain. Once the cultures reached the desired OD, a 500-μl aliquot was removed and resuspended in 1 ml of bacterial RNA Protect reagent (Qiagen, Hilden, Germany). Bacterial pellets were then frozen until RNA extraction.
RNA was extracted using the RNA Protect bacteria and RNeasy minikit (Qiagen) according to the manufacturer's instructions, with the following modifications: the lysis buffer was supplemented with 30 μg/ml lysozyme and 100 U/ml mutanolysin (Sigma), and the samples were incubated for 30 min with gentle shaking. RNase-free DNase I (Qiagen) was used to eliminate any residual DNA. Following this, 3 μg of RNA was reverse transcribed into single-stranded cDNA using the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The generated cDNA was then stored at −80°C until further analysis.
Real-time PCR was performed in an ABI Prism 7500 Fast real-time PCR system (Applied Biosystems). Amplification was carried out in a 25-μl final volume, which contained 1 μl of cDNA as a template, 200 nM of each primer, and 12.5 μl of SYBR green PCR Master Mix (Applied Biosystems). Thermal cycling consisted of an initial cycle at 95°C for 10 min followed by 35 cycles at 95°C for 15 s and 60°C for 1 min, followed by melting-curve analysis. Expression levels were determined by relative quantification using the threshold cycle method in which the expression level in the control culture (without bile) is arbitrarily set to 1 and the expression levels in the samples are calculated relative to that control. The 16S rRNA was used as an endogenous control using previously described primers (13). All experiments were carried out at least in duplicate.
DNA and protein sequences were analyzed using the computer program Clone Manager 5 (Scientific and Educational Software). Homology searches and multiple sequence alignments (clustalw) were carried out using the BLAST server of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and the software available on the web page of the Pôle-Bioinformatique Lyonnais (http://pbil.univ-lyon1.fr/).
Cloning of the BL0920 gene in Escherichia coli.
To characterize the potential role of the BL0920 gene in bile resistance, the coding region of this gene was PCR amplified with primers f-GTGACCTCACAGAAAAACGCG and r-TCACCGATCATCCTTTGGG and ligated into pETblue-1 vector according to the instructions of the manufacturer (Novagen, Darmstadt, Germany). The ligation mix was introduced into NovaBlue competent cells by transformation using the Perfectly Blunt cloning kit (Novagen) allowing white/blue screening of transformants. Plasmid DNA was extracted from a positive NovaBlue transformant and checked for its expected DNA content. This plasmid, designated pETBL0920, was introduced into Tuner(DE3)pLacI competent E. coli cells (tDE3 cells) and used for induced expression of the BL0920 gene according to the recommended protocol of the manufacturer (Novagen).
Evaluation of survival in the presence of bile.
To test the effect of BL0920 gene expression on bile resistance, an overnight culture of tDE3 cells carrying pETBL0920 was used to inoculate (1%) LB broth supplemented with 50 μg/ml ampicillin, 34 μg/ml chloramphenicol, and 0.5% glucose. Cells were incubated at 37°C with constant stirring until an OD600 of 0.3 was reached. The culture was then divided into three aliquots, and IPTG (isopropyl-β-d-thiogalactopyranoside; inducer) was added at a final concentration of 0 (control), 50, or 100 μM, respectively. Incubation was then continued for 120 min to allow expression of the BL0920 gene. Cells from each of these three test cultures were harvested by centrifugation, washed twice with phosphate-buffered saline, and resuspended in double the original volume of LB containing antibiotics, 3% bile, and the corresponding concentration of inducer (IPTG; 0, 50, or 100 μM). Cultures were incubated at 37°C, and samples were taken at 0, 90, and 180 min to determine survival in the presence of bile by plate counting on LB medium. In addition, the effect of these various inducing conditions on the expression level of the gene BL0920 was determined by real-time reverse transcription-PCR (RT-PCR) using the primers BL0920F and BL0920R (Table 1). The rrsA gene was used as an internal control (16).
Transport of fluorescent bile salts by BL0920-expressing cells.
To evaluate the role of BL0920 in bile extrusion, we used fluorescently tagged ursodeoxycholic acid (UDCA) (15). In short, tDE3 cells carrying empty pETblue-1 or the plasmid containing the BL0920 gene (pETBL0920) were grown in LB broth, supplemented with antibiotics and glucose as indicated above, to an OD600 of 0.4. The cultures were then induced by adding IPTG to a final concentration of 100 μM and incubated at 37°C for 90 min. Following this induction period, cultures were harvested, washed with 50 mM KPi (pH 7.5) buffer, and resuspended in the same buffer to an OD600 of 14. To these resuspended cultures, 100 μM (final concentration) of fluorescent UDCA was added, which was then followed by incubation at 37°C for 70 min in order to load the cells with UDCA. After this latter incubation, cells were washed twice and resuspended in 50 mM KPi (pH 7.5) containing glucose (10 mM). The suspension was incubated at 37°C for 5 min, and 1-ml samples of this suspension were taken at 0 and 5 min. The cells in these samples were harvested by centrifugation, and the cell pellet was resuspended in 300 μl of the same buffer. The fluorescence of cell suspensions and supernatants was then determined (absorbance, 494 nm; emission, 521) in a Cary Eclipse fluorescence spectrophotometer (Varian Iberica SA, Madrid, Spain).
Nucleotide sequence accession numbers.
The accession numbers of the BL0920 and Bbr0838 genes are NP_696096 and FJ660413, respectively.
RESULTS
Effect of bile on the expression of known and putative MDR genes.
We determined subinhibitory (i.e., not affecting growth rate) concentrations of bile for the different bifidobacterial strains. Bile concentrations of 0.1, 0.08, and 0.05% were found to be subinhibitory for B. longum, B. breve, and B. animalis, respectively.
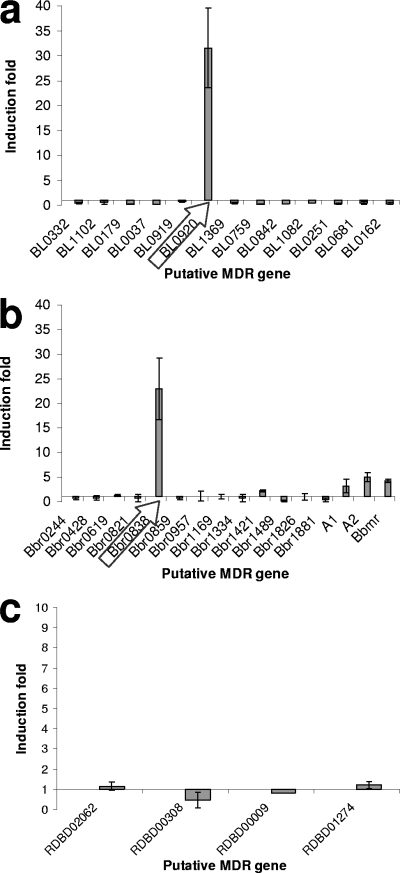
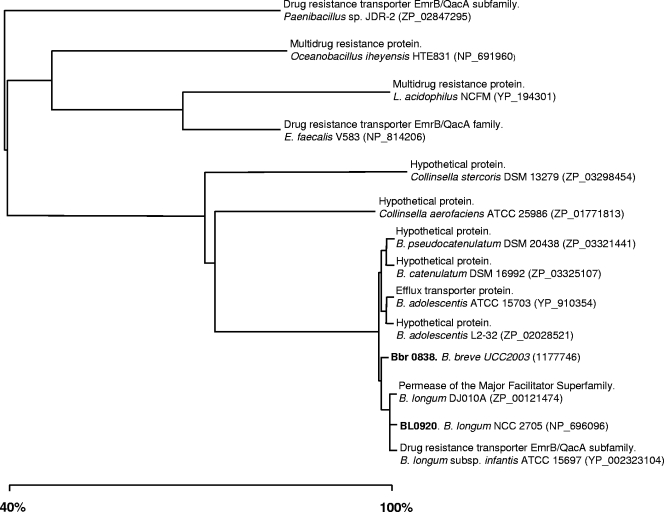
The effect of these bile concentrations on the expression levels of the selected MDR-encoding genes was tested using RT-PCR. All the designed primers (Table 1) used for the RT-PCR experiments were found to be specific, producing a single amplification product in each case (data not shown). The RT-PCR results allowed us to identify two open reading frames (ORFs) whose expression was very strongly upregulated by bile. The expression of the BL0920 gene from B. longum NCC2705 and the Bbr0838 gene from B. breve UCC2003 was induced more than 20 times in the presence of bile salts (Fig. 1). Interestingly, the deduced protein products of the BL0920 and Bbr0838 genes display a very high sequence similarity (>99% amino acid identity) (Fig. 2) and are thus assumed to be homologs of each other. For this reason, the remainder of this work focused on the characterization of the BL0920 gene-encoded protein.
FIG. 1.
Effect of subinhibitory concentrations of bile (0.1, 0.08, and 0.05%, respectively) on the transcription of putative MDR-encoding genes in B. longum subsp. longum NCC2705 (a), B. breve UCC2003 (b), and B. animalis subsp. lactis BB-12 (c) relative to the transcription of such genes in the absence of bile. The horizontal axis is placed at 1, which is the ratio reflecting the change in the expression level.
FIG. 2.
Phylogenetic relationship analysis of the permease domains of BL0920, Bbr0838, and other highly similar putative MDR proteins. The tree was constructed using the fast minimum evolution method and the Grishin distance model (12). Database accession numbers are given in parentheses.
The BL0920 gene product is a putative 683-amino-acid membrane protein of the major facilitator superfamily (4). A topological analysis at the Expasy proteomic server (10) predicted 12 to 14 transmembrane segments and a large C-terminal loop containing a cystathionine-beta-synthase (CBS) domain (Pfam PF00571). CBS domains, which bind ligands containing adenosyl groups, are known to play an important regulatory functional role in certain bacterial transporters (24). The predicted permease domain (represented by 471 amino acids at the N terminus of the protein) was subjected to BlastP analysis (1). Results indicate a very high similarity (>95% at amino acid level) to putative efflux transporters from B. longum subsp. longum, Bifidobacterium longum subsp. infantis, B. breve, Bifidobacterium adolescentis, Bifidobacterium catenulatum, and Bifidobacterium pseudocatenulatum (Fig. 2), species that are natural inhabitants of the intestinal tract of humans and other mammals. Interestingly, Bifidobacterium dentium and B. animalis subsp. lactis, which are not considered to be autochthonous members of the intestinal microbiota, do not possess BL0920 gene homologs. These two species, however, do possess other means of conferring bile resistance, namely, the bile salt hydrolase, which is constitutively expressed in B. animalis subsp. lactis BB-12 (9). Remarkably, the majority of the presumed BL0920 gene homologs found in genomes from other species belong to bacteria, such as Collinsella, Lactobacillus, or Enterococcus, which have frequently been associated with the intestine.
Interestingly, the expression of the previously characterized B. breve UCC2003 MDR transporters, BbmA/BbmB and BbmR, was found to be induced to a much lower degree in the presence of bile salts (3.21 ± 1.3 for bbmA, 5.00 ± 0.9 for bbmB, and 4.23 ± 0.3 for bbmR). None of the other putative B. longum or B. breve MDR-encoding genes was found to be significantly upregulated by bile, while in B. animalis subsp. lactis no induction was observed for any of the genes tested.
Role of BL0920 in bile tolerance.
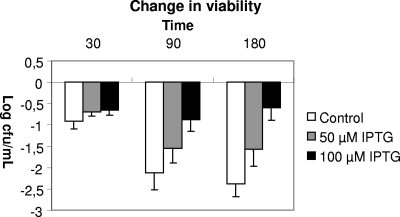
IPTG-induced cultures of E. coli tDE3 containing plasmid pETBL0920 showed an improved survival during exposure to 3% bile compared to uninduced cultures (Fig. 3). The highest concentration of inducer (IPTG) used resulted in the highest level of survival under conditions of bile exposure. The transcription levels of the BL0920 gene under the three different induction conditions used (0, 50, or 100 μM IPTG) were determined by RT-PCR. The results showed that the expression of the BL0920 gene was induced 1.6 and 2.5 times with 50 and 100 μM IPTG, respectively, relative to the expression of this gene in the noninduced culture. Rather high levels of BL0920 gene transcription (when present in pETBL0920) were also detected in the absence of inducer (0 μM IPTG), probably due to “leakage” of the expression system used. It is worthwhile to remark that E. coli cells carrying the BL0920 gene in plasmid pETBL0920, even under uninduced conditions, exhibit a significantly reduced growth rate, suggesting that the production of the BL0920 protein is toxic for the cells.
FIG. 3.
Bile-mediated effects on the culture viability following 30, 90, and 180 min of exposure to medium containing 3% bile. Results were calculated as change in viability relative to culture viability immediately prior to the addition of bile.
Extrusion of bile salts mediated by BL0920.
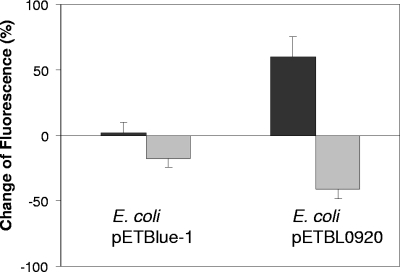
Fluorescence levels in tDE cells expressing the BL0920 gene were significantly lower than those obtained with the cells carrying the empty vector (pETblue-1), indicating that UDCA had been exported from cells to a much higher degree in the former strain (Fig. 4). These results were corroborated by those of the culture supernatants in which fluorescence levels were significantly higher for supernatants of tDE3 cells carrying pETBL0920 than for the control strain. These data show that the strain expressing BL0920 actively expels UDCA, which consequently accumulates in the supernatant, thereby substantiating the role of BL0920 in bile resistance by exporting the compound.
FIG. 4.
Fluorescence levels in cells and supernatants of tDE3 cells carrying the empty pETblue-1 plasmid or the same plasmid containing the BL0920 gene (plasmid pETBL0920), after cells were loaded with fluorescent UDCA followed by 5 min of incubation (37°C) in the presence of glucose. Dark gray bars represent the fluorescence in the supernatants, and light gray bars represent the fluorescence in the cells. Values are expressed as percent change relative to the fluorescence values at time zero.
DISCUSSION
In the present study we identified the BL0920 gene from B. longum and its homolog in B. breve, whose expression is heavily upregulated by subinhibitory concentrations of bile. Given the possible role of these genes in bile resistance and the consequent impact on colonization ability, we decided to further characterize the BL0920 gene from B. longum NCC2705, which is the more highly induced of the two homologs, at least under our experimental conditions. The gene showed high homology with putative efflux transporters from a variety of other intestinal microorganisms. When the gene was cloned into E. coli, a stepwise induction of the BL0920 gene was found to result in increased survival of the resulting recombinant strain in the presence of high concentrations of bile, while it was also shown to be capable of exporting UDCA from the cell. These findings, together with the significant transcriptional upregulation of this gene observed in B. breve and B. longum, clearly point to a role of the gene in bile resistance/tolerance in bifidobacteria. Interestingly, Price et al. (33) previously characterized a cholate transporter, designated Ctr (BL1102) from B. longum, which was shown to confer resistance to cholate and other antimicrobials when cloned into E. coli. Our results show that the expression of ctr was not induced by bile, suggesting that its primary role may be different from bile resistance. Although we observed a significant increase of bile tolerance and export when the BL0920 gene was expressed in E. coli, it is important to underline that other MDR systems involved in bile resistance may be acting in E. coli (23). This makes it difficult to directly correlate the phenotype observed with the physiological values of resistance and export in bifidobacteria.
An approximately three- to fivefold induction of transcription of previously identified B. breve MDR protein-encoding genes, the BbmA/BbmB genes (A1 and A2, respectively) and the BbmR gene (25, 26), was also observed. Expression of the BbmR gene in Lactococcus lactis did not confer bile resistance (26), indicating that, at least in this heterologous host, the BbmR gene is insufficient on its own to confer bile resistance.
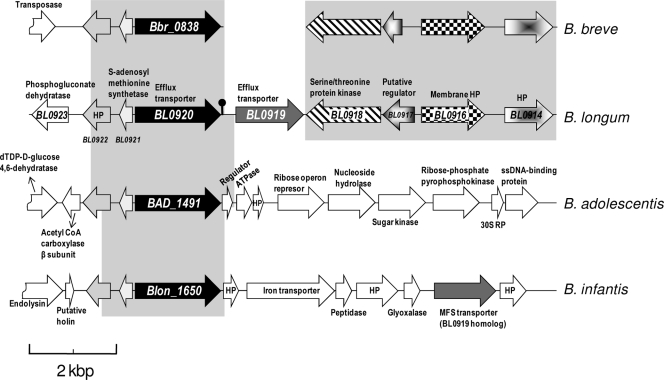
In the genome of B. longum NCC2705, the BL0920 gene is adjacent to a gene predicted to encode another putative efflux transporter, the BL0919 gene (Fig. 5). However, we did not observe transcriptional induction of the BL0919 gene in the presence of bile, suggesting that these two neighboring genes are subject to different transcriptional controls. In a previous study the BL0919 gene was found to be upregulated in galacto-oligosaccharide-containing medium compared to glucose medium, suggesting a different physiological role for this transporter (11). Downstream of the BL0919 and BL0920 genes, on the opposite strand, two genes are present specifying a putative two-component system, composed of a putative transcriptional regulator (BL0917) and a serine/threonine kinase with a transmembrane segment (BL0918). A recent publication has reported on a bile-inducible operon in Lactobacillus acidophilus, containing eight putative genes, encoding, among others, a major facilitator superfamily transporter involved in bile resistance and a two-component regulatory system (30). This underlines the potential role of two-component systems in regulating bile-induced transcription. However, although this may suggest an involvement of the two-component system in bile-mediated gene regulation in bifidobacteria, this has yet to be experimentally confirmed. The bioinformatic analysis of the sequences surrounding the BL0920 gene and comparison of the BL0920 gene sequence with the corresponding DNA regions in the genomes of B. breve, B. longum subsp. infantis, and B. adolescentis showed that (i) there is a common region (>95% homology) of about 3 kb within the four genomes, which includes the BL0920 gene (or its homologs) and the two upstream genes (BL0921 and BL0922); (ii) the BL0919 gene is present only in B. longum subsp. longum and B. longum subsp. infantis, although in the second genome the BL0920 and BL0919 genes are not consecutive genes, and five putative ORFs are intercalated between them; and (iii) the regions downstream from the BL0919 gene are similar in B. breve and B. longum (>85% homology), but B. breve seems to have segregated the BL0919 gene. A strong terminator (dyad symmetry) was found just downstream from the BL0920 gene; thus, it seems that this transporter is transcribed independently of the rest of the adjacent genes.
FIG. 5.
Organization of the B. longum subsp. longum NCC2705 genomic region containing the BL0920 gene and its flanking sequences and the corresponding sequences in B. breve UCC2003, B. adolescentis ATCC 15703, and B. longum subsp. infantis ATCC 15697. The pin-like symbol indicates a Rho-independent transcription terminator. The arrows indicate the relative positions and direction of transcription of the ORFs. White arrows indicate ORFs without significant homology. Arrows with the same filling pattern are considered to encode homologous proteins. Gray-shaded areas indicate DNA homologous sequences. HP, hypothetical protein; MFS, major facilitator superfamily; ssDNA, single-stranded DNA; CoA, coenzyme A.
The low bile concentrations used in this study are similar to those present in the colon, which is the normal habitat of bifidobacteria (2, 41). Under these conditions the BL0920 gene may play an important role in bifidobacterial resistance to bile, allowing the survival of the strains harboring it and perhaps increasing their colonization ability. Bile-mediated transcriptional upregulation of the genes encoding MDR transporters, which then confer bile tolerance, has been demonstrated for different enteric microorganisms (6, 18, 22). Furthermore, some of these transporters have been shown to be implicated in pathogenicity and colonization ability (21, 34). In the gram-positive microorganism Lactococcus lactis the MDR transporter lmrCD was found to be upregulated by cholate, an important bile salt. This transporter exports cholate from the cells, thus being the major contributor to cholate resistance in L. lactis (45). Interestingly, a recent metagenomic study identified commonly enriched gene sets in the human gut microbiome. The authors reported the multidrug efflux pumps (COG 0534) to be among the gene families that appear to be enriched in this environment (17).
In summary, we identified a new bile transporter from B. longum NCC2705, BL0920, and its homolog in B. breve UCC2003. Expression of the BL0920 gene in E. coli conferred resistance to bile, likely mediated by active efflux from the cells. Therefore, we propose to rename the BL0920 gene (and its homolog the Bbr0838 gene in B. breve) as betA (bile efflux transporter). To the best of our knowledge, this is the first bile efflux pump reported in bifidobacteria whose expression is induced by bile. These observations suggest an important role of the gene in bile resistance, which may be of crucial importance for the intestinal colonization ability of bifidobacteria.
Acknowledgments
This work was supported by grant AGL2007-61805 from the Spanish Ministry of Education and Science and by a CSET grant to support the Alimentary Pharmabiotic Centre from Science Foundation Ireland. M.G. was the recipient of a Ramón y Cajal Contract from the Spanish Ministry of Science and Education (MEC).
We acknowledge the generous gift of the strain B. longum NCC2705 from Fabrizio Arigoni (Nestlé Research Centre, Lausanne, Switzerland). We thank Sinead Leahy and Mary O'Connell-Motherway for the provision of unpublished B. breve UCC2003 genome data.
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begley, M., C. G. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29:625-651. [DOI] [PubMed] [Google Scholar]
- 3.Bron, P. A., M. Marco, S. M. Hoffer, E. Van Mullekom, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 186:7829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busch, W., and M. H. Saier, Jr., for the International Union of Biochemistry and Molecular Biology (IUBMB). 2004. The IUBMB-endorsed transporter classification system. Mol. Biotechnol. 27:253-262. [DOI] [PubMed] [Google Scholar]
- 5.Cerda-Maira, F. A., C. S. Ringelberg, and R. K. Taylor. 2008. The bile response repressor BreR regulates expression of the Vibrio cholerae breAB efflux system operon. J. Bacteriol. 190:7441-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee, A., S. Chaudhuri, G. Saha, S. Gupta, and R. Chowdhury. 2004. Effect of bile on the cell surface permeability barrier and efflux system of Vibrio cholerae. J. Bacteriol. 186:6809-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elkins, C. A., and D. C. Savage. 2003. CbsT2 from Lactobacillus johnsonii 100-100 is a transport protein of the major facilitator superfamily that facilitates bile acid antiport. J. Mol. Microbiol. Biotechnol. 6:76-87. [DOI] [PubMed] [Google Scholar]
- 8.Florez, A. B., C. G. de los Reyes-Gavilán, A. Wind, B. Mayo, and A. Margolles. 2006. Ubiquity of multidrug resistance genes in Lactococcus lactis strains isolated between 1936 and 1995. FEMS Microbiol. Lett. 263:21-25. [DOI] [PubMed] [Google Scholar]
- 9.Garrigues, C., B. Stuer-Lauridsen, and E. Johansen. 2005. Characterisation of Bifidobacterium animalis subsp. lactis BB-12 and other probiotic bacteria using genomics, transcriptomics and proteomics. Aust. J. Dairy Technol. 60:84-92. [Google Scholar]
- 10.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González, R., E. S. Klaassens, E. Malinen, W. M. de Vos, and E. E. Vaughan. 2008. Differential transcriptional response of Bifidobacterium longum to human milk, formula milk, and galactooligosaccharide. Appl. Environ. Microbiol. 74:4686-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grishin, N. V. 1995. Estimation of the number of amino acid substitutions per site when the substitution rate varies among sites. J. Mol. Evol. 41:675-679. [DOI] [PubMed] [Google Scholar]
- 13.Gueimonde, M., S. Tölkkö, T. Korpimäki, and S. Salminen. 2004. New real-time quantitative PCR procedure for quantification of bifidobacteria in human fecal samples. Appl. Environ. Microbiol. 70:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofman, A. F. 1998. Bile secretion and the enterohepatic circulation of bile acids, p. 937-948. In M. Feldman, B. F. Scharschmidt, and M. H. Sleisenger (ed.), Gastrointestinal and liver disease. W. B. Saunders, Philadelphia, PA.
- 15.Jean-Louis, S., S. Akare, M. A. Ali, E. A. Mash, E. Meuillet, and J. D. Martinez. 2006. Deoxycholic acid induces intracellular signalling though membrane perturbations. J. Biol. Chem. 281:14948-14960. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, A., H. Hirakawa, T. Hirata, K. Nishino, and A. Yamaguchi. 2006. Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J. Bacteriol. 188:5693-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurokawa, K., T. Itoh, T. Kuwahara, K. Oshima, H. Toh, A. Toyoda, H. Takami, H. Morita, V. K. Sharma, T. P. Srivastava, T. D. Taylor, H. Noguchi, H. Mori, Y. Ogura, D. S. Ehrlich, K. Itoh, T. Takagi, Y. Sakari, T. Hayashi, and M. Hattori. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiome. DNA Res. 14:169-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacroix, F. J., A. Cloeckaert, O. Grepinet, C. Pinault, M. Y. Popoff, H. Waxin, and P. Pardon. 1996. Salmonella typhimurium acrB-like gene: identification and role in resistance to biliary salts and detergents and in murine infection. FEMS Microbiol. Lett. 135:161-167. [DOI] [PubMed] [Google Scholar]
- 19.Lay, C., L. Riggotier-Gois, K. Holmstrom, M. Rajilic, E. E. Vaughan, W. M. de Vos, M. D. Collins, R. Thiel, P. Namsolleck, M. Blaut, and J. Doré. 2005. Colonic microbiota signatures across five northern European countries. Appl. Environ. Microbiol. 71:4153-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leahy, S. C., D. G. Higgins, G. F. Fitzgerald, and D. van Sinderen. 2005. Getting better with bifidobacteria. J. Appl. Microbiol. 98:1303-1315. [DOI] [PubMed] [Google Scholar]
- 21.Lin, J., and A. Martinez. 2006. Effect of efflux pump inhibitors on bile resistance and in vivo colonization of Campylobacter jejuni. J. Antimicrob. Chemother. 58:966-972. [DOI] [PubMed] [Google Scholar]
- 22.Lin, J., C. Cagliero, B. Guo, Y. W. Barton, M. C. Maurel, S. Payot, and Q. Zhang. 2005. Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni. J. Bacteriol. 187:7417-7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode stress-induced efflux system of Escherichia coli. Mol. Microbiol. 14:45-55. [DOI] [PubMed] [Google Scholar]
- 24.Mahmood, N. A., E. Biemans-Oldehinkel, J. S. Patzlaff, G. K. Schuurman-Wolters, and B. Poolman. 2006. Ion specificity and ionic strength dependence of the osmoregulatory ABC transporter OpuA. J. Biol. Chem. 281:29830-29839. [DOI] [PubMed] [Google Scholar]
- 25.Margolles, A., A. B. Florez, J. A. Moreno, D. van Sinderen, and C. G. de los Reyes-Gavilán. 2006. Two membrane proteins from Bifidobacterium breve UCC2003 constitute an ABC-type multidrug transporter. Microbiology 152:3497-3505. [DOI] [PubMed] [Google Scholar]
- 26.Margolles, A., J. A. Moreno, D. van Sinderen, and C. G. de los Reyes-Gavilán. 2005. Macrolide resistance mediated by a Bifidobacterium breve membrane protein. Antimicrob. Agents Chemother. 49:4379-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margolles, A., L. García, B. Sánchez, M. Gueimonde, and C. G. de los Reyes-Gavilán. 2003. Characterisation of a Bifidobacterium strain with acquired resistance to cholate—a preliminary study. Int. J. Food Microbiol. 82:191-198. [DOI] [PubMed] [Google Scholar]
- 28.Mazurkiewicz, P., K. Sakamoto, G. J. Poelarends, and W. N. Konings. 2005. Multidrug transporters in lactic acid bacteria. Mini Rev. Med. Chem. 5:173-181. [DOI] [PubMed] [Google Scholar]
- 29.Noriega, L., M. Gueimonde, B. Sánchez, A. Margolles, and C. G. de los Reyes-Gavilán. 2004. Effect of the adaptation to high bile salts concentrations on glycosidic activity, survival at low pH and cross-resistance to bile salts in Bifidobacterium. Int. J. Food Microbiol. 94:79-86. [DOI] [PubMed] [Google Scholar]
- 30.Pfeiler, E. A., M. A. Azcarate-Peril, and T. R. Klaenhammer. 2007. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 189:4624-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piddock, L. J. V. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4:629-636. [DOI] [PubMed] [Google Scholar]
- 32.Piddock, L. J. V. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price, C. E., S. J. Reid, A. J. M. Driessen, and V. R. Abratt. 2006. The Bifidobacterium longum NCIMB 702259 ctr gene codes for a novel cholate transporter. Appl. Environ. Microbiol. 72:923-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pumbwe, L., C. A. Skilbeck, V. Nakano, M. J. Avila-Campos, R. M. F. Piazza, and H. M. Wexler. 2007. Bile salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb. Pathog. 43:78-87. [DOI] [PubMed] [Google Scholar]
- 35.Ruas-Madiedo, P., A. Hernández-Barranco, A. Margolles, and C. G. de los Reyes-Gavilán. 2005. A bile salt-resistant derivative of Bifidobacterium animalis has an altered fermentation pattern when grown on glucose and maltose. Appl. Environ. Microbiol. 71:6564-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sánchez, B., C. G. de los Reyes-Gavilán, and A. Margolles. 2006. The F1F0-ATPase of Bifidobacterium animalis is involved in bile tolerance. Environ. Microbiol. 8:1825-1833. [DOI] [PubMed] [Google Scholar]
- 37.Sánchez, B., L. Ruiz, C. G. de los Reyes-Gavilán, and A. Margolles. 2008. Proteomics of stress response in Bifidobacterium. Front. Biosci. 13:6905-6919. [DOI] [PubMed] [Google Scholar]
- 38.Sánchez, B., M.-C. Champomier-Vergès, B. Stuer-Lauridsen, P. Ruas-Madiedo, P. Anglade, F. Baraige, C. G. de los Reyes-Gavilán, E. Johansen, M. Zagorec, and A. Margolles. 2007. Adaptation and response of Bifidobacterium animalis subsp. lactis to bile: a proteomic and physiological approach. Appl. Environ. Microbiol. 73:6757-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satokari, R. M., E. E. Vaughan, A. D. L. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanton, C., R. P. Ross, G. F. Fitzgerald, and D. van Sinderen. 2005. Fermented functional foods based on probiotics and their biogenic metabolites. Curr. Opin. Biotechnol. 16:198-203. [DOI] [PubMed] [Google Scholar]
- 41.Thomas, L. A., M. J. Veysey, G. French, P. B. Hylemon, G. M. Murphy, and R. H. Dowling. 2001. Bile acid metabolism by fresh human colonic contents: a comparison of caecal versus faecal samples. Gut 49:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie van Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 43.Whitehead, K., J. Versalovic, S. Roos, and R. A. Britton. 2008. Genomic and genetic characterization of bile stress response of probiotic Lactobacillus reuteri ATCC 55730. Appl. Environ. Microbiol. 74:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokota, A., M. Veenstra, P. Kurdi, H. van Veen, and W. N. Konings. 2000. Cholate resistance in Lactococcus lactis is mediated by an ATP-dependent multispecific organic anion transporter. J. Bacteriol. 182:5196-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaidi, A. H., P. J. Bakkes, J. Lubelski, H. Agustiandari, O. P. Kuipers, and A. J. M. Driessen. 2008. The ABC-type multidrug resistance transporter LmrCD is responsible for an extrusion-based mechanism of bile acid resistance in Lactococcus lactis. J. Bacteriol. 190:7357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]