Abstract
A large number of Streptomyces bacteria with antifungal activity isolated from samples collected in the Trondheim fjord (Norway) were found to produce polyene compounds. Investigation of polyene-containing extracts revealed that most of the isolates produced the same compound, which had an atomic mass and UV spectrum corresponding to those of candicidin D. The morphological diversity of these isolates prompted us to speculate about the involvement of a mobile genetic element in dissemination of the candicidin biosynthesis gene cluster (can). Eight candicidin-producing isolates were analyzed by performing a 16S rRNA gene-based taxonomic analysis, pulsed-field gel electrophoresis, PCR, and Southern blot hybridization with can-specific probes. These analyses revealed that most of the isolates were related, although they were morphologically diverse, and that all of them contained can genes. The majority of the isolates studied contained large plasmids, and two can-specific probes hybridized to a 250-kb plasmid in one isolate. Incubation of the latter isolate at a high temperature resulted in loss of the can genes and candicidin production, while mating of the “cured” strain with a plasmid-containing donor restored candicidin production. The latter result suggested that the 250-kb plasmid contains the complete can gene cluster and could be responsible for conjugative transfer of this cluster to other streptomycetes.
Actinomycete bacteria, especially those belonging to the family Streptomycetaceae, are well-known producers of secondary metabolites with diverse biological activities. Representatives of the genus Streptomyces produce a variety of antibiotics with antibacterial, antifungal, and antitumor activities. The majority of antibiotic-producing streptomycetes have been isolated from terrestrial environments, while antibiotic-producing streptomycetes from the marine sources remain largely unexplored. Therefore, studies of streptomycetes from the marine environment are important for unraveling their potential for antibiotic production. In addition, such studies might reveal the means by which antibiotic biosynthesis and resistance genes are spread in nature.
It is widely acknowledged that plasmids play an important role in genetic exchange between bacterial species. Conjugative plasmids are quite common in Streptomyces strains (13), and a number of these mobile genetic elements have been characterized in detail. The characterized mobile genetic elements include both circular plasmids, such as pIJ101 from Streptomyces lividans (14) and SCP2 from Streptomyces coelicolor (2, 35), and linear plasmids, such as SLP2 from S. lividans (6) and SCP1 from S. coelicolor (38, 39). The presence of a linear plasmid in Streptomyces was first reported in 1979, and the plasmid was the 17-kb pSLA2 plasmid of Streptomyces rochei (11). SCP1 of S. coelicolor was discovered in the early 1970s (38, 39), but because of its large size (356 kb), isolation of this plasmid with conventional techniques was not possible and therefore it was not recognized as a linear plasmid until pulsed-field gel electrophoresis (PFGE) was invented. Later, SCP1 was shown to harbor a complete set of genes for biosynthesis of the antibiotic methylenomycin (21; K. F. Chater, C. J. Bruton, S. J. O'Rouke, and A. W. Wietzorrek, 5 July 2001, Patent Cooperation Treaty international application WO/2001/048228), while another linear plasmid, found in S. rochei, has been shown to contain genes for biosynthesis of both lankamycin and lankacidin (16, 19, 28, 36). Other examples of plasmids include pPZG103 carrying oxytetracycline biosynthesis genes acquired from the chromosome of Streptomyces rimosus (10) and pKSL from Streptomyces lasaliensis, which might be involved in the production of lasalocid and/or echinomycin (17, 20).
Linear plasmids can be transferred between Streptomyces strains by means of conjugation, and SCP1 is an example of a conjugative linear plasmid as it is easily transferred from an SCP1+ strain to an SCP1− strain (39). Interspecific transfer to S. lividans and Streptomyces parvulus has also been reported for this plasmid, and it was demonstrated that the recipient strains had acquired the ability to produce and be resistant to methylenomycin (12, 21). Transfer of intact linear plasmids containing mercury resistance genes from two Streptomyces strains isolated from the marine environment to S. lividans, conferring mercury resistance to the initially mercury-sensitive recipient, has been reported by Ravel et al. (32). It has also been shown that interspecific transfer of linear plasmids is possible in sterile amended soil microcosms, suggesting that mercury resistance might be spread by plasmid transfer in polluted environments (31).
We report here isolation and screening of several thousand actinobacterial strains from the Trondheim fjord (Norway), which resulted in identification of producers of both known and potentially new polyene macrolides with antifungal activity. The ability to produce the polyene macrolide candicidin D was found to be widespread among the Trondheim fjord Streptomyces isolates. We also report that the candicidin biosynthesis genes (can) are present on a linear plasmid identified in one of these isolates, suggesting that the can genes might be spread by means of conjugation.
MATERIALS AND METHODS
Strain isolation and handling.
Sediment samples were collected from different sites in the Trondheim fjord (Table 1). Sediments from depths of 1 to 28 m were collected by scuba divers (site 1 [63°34′N, 10°37′E] and site 2 [63°26 ′N, 10°21′E]), while sediments from a depth of 450 m (site 3 [63°29′N, 10°18′E]) were obtained with a box corer. The upper 5-cm portions of the sediments were collected in ziplock bags (scuba diver) or with a sterile spade (box corer) and transferred to 1-liter sterile plastic containers. Approximately 10% of the container volume was filled with 60% sediment and 40% seawater from the sampling site. This was done in order to ensure aerobic conditions in storage during processing. Samples were processed on the day of sampling or on the day after sampling. Neuston layer samples were obtained from one site (63°58′N, 10°81′E) in the Trondheim fjord. The neuston layer (surface microlayer) was collected using Teflon plates as described by Kjelleberg et al. (22). The plates were immersed in water and gently lifted through the water surface, and then bacterioneuston was scraped off using a rubber edge. All samples were collected no further out than 2 to 3 m from the shore and with the water temperature between 4.3 and 5.8°C. Actinomycetes were isolated from the sediment and neuston layer samples using different types of pretreatment and selective media (details are described in the supplemental material). Bacterial strains used in this study are listed in Table 2.
TABLE 1.
Sample description
| Sample no. | Sampling location | Sample characteristics | No. of actinomycetes isolated |
|---|---|---|---|
| 1 | 63°34′N, 10°37′E | 1-m-deep sediment | 179 |
| 2 | 63°34′N, 10°37′E | 7.5-m-deep sediment | 276 |
| 3 | 63°26′N, 10°21′E | 4.5-m-deep sediment | 353 |
| 4 | 63°26′N, 10°21′E | 6-m-deep sediment | 1,213 |
| 5 | 63°26′N, 10°21′E | 27-m-deep sediment | 566 |
| 6 | 63°29′N, 10°18′E | 450-m-deep sediment | 696 |
| 7 | 63°58′N, 10°81′E | Neuston layer | 425 |
| Total | 3,708 |
TABLE 2.
Bacterial strains used in this study
| Species or strain | Phenotypea | Source or reference |
|---|---|---|
| Candida albicans | 5FCr FLCr ITRAr AMBs | CCUGb (strain 39343) |
| Candida glabrata | FLCr ITRAr AMBr 5FCs | CCUG (strain 39342) |
| Streptomyces sp. strain MP47-06 | Candicidin producer | This study |
| Streptomyces sp. strain MP47-91 | Candicidin producer | This study |
| Streptomyces sp. strain MP15-36 | Candicidin producer | This study |
| Streptomyces sp. strain MP18-04 | Candicidin producer | This study |
| Streptomyces sp. strain MPS08-39 | Candicidin producer | This study |
| Streptomyces sp. strain MPS08-73 | Candicidin producer | This study |
| Streptomyces sp. strain MPS05-43 | Candicidin producer | This study |
| Streptomyces sp. strain MPS07-63 | Does not produce polyenes | This study |
| Streptomyces sp. strain MPS07-67 | Putative producer of another polyene | This study |
| Streptomyces sp. strain MPS05-34 | Candicidin producer | This study |
| S. griseus IMRU 3570 | Candicidin producer | 4 |
| S. lividans TK64(pSET152) | Amr | 3 |
| Rifr mutant of Streptomyces sp. strain MPS07-63 | Rifr | This study |
5FC, flucytosine; FLC, fluconazole; ITRA, itraconazole; AMB, amphotericin B; Am, apramycin; Rif, rifampin.
CCUG, Culture Collection, Göteborg University.
Screening for antifungal activity.
The use of 96-well plates for isolation of actinomycetes and for testing of antifungal metabolite production, as well as the automatic handling of extract plates using a robotic liquid handling system, made it possible to screen a large number of isolates. All technical details related to the antifungal screening procedure are described in the supplemental material. Two fungal strains were used in the screen, strains of Candida albicans and Candida glabrata (Table 2). Including the amphotericin B-resistant organism C. glabrata provided an opportunity to identify potential polyene producers based on differences in inhibition of the two fungal strains. Growth inhibition of the fungal strains was evaluated by measuring the optical density at 660 nm.
LC-TOF MS analysis of bacterial extracts.
Liquid chromatography (LC)-mass spectrometry (MS) analysis was performed using an Agilent 1100 series LC with diode array detector (DAD) connected to an Agilent 6210 time of flight (TOF) MS. LC was performed using a Zorbax SB C18 column (2.1 by 150 mm) with a flow rate of 0.25 ml/min and the following mobile phases: 10 mM ammonium acetate in water (pH 4.0) and then an acetonitrile gradient from 0 to 2 min, 30% acetonitrile from 2 to 10 min, and 30 to 60% acetonitrile for 20 min. The TOF MS was operated in either negative or positive electron spray ionization mode with a gas temperature of 350°C, drying gas at a rate of 10 liters/min, and a nebulizer pressure of 50 lb/in2. A reference solution for correction of mass axes was continuously injected through a second nebulizer needle. The mass accuracy of the TOF MS is 3 ppm, and a compound database search based on accurate mass determination was performed using a range of ±5 ppm.
16S rRNA gene analysis.
Total DNA of the bacteria was isolated using a DNeasy blood and tissue kit (Qiagen). Primers BP_F27 (5′-AGAGTTTGATCMTGGCTCAG-3′) and BP_R1492 (5′-TACGGYTACCTTGTTACGACTT-3′) (26) were used to amplify 1,490 bp of the 16S rRNA gene. The 50-μl PCR mixture contained total DNA isolated from the different isolates (10 to 20 ng), 1× Expand high-fidelity buffer with MgCl2 (Roche), 400 nM of each primer, 200 μM of each deoxynucleoside triphosphate, and 2.6 U of Expand high-fidelity enzyme mixture (Roche). The reaction was performed using the following conditions: 94°C for 4 min, followed by 35 cycles of 45 s at 94°C, 20 s at 55°C, and 2 min at 66°C and then a final 5-min extension at 70°C. The 50-μl reaction mixture was subjected to gel electrophoresis, and the resulting DNA fragment (about 1,490 bp) was purified and then cloned in the pDrive vector (Qiagen). Purified plasmids were sent to MWG Biotech for sequencing using the pDrive-specific primers M13 forward (5′-GTAAAACGACGGCCAGT-3′) and M13 reverse (5′-AACAGCTATGACCATG-3′) described in the Qiagen PCR cloning handbook and primer 1100R (40) (5′-AGGGTTGCGCTCGTTG-3′). Sequences were aligned with each other and with the most homologous actinomycete 16S rRNA gene sequences from the GenBank database using the Molecular Evolutionary Genetics Analysis (MEGA) software, version 4.0.2 (37). As not all sequences retrieved from the GenBank database were the same length, the ends of the sequences were trimmed, leaving 1,476 bp as the basis for construction of a phylogenetic tree. The tree was constructed by the neighbor-joining method provided with the MEGA software using 2,000 bootstrap replicates.
Southern blot analyses.
Enzymatic manipulations and agarose gel electrophoresis were performed as described by Sambrook and Russell (34). Total DNA was isolated with a DNeasy blood and tissue kit (Qiagen). DNA fragments were isolated from agarose gels using the QIAEX II suspension (Qiagen). PCR primers were designed for the candicidin gene cluster of Streptomyces griseus IMRU 3570 (4) (primers pabAB1 [5′-GTCGAGAGCGCCTTCCTGAC-3′], pabAB2 [5′-TGTCCGCCTCGGCCATGTTG-3′], canRA1 [5′-ACCGGCGTGCGAGCAGCTAC-3′], canRA2 [5′-GCCGGCCTGGTCCAGGCTTC-3′], canP3_1 [5′-CGCACCAGACCGAGCATCAT-3′], and canP3_2 [5′-GACGGCACGGTGGAATCACT-3′]) and were purchased from MWG Biotech. pabAB and canP3 PCR products were cloned using a Qiagen PCR cloning kit (Qiagen) and were sequenced by MWG Biotech using the M13 primers described above. Probes were synthesized using a PCR digoxigenin probe synthesis kit (Roche Applied Science) and used for Southern blot analyses, which were performed as described by the manufacturer for the digoxigenin system (33). Strains used for the Southern blot analyses are listed in Table 2.
PFGE.
DNA plugs for PFGE were prepared by using method described by Kieser et al. (15), with some modifications. Strains were grown in tryptone soya broth (Oxoid) with 0.1% glycine. Mycelium was resuspended in 5 ml TE25-sucrose before it was mixed 50:50 with 1.5% InCert agarose (Cambrex) in phosphate-buffered saline (8 g/liter NaCl, 0.2 g/liter KCl, 1.44 g/liter Na2HPO4, 0.24 g/liter KH2PO4; pH 7.4). Plugs were incubated for 1 h at 37°C in Tris-EDTA-25% sucrose (15) containing 5 mg/ml lysozyme. Plugs were transferred to 0.5× Tris-borate-EDTA buffer 1 h before they were used. The PFGE was carried out using 1% SeaKem Gold agarose (Cambrex) in 0.5× Tris-borate-EDTA buffer at 6 V cm−1 and 16°C, with an initial switch time of 50 s and a final switch time of 90 s for 24 h. PFGE was performed with the CHEF-DR II pulsed-field electrophoresis system (Bio-Rad). The size markers used were yeast chromosome PFG marker and MidRange II PFG marker (both obtained from New England BioLabs).
“Curing” experiment.
A spore suspension of Streptomyces sp. strain MP15-36 was serially diluted, plated on soya flour mannitol agar (SFM), and incubated at 30°C and 37°C for 3 to 5 days. Production of heptaene macrolides on SFM usually results in a visible yellow color around the producing colonies, and the plates were checked every day for colorless colonies. On the plates incubated at 37°C, seven “white” colonies were identified, restreaked on new SFM plates together with two “yellow” colonies (as positive controls), and incubated at 25°C before extraction of colonies with dimethyl sulfoxide (DMSO). The extracts were scanned to obtain UV/visible spectra. Extracts from six of the seven “white” candidates did not produce the UV/visible spectrum characteristic of candicidin production. The two “yellow” colonies and one of the “white” colonies produced the usual UV/visible spectrum indicating candicidin production. A Southern blot analysis of total DNA isolated from the six non-candicidin-producing clones was performed using the pabAB probe. Five of the six candidate clones exhibited hybridization to the probe, indicating that only one candidate clone might have lost the plasmid as a result of growing at the elevated temperature. This clone was tested for the presence of the “lethal zygosis” phenotype (2) by plating a spore suspension on an SFM plate and then placing a drop of a spore suspension from the original plasmid-containing strain in the middle of the plate. After 3 days of incubation, a zone of inhibition (inhibition of sporulation) was clearly visible around the plasmid-containing strain growing in the middle of the plate, indicating that it had the “lethal zygosis” phenotype due to the plasmid transfer. Figure S1 in the supplemental material includes images showing the phenomenon described above.
Selection of the rifampin-resistant mutants.
For selection of rifampin-resistant mutants, a spore suspension (>107 CFU) of a strain was plated on ISP2 containing 2.5 to 5 μg/ml rifampin. The resulting rifampin-resistant colonies were transferred to ISP2 containing 5 to 10 μg/ml rifampin. Colonies able to tolerate the highest rifampin concentrations were chosen for the mating experiments.
Mating experiments.
Mating of strains was performed at 20°C or 30°C on ISP2 [S. lividans TK64(pSET152) and Rifr mutants], CP-6 [S. lividans TK64(pSET152)], and SFM [S. lividans TK64(pSET152) and Rifr mutants]. Approximately equal amounts of spores of the donor and the recipient (ca. 107 CFU) were plated on agar medium and incubated for 5 to 7 days until sporulation occurred. Spores were washed off with sterile water, filtered, and plated on media with antibiotic selection (apramycin or rifampin, depending on the recipient strain). After mating with S. lividans TK64(pSET152), serial dilutions of the apramycin-resistant colonies were plated on CP-6, grown until sporulation occurred, and replica plated onto a confluent lawn of the original recipient strain. Colonies that had the “lethal zygosis” phenotype (2) were extracted with DMSO and analyzed by UV/visible scanning for the spectrum characteristic of candicidin.
Candidates resulting from mating with the Rifr mutants were screened using colony extraction and subsequent UV/visible scanning to identify candicidin producers. Candidates that were potential candicidin producers were then checked by Southern blot analysis for the presence of can genes.
Nucleotide sequence accession numbers.
DNA sequences have been deposited in the GenBank database under accession numbers EU263060 to EU263067.
RESULTS AND DISCUSSION
Isolation and antifungal activity screening of actinomycetes from the Trondheim fjord reveal a large number of polyene producers.
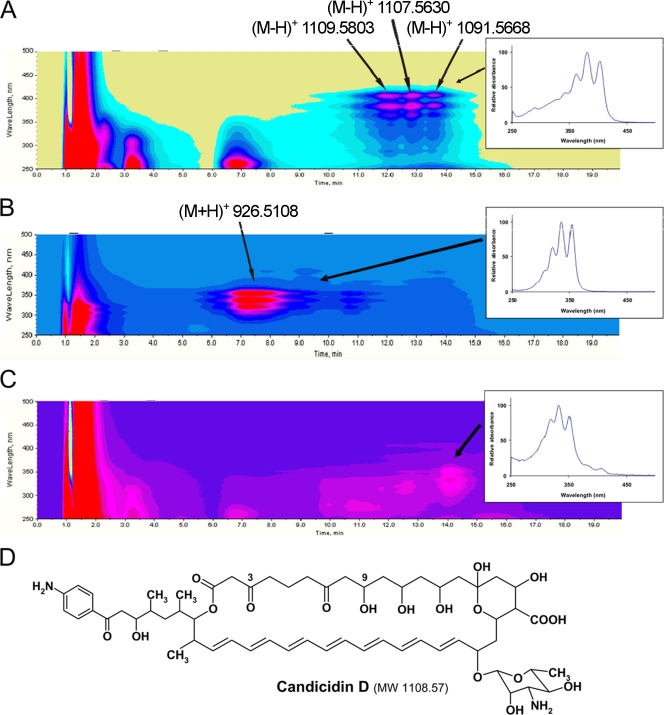
Actinomycete bacteria were isolated from marine sediments collected at different locations and different depths (range, 1 m to 450 m [Table 1 shows characteristics of the samples]), as well as from the neuston layer in the Trondheim fjord (Norway) using different selective media (see Materials and Methods and the supplemental material). A total of 3,708 isolates obtained from the samples were cultivated on different solid production media in 96-well plates, and extracts were screened for antifungal activity against C. albicans and/or C. glabrata. This screen revealed 1,044 isolates with antifungal activity against one or both of the test organisms, and extracts of these isolates were characterized by UV/visible spectrophotometry in order to identify polyene macrolide-like spectra. Polyene macrolide compounds have a characteristic three-peak UV absorbance spectrum, and the wavelengths of the peak maxima are determined by the number of conjugated double bonds (29). The UV absorbance spectrum can therefore be used to identify potential polyene macrolide-producing isolates. The UV/visible scans indicated that there was polyene production in 655 of the 1,044 isolates. The isolates capable of producing a putative heptaene compound(s) with UV absorption maxima of 366, 388, and 412 nm clearly dominated the group of polyene producers, accounting for more than 70% of the polyene-like spectra. Extracts from 62 polyene-producing actinomycetes, representing the isolates from different sources in the Trondheim fjord, were analyzed using LC-DAD-TOF MS. DAD isoplots of three of the extracts are shown in Fig. 1. The isoplot in Fig. 1A represents the putative heptaene compound(s) with UV absorption maxima of 366, 388, and 412 nm and shows three putative heptaene compounds eluting at 11.5 to 14.0 min. These three compounds have almost identical UV spectra, and only the profile of the second compound is shown in the inset in Fig. 1A. Accurate masses of these three compounds were determined by TOF MS using negative electron spray ionization, and the molecular ions are indicated in Fig. 1A. A search of the Dictionary of Natural Products database (http://dnp.chemnetbase.com/) with the corresponding accurate mass [(M-H+) 1107.5630, corresponding to an accurate molecular mass of 1,108.5703 Da] using a range of ±5 ppm resulted in a match with candicidin D (Fig. 1D), a known heptaene macrolide with antifungal activity produced by S. griseus (27). The first peak in Fig. 1A with molecular ion 1109.5803 may correspond to a candicidin derivative with an hydroxyl group at the C-3 position instead of a keto group. The last peak in the chromatogram (accurate molecular mass, 1,091.5668 Da) most probably corresponds to a candicidin analogue with one less oxygen due to the absence of an hydroxy group at C-9 (7). The accurate mass determination coupled with the heptaene-like UV spectrum strongly suggested that the compounds identified were indeed candicidin derivatives. Of the 62 extracts analyzed by LC-DAD-TOF MS, 52 produced this UV and mass chromatography profile.
FIG. 1.
(A to C) Diode array isoplots and UV/visible spectra of major compounds of three actinomycete extracts (insets). The wavelength is expressed as a function of elution time. See text for details. (D) Chemical structure of the polyene macrolide candicidin D. MW, molecular weight.
The DAD plot in Fig. 1B shows the results for a putative pentaene, as the UV absorption maxima are between 320 and 350 nm (29). TOF MS analysis of the extract showed that the accurate molecular mass of this possible pentane compound is 925.5035 Da. The Dictionary of Natural Products database search resulted in only one hit, nystatin A1, which is a tetraene and not a pentaene. The compound in question might therefore be an unknown pentaene with the same accurate molecular mass as nystatin A1. The DAD plot in Fig. 1C shows the results for a putative pentaene compound, and TOF MS analysis detected accurate molecular masses of 911.6168, 947.5951, and 1025.6118 Da associated with this compound. None of these molecular masses had any hits in the Dictionary of Natural Products database search using a range of ±5 ppm. In conclusion, the extracts described in Fig. 1B and 1C probably contained potentially new pentaene macrolides.
A group of streptomycetes isolated from the Trondheim fjord harbors the candicidin biosynthesis gene cluster.
The morphological diversity of the actinomycetes isolated from different locations in the Trondheim fjord and found to synthesize candicidin prompted us to investigate the molecular taxonomy of a limited number of these isolates using 16S rRNA gene analysis. PCR amplification, cloning, and sequencing of the 16S rRNA gene fragments from eight candicidin producers (Streptomyces sp. strains MP47-06, MP47-91, MP18-04, MP15-36, MPS08-73, MPS08-39, MPS05-43, and MPS05-34) exhibiting different morphological characteristics were performed. The known candicidin producer S. griseus IMRU 3570 (4) was also included in the analysis. The 16S rRNA gene sequence analysis demonstrated that seven of the eight isolates and S. griseus IMRU 3570 shared 16S rRNA gene sequence identities of 99.4 to 100%, corresponding to 0 to 9 nucleotide differences at 1,490 locations. Only one strain (MPS05-43) differed significantly from the other strains, and the levels of 16S rRNA gene sequence identity of this strain with the other seven isolates ranged from 95.4 to 95.6%. A phylogenetic tree was constructed for all the sequences and the most homologous sequences revealed by BLAST searches (see Fig. S2 in the supplemental material). Previous studies have indicated that some Streptomyces strains can have very similar 16S rRNA gene sequences and still be classified as members of different species based on DNA-DNA hybridization and phenotypic data (9, 25).
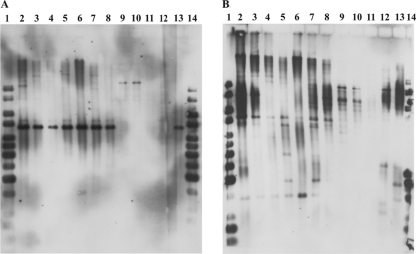
Although the LC-TOF MS data strongly suggested that the polyene antifungal synthesized by these streptomycetes is candicidin D, we decided to verify this using molecular probes specific for the candicidin biosynthesis gene cluster of the known candicidin producer S. griseus IMRU 3570 (4). PCR primers were designed so that they amplified ca. 400- to 500-bp fragments of the genes canRA (ABC transporter), pabAB (p-aminobenzoic acid synthetase), and canP3 (polyketide synthase). Total DNAs isolated from two (Streptomyces sp. strains MP47-06 and MP47-91) of the eight candicidin-producing strains were used as templates in this experiment. PCRs resulted in successful amplification of the expected fragments with both templates, confirming that at least two of the isolates chosen contain the candicidin biosynthesis gene cluster. The PCR fragments of pabAB and canP3 from Streptomyces sp. strain MP47-91 were sequenced, and the sequences were confirmed to be 99% identical to the corresponding S. griseus IMRU 3570 sequences. Next, total DNA was isolated from the other six candicidin producers (Streptomyces sp. strains MP18-04, MP15-36, MPS08-73, MPS08-39, MPS05-43, and MPS05-34), one nonproducer (Streptomyces sp. strain MPS07-63), one putative producer of a different polyene macrolide (Streptomyces sp. strain MPS07-67), and S. griseus IMRU 3570. A series of Southern blot analyses of BamHI-digested total DNAs with two different can probes (pabAB and canP3) clearly showed that all of the candicidin-producing strains tested harbor the candicidin biosynthesis gene cluster (Fig. 2A and B).
FIG. 2.
Southern blot analyses with the pabAB (A) and canP3 (B) probes. Lanes 1 and 14, Fermentas GeneRuler 1-kb DNA ladder, lane 2, S. griseus; lane 3, Streptomyces sp. strain MP47-06); lane 4, Streptomyces sp. strain MP47-91; lane 5, Streptomyces sp. strain MP18-04; lane 6, Streptomyces sp. strain MP15-36; lane 7, Streptomyces sp. strain MPS08-73; lane 8, Streptomyces sp. strain MPS08-39; lanes 9 and 10, Streptomyces sp. strain MPS05-43; lane 11, Streptomyces sp. strain MPS07-63; lane 12, Streptomyces sp. strain MPS07-67; lane 13, Streptomyces sp. strain MPS05-34. DNA was digested with BamHI.
Seven of the candicidin producers (Streptomyces sp. strains MP47-06, MP47-91, MP18-04, MP15-36, MPS08-73, MPS08-39, and MPS05-34) displayed a hybridization pattern with pabAB resembling that of S. griseus, which indicated that the pabAB-containing region is highly conserved among these isolates as the restriction fragment size is the same. However, one isolate, Streptomyces sp. strain MPS05-43, displayed a totally different hybridization pattern (Fig. 2, lanes 9 and 10), suggesting that the organization of the pabAB-containing region in this strain is different. Interestingly, this strain was also the strain shown to be the strain most phylogenetically distinct from all the other strains and from S. griseus IMRU 3570. DNAs from the non-candicidin-producing strain (Streptomyces sp. MPS07-63) and from the putative producer of a different polyene macrolide (Streptomyces sp. strain MPS07-67) did not show any hybridization with the pabAB probe, confirming that this analysis was specific enough for the can cluster. The canP3 probe was found to be less specific, which is not surprising considering that streptomycetes often have more than one polyketide synthase cluster and that polyketide synthase genes in general share at least some homology. Hybridization of the canP3 probe to the total DNA of the putative producer of a different polyene macrolide (Streptomyces sp. strain MPS07-67) proves this point.
Candicidin biosynthesis gene cluster is located on a linear plasmid in one of the isolates.
So far, the ability to produce candicidin has been described for S. griseus (27), Streptomyces acrimycini (1), Streptomyces levoris (24), Streptomyces helvoloviolaceus (23), Streptomyces sp. strain FR-008 (7), S. coelicolor JI2159 (8), S. coelicolor JI1157 (8), S. griseus JI2212 (8), and Streptomyces albus G (8), which seem to have originated from different sources. The candicidin producers identified in this study, however, all originated from the Trondheim fjord sediments and neuston layer. The eight strains selected for further study differed morphologically, but sequencing of the 16S rRNA genes showed that only one strain (MPS05-43) was phylogenetically distinct from all the other strains. It is therefore possible that the other seven isolates are closely related and that the can cluster might either have originated from a common ancestor or have been transferred among the strains by a mobile genetic element.
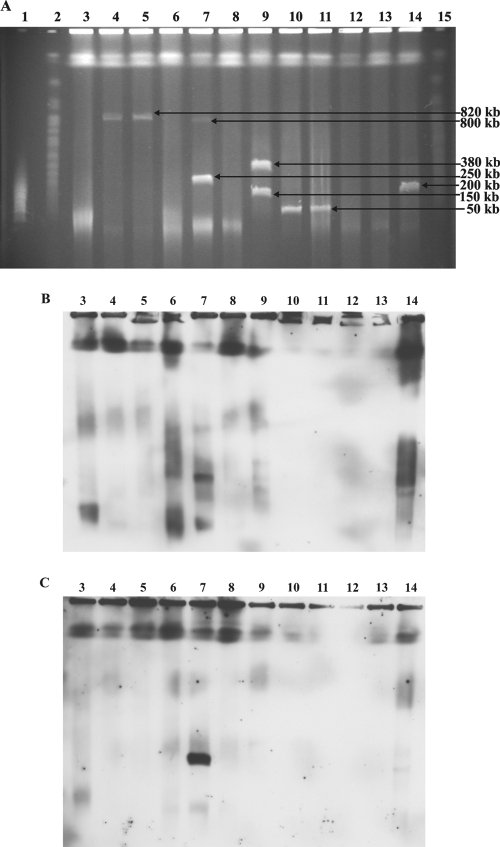
Considering the size of the can gene cluster (ca. 140 kb), a linear plasmid seemed to be a plausible candidate for such a mobile element. To investigate the presence of linear plasmids in the eight isolates that harbor the can gene cluster, their total DNAs were subjected to PFGE. Also, S. griseus IMRU 3570, the non-candicidin-producing strain (Streptomyces sp. strain MPS07-63), and the putative producer of a different polyene macrolide (Streptomyces sp. strain MPS07-67) were included in the analysis. PFGE revealed the presence of presumably linear plasmids ranging in size from ca. 50 kb to 820 kb in six of the candicidin-producing strains (Fig. 3A). No linear plasmids were detected in S. griseus IMRU 3570 and isolates MP18-04, MPS08-73, MPS07-63, and MPS07-67. The gel from the PFGE was used for a Southern blot analysis with the pabAB and canP3 probes. Both probes hybridized to the same ca. 250-kb plasmid present in Streptomyces sp. strain MP15-36 (Fig. 3B and C). In addition, the probes hybridized to the chromosomal DNA of the other seven candicidin producers, as well as to that of S. griseus. Integration into chromosomal DNA has been reported for at least two linear plasmids. SCP1 is able to integrate into the chromosome of S. coelicolor (18), and pPZG101 can integrate into the chromosome of S. rimosus (10). It is therefore possible that the can plasmid might be integrated into the chromosomes of the candicidin-producing strains that did not harbor this plasmid or that these strains are strains with chromosomal can genes.
FIG. 3.
(A) PFGE, (B) Southern hybridization with the pabAB probe, and (C) Southern hybridization with the canP3 probe for candicidin-producing strains. Lane 1, MidRange II PFG marker; lanes 2 and 15, yeast chromosome PFG marker (markers are not shown for Southern hybridization); lane 3, S. griseus; lane 4, Streptomyces sp. strain MP47-06; lane 5, Streptomyces sp. strain MP47-91; lane 6, Streptomyces sp. strain MP18-04; lane 7, Streptomyces sp. strain MP15-36; lane 8, Streptomyces sp. strain MPS08-73; lane 9, Streptomyces sp. strain MPS08-39; lanes 10 and 11, Streptomyces sp. strain MPS05-43; lane 12, Streptomyces sp. strain MPS07-63; lane 13, Streptomyces sp. strain MPS07-67; lane 14, Streptomyces sp. strain MPS05-34.
Mating experiments with S. lividans and the non-candicidin-producing Trondheim fjord isolate failed to demonstrate can plasmid transfer.
To test the can plasmid's interspecific transfer ability, a mating experiment was conducted. S. lividans TK64 carrying a streptomycin resistance marker has been used successfully in the past for mating experiments. The can plasmid-containing strain (Streptomyces sp. strain MP15-36) was tested for sensitivity to streptomycin and apramycin. As MP15-36 was resistant to streptomycin but not apramycin, S. lividans TK64 could not be used. Instead, S. lividans TK64(pSET152) was used as the recipient in the mating experiments (see Materials and Methods). As it was not possible to select the transconjugants directly, the plates were replicated on CP-6-apramycin plates on a layer of S. lividans TK64(pSET152) to see if the “lethal zygosis” phenotype (“pock” formation) characteristic of plasmid transfer in Streptomyces (2) could be observed. “Pock” formation was indeed observed in this experiment, and 10 candidates were chosen for a Southern blot analysis. DMSO extracts of these 10 candidates were also prepared to assess whether they were able to produce candicidin. UV/visible scans of the extracts did not show any trace of candicidin, and the pabAB probe did not hybridize to the total DNA from any of the candidates (data not shown). The mating experiments were repeated using ISP2 and SFM, but no positive results were obtained.
Next, we decided to try mating with one of the can-negative Trondheim fjord isolates (Streptomyces sp. strain MPS07-63), assuming that the “native” strain might be able to accept and stabilize the plasmid. The strain chosen was the strain that was negative with both the pabAB and canP3 probes in the Southern blot analysis described above. Also, the strain did not carry any linear plasmids, which was confirmed by PFGE, and comparison of 16S rRNA gene sequences from MP15-36 and MPS07-63 showed that these two strains are closely related, as the sequences were identical. To have a means of selecting for transconjugants after mating, rifampin-resistant mutants were obtained (see Materials and Methods). Two of the mutants were chosen for mating with the can plasmid strain on both ISP2 and SFM agar. After selection for the Rifr strain, 100 random colonies were screened by extraction with DMSO, followed by UV/visible scanning. Two putative transconjugants were chosen for Southern blot analyses with the pabAB and canP3 probes as the UV/visible scans indicated that a heptaene was present in the extracts, but again the results were negative (data not shown).
There are several possible explanations for why these mating experiments were unsuccessful. Assuming that the can plasmid is responsible for the abundance of candicidin producers in our strain collection, one would expect the transfer frequency of the plasmid to be quite high. However, the transfer frequency might be much lower than anticipated, and without a selectable marker, detecting the transconjugants is quite difficult. Also, for the Rifr strain the initial screening of putative transconjugants was dependent on actual production of candicidin (see Materials and Methods). It is possible that the plasmid might transfer at a higher rate but the transconjugants are not able to produce candicidin and are therefore not recognized. Also, we cannot exclude the possibility that the plasmid is not stable in the recipient hosts chosen and that it is therefore easily lost.
can plasmid can be reintroduced into the “cured” strain, restoring candicidin production.
In order to demonstrate transfer of the can plasmid, a “curing” experiment with the can plasmid strain was performed in order to obtain a suitable recipient for mating. Loss of linear plasmids as a result of growth at a higher temperature has been demonstrated for Streptomyces (30). A “curing” experiment with Streptomyces sp. strain MP15-36 was carried out (see Materials and Methods) by incubating the strain at 37°C. Candidates that did not produce candicidin were checked for loss of the plasmid by a Southern blot analysis using the pabAB probe, and one putative plasmid-free candidate was obtained. A Rifr mutant of the potential plasmid-free strain was obtained for easier selection of transconjugants (see Materials and Methods), and mating between the original plasmid-containing strain and the Rifr plasmid-free strain was performed on SFM agar. Twelve rifampin-resistant colonies were selected, and a Southern blot analysis of their total DNAs was performed using the pabAB probe. This probe hybridized to the DNAs from all candidates (data not shown), and three of them were subsequently tested for candicidin production by LS-TOF MS. A molecule with the mass of candicidin was shown to be present in the extracts of all three candidates tested, indicating that the plasmid was successfully transferred to the plasmid-free strain at a high frequency, restoring the ability to produce candicidin.
Conclusion.
In this study we investigated the antibiotic production potential of 3,708 isolates derived from marine sediment and neuston layer samples. We showed that many of these isolates produce compounds with antifungal activity and that a high proportion of Streptomyces bacteria isolated from different sources in the Trondheim fjord produce the antifungal polyene macrolide candicidin. Genes involved in the biosynthesis of candicidin were shown to be present on a linear plasmid in one of the Streptomyces strains isolated from the Trondheim fjord sediment. This strain could be cured of the plasmid, resulting in loss of candicidin production, while reintroduction of the plasmid by means of conjugation restored production. Although the can plasmid could not be transferred to other Streptomyces strains in the laboratory, it is possible that this plasmid is responsible for spreading the candicidin biosynthesis gene cluster to other strains in its natural environment.
Supplementary Material
Acknowledgments
We thank G. Johnsen for helping with collection of the sediment samples, J. A. Gil for the gift of S. griseus IMRU 3570, and K. F. Degnes for helping with some of the analyses.
This work was supported by the Research Council of Norway and the Norwegian University of Science and Technology.
Footnotes
Published ahead of print on 13 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Asturias, J. A., J. F. Martin, and P. Liras. 1994. Biosynthesis and phosphate control of candicidin by Streptomyces acrimycini JI2236: effect of amplification of the pabAB gene. J. Ind. Microbiol. 13:183-189. [DOI] [PubMed] [Google Scholar]
- 2.Bibb, M. J., R. F. Freeman, and D. A. Hopwood. 1977. Physical and genetical characterisation of a second sex factor, SCP2, for Streptomyces coelicolor A3(2). Mol. Gen. Genet. 154:155-166. [Google Scholar]
- 3.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. Nagaraja Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Campelo, A. B., and J. A. Gil. 2002. The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology 148:51-59. [DOI] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Chen, C. W., T. W. Yu, Y. S. Lin, H. M. Kieser, and D. A. Hopwood. 1993. The conjugative plasmid SLP2 of Streptomyces lividans is a 50 kb linear molecule. Mol. Microbiol. 7:925-932. [DOI] [PubMed] [Google Scholar]
- 7.Chen, S., X. Huang, X. Zhou, L. Bai, L. He, K. J. Jeong, S. Y. Lee, and Z. Deng. 2003. Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem. Biol. 10:1065-1076. [DOI] [PubMed] [Google Scholar]
- 8.Gil, J. A., L. M. Criado, T. Alegre, and J. F. Martín. 1990. Use of a cloned gene involved in candicidin production to discover new polyene producer Streptomyces strains. FEMS Microbiol. Lett. 58:15-18. [DOI] [PubMed] [Google Scholar]
- 9.Goodfellow, M., Y. Kumar, D. P. Labeda, and L. Sembiring. 2007. The Streptomyces violaceusniger clade: a home for streptomycetes with rugose ornamented spores. Antonie van Leeuwenhoek 92:173-199. [DOI] [PubMed] [Google Scholar]
- 10.Gravius, B., D. Glocker, J. Pigac, K. Pandza, D. Hranueli, and J. Cullum. 1994. The 387 kb linear plasmid pPZG101 of Streptomyces rimosus and its interactions with the chromosome. Microbiology 140:2271-2277. [DOI] [PubMed] [Google Scholar]
- 11.Hayakawa, T., T. Tanaka, K. Sakaguchi, N. Otake, and H. Yonehara. 1979. A linear plasmid-like DNA in Streptomyces sp. producing lankacidin group antibiotics. J. Gen. Appl. Microbiol. 25:255-260. [Google Scholar]
- 12.Hopwood, D. A., and H. M. Wright. 1973. Transfer of a plasmid between Streptomyces species. J. Gen. Microbiol. 77:187-195. [Google Scholar]
- 13.Hopwood, D. A., and T. Kieser. 1993. Conjugative plasmids of Streptomyces, p. 293-311. In D. B. Clewell (ed.) Bacterial conjugation, 1st ed. Plenum Press, New York, NY.
- 14.Kieser, T., D. A. Hopwood, H. M. Wright, and C. J. Thompson. 1982. pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol. Gen. Genet. 185:223-238. [DOI] [PubMed] [Google Scholar]
- 15.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 16.Kinashi, H., E. Mori, A. Hatani, and O. Nimi. 1994. Isolation and characterization of linear plasmids from lankacidin-producing Streptomyces species. J. Antibiot. 47:1447-1455. [DOI] [PubMed] [Google Scholar]
- 17.Kinashi, H., M. Shimaji, and A. Sakai. 1987. Giant linear plasmids in Streptomyces which code for antibiotic biosynthesis genes. Nature 328:454-456. [DOI] [PubMed] [Google Scholar]
- 18.Kinashi, H., M. Shimaji-Murayama, and T. Hanafusa. 1992. Integration of SCP1, a giant linear plasmid, into the Streptomyces coelicolor chromosome. Gene 115:35-41. [DOI] [PubMed] [Google Scholar]
- 19.Kinashi, H., S. Fujii, A. Hatani, T. Kurokawa, and H. Shinkawa. 1998. Physical mapping of the linear plasmid pSLA2-L and localization of the eryAI and actI homologs. Biosci. Biotechnol. Biochem. 62:1892-1897. [DOI] [PubMed] [Google Scholar]
- 20.Kinashi, H., S. L. Otten, J. S. Duncan, and C. R. Hutchinson. 1988. Frequent loss and restoration of antibiotic production by Streptomyces lasaliensis. J. Antibiot. 41:624-637. [DOI] [PubMed] [Google Scholar]
- 21.Kirby, R., and D. A. Hopwood. 1977. Genetic determination of methylenomycin synthesis by the SCP1 plasmid of Streptomyces coelicolor A3(2). J. Gen. Microbiol. 98:239-252. [DOI] [PubMed] [Google Scholar]
- 22.Kjelleberg, S., T. A. Stenström, and G. Odham. 1979. Comparative study of different hydrophobic devices for sampling lipid surface films and adherent microorganisms. Mar. Biol. 53:21-25. [Google Scholar]
- 23.Konev, Y. E., V. A. Tsyganov, E. D. Etingov, N. M. Zaval'naya, and A. I. Filippova. 1973. Isolation and identification of heptaene antibiotic produced by Streptomyces helvoloviolaceus. Antibiotiki 18:354-358. [PubMed] [Google Scholar]
- 24.Korenyako, A. I., Y. M. Khokhlova, N. I. Nikitina, and A. V. Puchnina. 1961. Streptomyces: producers of heptaene antibiotics. Mikrobiologiya 30:633-641. [PubMed] [Google Scholar]
- 25.Kumar, Y., and M. Goodfellow. 2008. Five new members of the Streptomyces violaceusniger 16S rRNA gene clade: Streptomyces castelarensis sp. nov., comb. nov., Streptomyces himastatinicus sp. nov., Streptomyces mordarskii sp. nov., Streptomyces rapamycinicus sp. nov. and Streptomyces ruanii sp. nov. Int. J. Syst. Evol. Microbiol. 58:1369-1378. [DOI] [PubMed] [Google Scholar]
- 26.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics, 1st ed. John Wiley & Sons, Chichester, United Kingdom.
- 27.Lechevalier, H., R. F. Acker, C. T. Corke, C. M. Haenseler, and S. A. Waksman. 1953. Candicidin, a new antifungal antibiotic. Mycologia 45:155-171. [Google Scholar]
- 28.Mochizuki, S., K. Hiratsu, M. Suwa, T. Ishii, F. Sugino, K. Yamada, and H. Kinashi. 2003. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 48:1501-1510. [DOI] [PubMed] [Google Scholar]
- 29.Omura, S., and H. Tanaka. 1984. Production, structure, and antifungal activity of polyene macrolides, p. 351-404. In S. Omura (ed.), Macrolide antibiotics. Chemistry, biology and practice, 1st ed. Academic Press, Inc., Orlando, FL.
- 30.Pang, X., Y. Sun, J. Liu, X. Zhou, and Z. Deng. 2002. A linear plasmid temperature-sensitive for replication in Streptomyces hygroscopicus 10-22. FEMS Microbiol. Lett. 208:25-28. [DOI] [PubMed] [Google Scholar]
- 31.Ravel, J., E. M. H. Wellington, and R. T. Hill. 2000. Interspecific transfer of Streptomyces giant linear plasmids in sterile amended soil microcosms. Appl. Environ. Microbiol. 66:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravel, J., H. Schrempf, and R. T. Hill. 1998. Mercury resistance is encoded by transferable giant linear plasmids in two Chesapeake Bay Streptomyces strains. Appl. Environ. Microbiol. 64:3383-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roche Applied Science. 2003. DIG application manual for filter hybridization. Roche Applied Science, Indianapolis, IN.
- 34.Sambrook, J., and D. W. Russell. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Schrempf, H., H. Bujard, D. A. Hopwood, and W. Goebel. 1975. Isolation of covalently closed circular deoxyribonucleic acid from Streptomyces coelicolor A3(2). J. Bacteriol. 121:416-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suwa, M., H. Sugino, A. Sasaoka, E. Mori, S. Fujii, H. Shinkawa, O. Nimi, and H. Kinashi. 2000. Identification of two polyketide synthase gene clusters on the linear plasmid pSLA2-L in Streptomyces rochei. Gene 246:123-131. [DOI] [PubMed] [Google Scholar]
- 37.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 38.Vivian, A. 1971. Genetic control of fertility in Streptomyces coelicolor A3(2): plasmid involvement in the interconversion of UF and IF strains. J. Gen. Microbiol. 69:353-364. [DOI] [PubMed] [Google Scholar]
- 39.Vivian, A., and D. A. Hopwood. 1970. Genetic control of fertility in Streptomyces coelicolor A3(2): the IF fertility type. J. Gen. Microbiol. 64:101-117. [DOI] [PubMed] [Google Scholar]
- 40.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.