Abstract
Microorganisms can account for up to 60% of the fresh weight of marine sponges. Marine sponges have been hypothesized to serve as accumulation spots of particular microbial communities, but it is unknown to what extent these communities are directed by the organism or the site or occur randomly. To address this question, we assessed the composition of specific bacterial communities associated with Aplysina fulva, one of the prevalent sponge species inhabiting Brazilian waters. Specimens of A. fulva and surrounding seawater were collected in triplicate in shallow water at two sites, Caboclo Island and Tartaruga beach, Búzios, Brazil. Total community DNA was extracted from the samples using “direct” and “indirect” approaches. 16S rRNA-based PCR-denaturing gradient gel electrophoresis (PCR-DGGE) analyses of the total bacterial community and of specific bacterial groups—Pseudomonas and Actinobacteria—revealed that the structure of these assemblages in A. fulva differed drastically from that observed in seawater. The DNA extraction methodology and sampling site were determinative for the composition of actinobacterial communities in A. fulva. However, no such effects could be gleaned from total bacterial and Pseudomonas PCR-DGGE profiles. Bacterial 16S rRNA gene clone libraries constructed from directly and indirectly extracted DNA did not differ significantly with respect to diversity and composition. Altogether, the libraries encompassed 15 bacterial phyla and the candidate division TM7. Clone sequences affiliated with the Cyanobacteria, Chloroflexi, Gamma- and Alphaproteobacteria, Actinobacteria, Bacteroidetes, and Acidobacteria were, in this order, most abundant. The bacterial communities associated with the A. fulva specimens were distinct and differed from those described in studies of sponge-associated microbiota performed with other sponge species.
The phylum Porifera (sponges) consists of benthic (sessile) organisms that occur primarily in marine environments at different depths (26). Sponges are classified into three groups, namely, the Calcarea (calcareous sponges), Hexactinellida (glass sponges), and Demospongiae (5, 26). The group Demospongiae, also called demosponges, encompasses 95% of the ca. 5,500 living sponge species described thus far (5). As typical filter feeders, demosponges are the prime bacterial filters of the sea. They are capable of pumping thousands of liters of water per day (23), using prokaryotic microorganisms as the main source of food (1, 43, 47). In addition to demosponges feeding on microorganisms, the presence of bacteria in high density in internal sponge layers (mesohyl) indicates that a selective process favoring particular prokaryotes, involving microbe-sponge interactions, is likely to occur (64). Furthermore, the dawn of the interactions between Prokarya and higher organisms may actually lie in the demosponges, whose origin is estimated to date back to 550 million years ago (5, 33).
Putative interactions between demosponges and microorganisms, presumably mostly consisting of Bacteria and Archaea, were first demonstrated by transmission electron microscopy (TEM), where high amounts of microorganisms were observed in the mesohyl (1, 14, 16, 64). Hence, these bacterium-rich sponges have been termed “bacteriosponges” (46). While investigating 11 taxonomically different demosponges using TEM, Vacelet and Donadey (64) identified two different sponge types in respect of their association with bacteria. Sponges with thick mesohyl contained abundant, dense, and morphologically diverse microbial communities (i.e., bacteriosponge), while those with a well-developed aquiferous system and low-density mesohyl contained few bacterial cells and typically only single bacterial morphotypes. The two types have recently been called “high-microbial-abundance” (HMA) and “low-microbial-abundance” (LMA) sponges, respectively (23). In HMA sponges, bacterial densities may reach 108 to 1010 bacterial cells per g (wet weight) of sponge, exceeding seawater concentrations by 2 to 4 orders of magnitude (15, 23). Based on the analysis of 16S rRNA genes, over 15 bacterial phyla have thus far been reported to occur in association with marine sponges (11, 23, 56). Among these are typical sponge-associated bacteria such as members of the Cyanobacteria, Chloroflexi, Proteobacteria, Acidobacteria, Verrucomicrobia, and the candidate phyla “Poribacteria” and TM6 (14, 30, 51, 56, 60, 68).
Increasing research interest in the sponge-associated microbiota has emerged in the past few years, mainly due to the in spongium production of an enormous diversity of biologically active secondary metabolites (56). Recent studies suggest that certain bioactive compounds retrieved from marine sponges—such as complex polypeptides and nonribosomal peptides—are likely to be synthesized by the symbiont bacteria (27, 41, 42). Such bioactive secondary metabolites offer great promise for use in biotechnology and medicine (3, 22, 27, 41, 42, 51, 59). In particular, cytotoxic compounds, i.e., antitumoral substances and polyketides, may find application in anticancer therapies (13, 42, 51). Recent investigations revealed the presence of dibromotyrosine-derived metabolites in Aplysina fulva (Pallas, 1766) specimens collected along the Brazilian shore (39). However, a putative role of microbial symbionts in the production of such metabolites, commonly found to display biological activity, remains to be evaluated.
Despite the global-scale occurrence of sponges in Earth's marine ecosystems, the investigation of their associated bacterial communities has thus far been restricted only to certain areas (1, 11, 13, 14, 27, 54, 58, 68). To our knowledge, no studies have been conducted, to date, on sponge-associated microbes in subtropical, South Atlantic open shore waters. In the present study, we assess the diversity and composition of the bacterial community associated with the demosponge A. fulva collected at two different sites at the Brazilian shore. A suite of tools, ranging from plate count estimations and TEM to sponge DNA-based analyses of bacterial 16S rRNA genes, was used. We hypothesized that a distinct bacterial community occurs in A. fulva, which is different from that in the surrounding bulk water, as well as from those in other sponge species.
MATERIALS AND METHODS
Field design and sampling.
Specimens of the Atlantic sponge A. fulva were collected in triplicate by snorkeling in shallow coastal waters at depths ranging from 2 to 5 m at Caboclo Island (22°45′18.81"S, 41°53′23"W) and Tartaruga beach (22°45′20.67"S, 41°54′13.53"W) in Búzios, Rio de Janeiro, Brazil, in March 2007. The sampling sites were ∼3 km from each other. The individual samples were placed separately in 50-ml Falcon tubes containing seawater, stored on ice, and taken to the laboratory within 24 h. In the lab, subsamples of ∼10 g of sponge taken from each individual specimen were used. For each sponge sample, a corresponding 200-ml bulk sample of the surrounding seawater was taken in Falcon tubes and stored on ice.
Plate counting.
Per sample, 1 g of A. fulva was cut into small pieces and ground with mortar and pestle. In addition, 1 ml of corresponding seawater was used. The material was transferred to tubes containing 9-ml sterile saline and glass beads and vortex mixed for 1 min. Tubes were allowed to stand for 1 min. From the resulting supernatant, serial 10-fold dilutions were made and plated in triplicate onto R2A (Difco, Sparks, MD). Plates were incubated at 28°C for 7 days, after which the population sizes of culturable heterotrophic bacteria were estimated. Log-transformed CFU values were compared by using two-sided t tests.
Electron microscopy.
Aliquots of sponge material (two per site) were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 1 h at room temperature, and then small pieces (1-2 mm) were cut and washed in the same buffer, postfixed in buffered 1% OsO4 (pH 7.2), dehydrated in an acetone series, and embedded in Polybed 812 (Polysciences, Inc., PA). Ultrathin sections were stained with saturated uranyl acetate for 20 min and lead citrate for 1 min and observed with a transmission electron microscope (FEI Morgagni, Eindhoven, The Netherlands) operating at an accelerating voltage of 80 kV.
Total community DNA extraction.
Two different strategies of sample handling prior to DNA extraction from sponge were used, hereafter referred to as the “direct” and “indirect” methods of DNA extraction. In the former method, DNA is directly extracted from the sponge without any prior treatment, whereas the latter method relies on an attempt to first separate microbial cells from the sponge matrix prior to DNA isolation. Direct DNA extraction was carried out with 0.6 g of A. fulva ground in liquid nitrogen using a mortar and pestle. The resulting material was used for DNA extraction with the FastDNA SPIN for Soil kit (MP Biomedicals, LLC, Illkirch, France). Briefly, the chosen DNA extraction kit works on the basis of a harsh mechanical lysis (bead beating), followed by homogenization and protein solubilization steps, after which genomic DNA is purified from contaminants through the use of a silica matrix. To process the sponge samples, a slight modification of the kit protocol was introduced. Longer centrifugation periods (6 min instead of 1 min) were required for the step in which DNA bound to the silica matrix is separated from debris through a filter membrane. For the indirect DNA extraction method, 6.0 g of sponge was ground for 1 min with a mortar and pestle in 10 ml of sterile saline. To further disperse the sponge matrix and detach the microbial cells from it, the resulting material was transferred to Erlenmeyer flasks containing 35 ml of sterile saline and glass beads (4 mm in diameter). The flasks were shaken for 20 min at 200 rpm. The homogenate was filtered over filter paper, and the collected filtrate centrifuged at 500 × g for 2 min to enable further removal of sponge debris. The supernatant was then transferred to sterile centrifuge tubes and subjected to a final centrifugation step of 10,000 × g for 30 min. The resulting “microbial pellets” were then used as starting material for DNA extraction with a FastDNA SPIN for Soil Kit as described above. Both methods were found to yield considerable amounts of clean DNA from A. fulva (estimated in 100 ng per g of fresh sponge). Seawater samples were filtered through 0.2-μm-pore-size nitrocellulose filters (Millipore, São Paulo, Brazil) in sterile conditions using a vacuum pump. Filters were cut into small pieces and directly used for DNA extraction with the FastDNA SPIN for Soil Kit using the manufacturer's protocol.
Bacterial PCR for DGGE analysis.
The composition of the total bacterial communities associated with A. fulva (as well as in the bulk water) was assessed by PCR-denaturing gradient gel electrophoresis (PCR-DGGE). 16S rRNA genes were amplified by PCR using the primer pair F968-GC and R1401-1a (4). The reaction mixture (25 μl) was prepared containing 1 μl of template DNA (1 to 5 ng), 1× Stoffel buffer (10 mM KCl, 10 mM Tris-HCl [pH 8.3]; Applied Biosystems, Foster, CA), 0.2 mM deoxynucleoside triphosphates (dNTPs), 3.5 mM MgCl2, 1% (wt/vol) formamide, 0.4 μM concentrations of both primers, 0.01 mg of T4 gene 32 protein/ml, and 2.5 U of Taq DNA polymerase (Stoffel fragment; Applied Biosystems, Foster City, CA). After an initial denaturation at 94°C for 5 min, 35 cycles of 1 min at 95°C, 1 min at 53°C, and 2 min at 72°C were performed. A final extension step of 10 min at 72°C was used to finish the reaction. All PCR amplifications were performed in a GeneAmp PCR system 9700 cycler (Applied Biosystems). All amplicons were checked after electrophoresis in 1% agarose gels after staining with ethidium bromide and visualized under UV light.
PCR of specific bacterial groups for DGGE analysis.
A nested PCR approach was applied for the generation and analysis of Pseudomonas specific 16S rRNA gene fragments. The amplification was carried out using the primers F311Ps and R1459Ps (36) with the reaction mixture and PCR conditions described by Costa et al. (8), except for the number of cycles (28 cycles in the present study). The amplicons were used as templates (2 μl) for the generation of bacterial 16S rRNA gene fragments as described above, except for the number of cycles (25 cycles in the present study). A seminested PCR strategy was used for the assessment of actinobacterial communities by DGGE. The primer pair F243 (24) and R1401-1a (4) was used in the first step to generate actinobacterium-specific amplicons. The reaction mixture (25 μl) consisted of 1 μl of template DNA (1 to 5 ng), 1× Stoffel buffer, 0.2 mM dNTPs, 3.75 mM MgCl2, 5% (vol/vol) dimethyl sulfoxide, 0.2 μM concentrations of both primers, and 2.5 U of Taq DNA polymerase. After 5 min of denaturation at 94°C, 30 cycles of 1 min at 94°C, 1 min at 63°C, and 2 min at 72°C were performed, followed by a 10-min final extension at 72°C. The amplicons were used as templates in a subsequent PCR for DGGE analysis using the primer pair F968-GC/R1401-1a, as described previously except for the number of cycles (25 cycles in the present study).
DGGE profiling.
All DGGE assays were performed in a PhorU-2 gradient system (Ingeny International, Goes, The Netherlands). The total bacterial and group-specific 16S rRNA gene amplicons were applied, in even concentrations, onto polyacrylamide gels containing 46.5 to 65% gradients of denaturants (100% denaturants defined as 7 M urea and 40% formamide) and 6 to 9% gradients of acrylamide. Mixtures of PCR products from five bacterial species (i.e., Arthrobacter, Burkholderia, Enterobacter, Listeria, and Rhizobium spp.) were applied at the edges of the gels as markers. Electrophoresis was performed in 0.5× Tris-acetate-EDTA buffer (pH 8.0) at 58°C and 140 V for 16 h. The gels were silver stained (25) and air dried, after which they were examined.
Analysis of DGGE profiles.
DGGE profiles were analyzed with GelCompar II 4.06 (Applied Maths, Kortrijk, Belgium) as described by Rademaker et al. (45). Images were normalized using the markers, and the Pearson correlation coefficient (r) for each pair of lanes within a gel was calculated as a measure of similarity between the community profiles (7). Cluster analysis was performed by applying the unweighted pair group method with mathematical averages (UPGMA) using the similarity matrix generated. Besides cluster analysis, we used constrained (i.e., canonical) ordination techniques to infer the extent to which each of the parameters investigated in the present study (i.e., sponge versus bulk water, sampling site and DNA extraction strategy) contributes to the observed variability in the DGGE profiles. To this end, the positions and intensities of each band in the DGGE profiles were first estimated using the software GelcomparII. Automatic assignment of band positions was checked manually and corrected when needed. After processing, each gel was represented by a datasheet containing a list of bands and their relative abundances (i.e., intensities) per profile (i.e., sample) (7). Detrended correspondence analysis (DCA) was first run for each of the analyzed gels using Canoco for Windows 4.5 (Microcomputer Power, Ithaca, NY). DCA provides an estimation of so-called “lengths of gradient” of the overall data set (PCR-DGGE ribotype variation, in the present study). This serves as a practical means of selecting the appropriate constrained ordination method for a given data set, i.e., either redundancy analysis (RDA; based on a linear response model of biological data to environmental parameters) or canonical correspondence analysis (CCA; based on unimodal responses of biological data to environmental parameters), as in Canoco for Windows 4.5 (32). Thus, based on the lengths of gradient obtained by DCA, we selected the appropriate constrained canonical model for each data set (i.e., DGGE gel). RDA and/or CCA were run in Canoco for Windows 4.5 using default parameters (32). Moreover, Shannon diversity indices were calculated for each DGGE profile analyzed with Canoco for Windows 4.5 and compared by using a two-sided t test.
Identification of dominant bands in Pseudomonas DGGE profiles of A. fulva.
Bands of interest were cut out from the Pseudomonas-specific DGGE profiles. The gel slices were transferred to Eppendorf tubes containing 50 μl of sterile MilliQ water, cut into smaller pieces to facilitate the elution of DNA, and incubated overnight at 4°C. To reamplify the excised bands, PCR was run on 8 μl of the eluted suspension as explained above. The resulting amplicons were analyzed via DGGE as described above. The amplicons were then loaded onto DGGE gels with the original community DNA sample to check for their electrophoretic mobility. Excised bands displaying the same melting behavior as the original bands in the community profiles were used as templates for a further PCR amplification (forward primer without GC clamp) prior to cloning. The reaction mixture (50 μl) contained 1.5 μl of eluted band suspension as DNA template (1 to 5 ng). PCR was carried out as detailed above. After purification, PCR products were ligated into pGEM-T Easy vectors (pGEM-T Vector System II; Promega, Madison, WI), followed by transformation of competent Escherichia coli JM109 cells (Promega) according to the manufacturer's instructions. White colonies were picked and heat lysed (15 min at 95°C). The lysed cells were subjected to PCR amplification with the primers F968-GC and R1401-1a, and the amplicons were compared to the community patterns on DGGE. Cloned inserts whose electrophoretic mobility on DGGE matched that of the original band were selected for sequencing in an Applied Biosystems 3130 genetic analyzer as explained below.
Cloning and sequencing of bacterial 16S rRNA genes amplified from sponge-extracted DNA.
Two clone libraries of bacterial 16S rRNA gene fragments were generated to cross-compare the two DNA extraction strategies. Nearly complete 16S rRNA gene fragments were obtained by using the primers F8 (5′-AGA GTT TGA TCM TGG CTC AG-3′) and R1492 (5′-GGT TAC CTT GTT ACG ACT T-3′). The PCR mixtures (25 μl) contained 1 μl of template DNA (1-5 ng), 1× Stoffel buffer, 0.2 mM dNTPs, 3.75 mM MgCl2, 0.1 bovine serum albumin, 2% (vol/vol) dimethyl sulfoxide, 0.2 μM concentrations of both primers, and 2.5 U of Taq DNA polymerase. After initial denaturation at 94°C for 5 min, 30 cycles of 1 min at 94°C, 1 min at 56°C, and 2 min at 72°C were performed, followed by a final extension for 10 min at 72°C.
Amplicons from the same DNA extraction method were pooled and purified by using a QIAquick PCR purification Kit (Qiagen GmbH, Hilden, Germany). The products were subjected to cloning as described above. The inserts were PCR amplified using primers T7 and SP6 (Promega) according to the supplier's recommendations. Cloned fragments of the right size (ca. 1,500 bp) were purified in Sephadex (GE Healthcare Bio-Science AB, Uppsala, Sweden) columns and subjected to sequencing with the chain termination method. The sequencing reaction (20 μl) contained 1× sequencing buffer (Applied Biosystems), 2 μl of BigDye terminator mix (Applied Biosystems), and a 0.1 μM concentration of primer F8. Ready-to-sequence DNA templates were isopropanol cleaned and resuspended in 10 μl of HiDi formamide (Applied Biosystems). After denaturation at 95°C for 3 min, sequencing of PCR products was performed with a 3130XL genetic analyzer (Applied Biosystems). The standard running procedures recommended by the manufacturer were used. The resulting 16S rRNA gene sequences were submitted to the EMBL database under accession numbers FM160745 to FM160915 and FM160916 to FM160942 for bacterial and Pseudomonas DGGE band clone libraries, respectively.
Assessment of 16S rRNA gene sequence diversity.
Totals of 79 and 94 sequences were obtained from, respectively, the 16S rRNA gene libraries representing the direct and the indirect DNA extraction methods. Prior to the analyses, all sequences were checked for chimera formation using Bellerophon v.3 (http://greengenes.lbl.gov). One sequence from each clone library was found to be chimeric and thus removed from further analyses. The remaining sequences were classified using the Ribosomal Database Project II (RDP) classifier with a confidence threshold of 70% (analysis run at the phylum level). Furthermore, the sequences were used to test whether the two libraries were different using the program LIBSHUFF (53). To assess bacterial diversity and richness, sequences were aligned by using CLUSTAL W. Pairwise sequence similarities were calculated with DNADIST (http://cmgm.stanford.edu/phylip/dnadist.html) using the Kimura two-parameter algorithm (29). Using the generated similarity matrix, the sequences were assigned to operational taxonomic units (OTUs) using DOTUR (52). The frequency data assigned to a “unique” OTU at the “species” (97% similarity criterion) and “genus” (95% similarity criterion) levels were used to yield rarefaction curves, Shannon diversity values, and Chao1 richness indices. Finally, we made an attempt to adequately place all of the clone sequences retrieved in the present study in their most likely phylogenetic context. Details on the extensive phylogenetic inference and tree drawing procedures used to this end are provided in the supplemental material.
RESULTS
Sponge sampling and identification.
Greenish and/or brownish body parts (each sample being about 5 to 10 cm long and 2 cm in diameter) were severed from replicate individual sponges collected in shallow waters (2- to 5-m depth) at both Caboclo Island and Tartaruga beach. After transport to the laboratory, all material was microscopically identified as belonging to A. fulva using the morphological criteria established by van Soest (26).
Enumeration of culturable bacteria.
The CFU counts of heterotrophic bacteria on R2A were generally low in the control bulk water at the two sites (range, log 2.91 to log 3.46/ml, giving an average of log 3.19/ml). These counts were statistically similar between the sites. In contrast, CFU counts in all sponge specimens sampled at both sites were significantly (P < 0.05) higher (average of log 5.23/g of fresh sponge) than those observed in bulk seawater. Specifically, log values of 5.09 and 5.38 CFU/g of sponge were found, on average, in the specimens collected at Caboclo Island and Tartaruga beach, respectively. These abundance values were statistically similar.
Electron microscopy of A. fulva-associated bacteria.
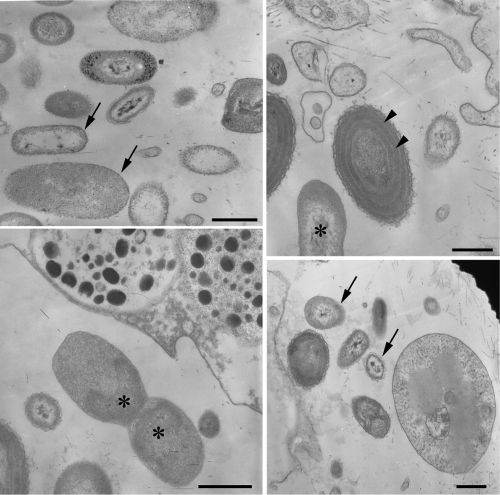
TEM of the A. fulva samples revealed a range of different morphotypes of bacteria within the sponge body (Fig. 1). Overall, by morphological criteria, large fractions of the observable cells were presumably of bacterial or archaeal origin. The different morphotypes varied in cell shape, surface structure, and the presence or not of an observable nucleoid (Fig. 1). Long and short rods were both observed (Fig. 1) with the usual, presumably gram-negative, surface structure. In some cases, we observed microorganisms containing an internal membrane system similar to those detected in the Cyanobacteria (Fig. 1).
FIG. 1.
TEM of A. fulva mesohyl. Several bacterial morphotypes (arrows), including membrane-bound nucleoid-containing bacteria (asterisks), can be observed. Microorganisms with an internal membrane system can also be observed (arrowheads). Scale bars, 0.5 μm.
PCR-DGGE analysis of bacterial communities in A. fulva. (i) Total bacterial DGGE profiles.
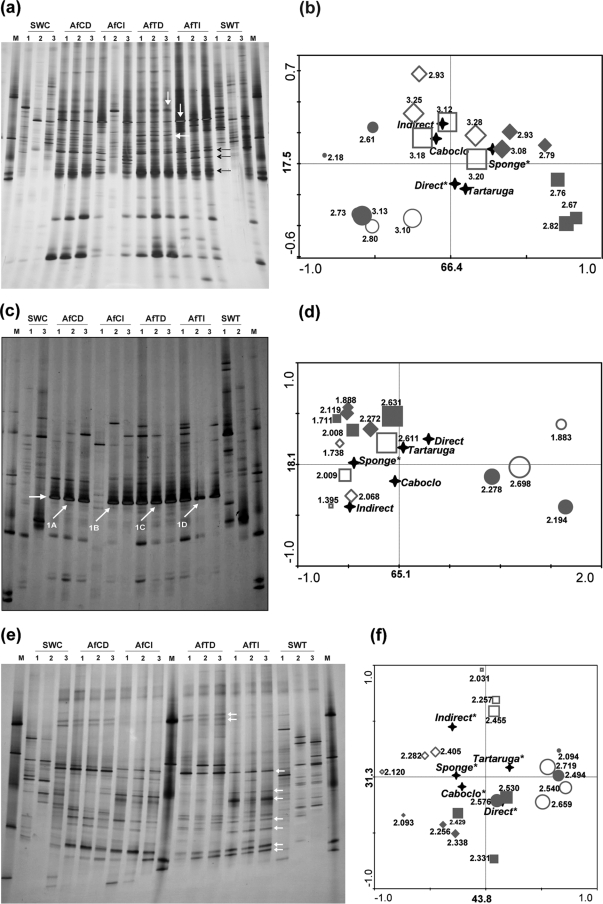
The A. fulva-associated bacterial DGGE profiles consistently encompassed a few (four to five) strong bands in addition to a large number of fainter bands (16 to 43) (Fig. 2a). The profiles were grossly similar across the replicates, whereas bulk water replicates revealed considerable variation. Exceptions to this rule were the A. fulva samples from Caboclo Island treated with the indirect DNA extraction method, whose DGGE fingerprints displayed considerable within-replicate variation (Fig. 2a). A drastic shift in community structure was apparent from a comparison of the profiles of sponge and seawater samples both by visual (Fig. 2a) and statistical means (i.e., cluster [see Fig. S1 in the supplemental material] and ordination analysis). Ordination via RDA of the DGGE band data and environmental variables is shown in Fig. 2b. The sponge and seawater samples were mainly discriminated along the horizontal axis of the diagram, which accounts for more than 65% of the overall variation. The main discriminating factor (P < 0.05) was the presence of the sponge, followed by the DNA extraction method. Indirect DNA extraction yielded a higher diversity (P < 0.05) than direct extraction, as evidenced by the calculated Shannon diversity indices (Fig. 2b), probably due more to higher equitability among bands than to a higher number of ribotypes observed. RDA performed on the basis of the DGGE profiles of the sponge-associated communities revealed that DNA extraction methodology significantly (P < 0.05) affected the relative abundance and distribution of the ribotypes (data not shown). This was corroborated by the subclustering of the sponge samples as a function of DNA extraction strategy along the vertical axis of the ordination diagram (Fig. 2). The influence of sampling site on the structure of the A. fulva-associated bacterial communities was not significant at P < 0.05.
FIG. 2.
PCR-DGGE 16S rRNA gene fingerprints of A. fulva and seawater DNA samples generated with “total-community” bacterial primers (a) and specific primer systems for Pseudomonas (c) and Actinobacteria (e). The arrows in panels a and e indicate bands that occur in the sponge profiles but not in water. Some of these bands, especially those in panel e, are characteristic of a specific DNA extraction strategy. The arrows in panel c show bands that were cloned and sequenced. Corresponding ordination biplots of PCR-DGGE fingerprints and qualitative environmental variables are shown in panels b, d, and f. Symbols: ⧫, A. fulva from Caboclo Island extracted directly (AfCD); ⋄, A. fulva from Caboclo Island extracted indirectly (AfCI); ▪, A. fulva from Tartaruga beach extracted directly (AfTD); □, A. fulva from Tartaruga beach extracted indirectly (AfTI); •, surrounding seawater from Caboclo Island (SWC); ○, surrounding seawater from Tartaruga beach (SWT). Shannon's indices of diversity are shown for each sample next to symbols. Symbol sizes correspond to their estimated diversity indices. Labels displayed on the diagram axes refer to the percentage variations of PCR-DGGE ribotypes; environment correlation accounted for the respective axis. The “star” symbols represent the centroid positions of the environmental variables in the diagram. Variables that significantly (P < 0.05) influence the bacterial community composition are indicated by an asterisk.
(ii) Pseudomonas DGGE profiles.
The profiles of the A. fulva-associated microbial assemblages obtained with Pseudomonas primers revealed moderate levels of band richness (range, 10 to 24 bands), while the corresponding bulk water samples revealed slightly higher richness levels (17 to 28 bands) (Fig. 2c). As observed for the total bacterial community, a high degree of variability was detected among the DGGE profiles generated from seawater replicates. Reassuringly, a clear difference was observed between the seawater and the sponge DGGE profiles. UPGMA-assisted cluster analysis revealed two main clusters, one encompassing all DGGE profiles generated from A. fulva, irrespective of DNA extraction strategy and sampling site, and the other encompassing all seawater-derived profiles. The similarity level between the two clusters was only 10% (see Fig. S1 in the supplemental material). CCA applied on the same data set confirmed that the only factor that significantly influenced the DGGE profiles was the presence or not of the sponge (P < 0.05). This consistent selection for a distinct community by A. fulva is evident from the ordination diagram (Fig. 2d), in which 65% of the total variability is represented by the horizontal axis, which correlates with the factor “sponge.” In fact, the differentiation between sponge and seawater samples was primarily caused by the presence of a dominant band (arrows in Fig. 2c) in all A. fulva-derived profiles, whereas this band was not found in any seawater profiles. Given the apparent enrichment of this ribotype in A. fulva DGGE profiles, sequence analysis was carried out (see below).
(iii) Actinobacterial DGGE profiles.
The actinobacterial DGGE profiles showed moderate band richness (range, 14 to 21 bands) across the A. fulva samples and slightly higher richness in the seawater samples (12 to 28 bands) (Fig. 2e). As expected, the A. fulva-associated patterns clustered together and, apart from the patterns derived from corresponding seawater, with <10% similarity between the two clusters. Subclusters that corresponded to the factors DNA extraction and sampling site were also observed (see Fig. S1 in the supplemental material). Furthermore, ordination via RDA, representing 75% of the variation (Fig. 2d), confirmed the cluster analysis, in that all of the factors analyzed—i.e., sponge type, DNA extraction methodology, and site—significantly affected the actinobacterial profiles (P < 0.05). Especially remarkable was the high number of bands found in association with A. fulva that were detected or not as a function of DNA extraction methodology (Fig. 2e). Based on all DGGE profiles, the factors sponge and sampling site significantly affected the diversity of actinobacterial ribotypes, as assessed by a comparative analysis of Shannon diversity indices (Fig. 2f). DNA extraction methodology did not impact the apparent actinobacterial diversity in A. fulva (Fig. 2f).
Analysis of sequences of dominant DGGE bands generated with the Pseudomonas primers.
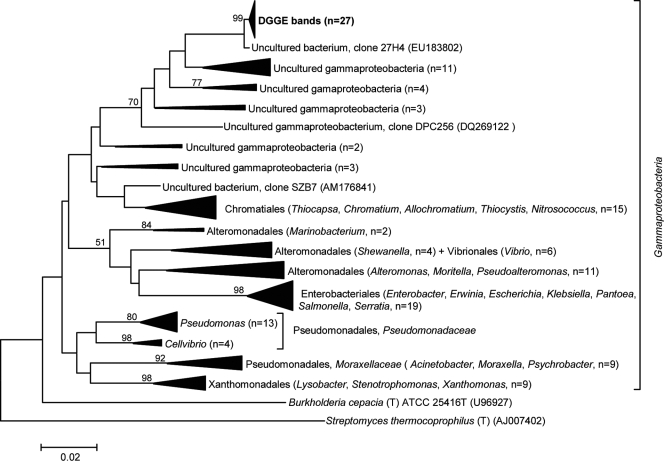
The labeled arrows in Fig. 2c (1A, 1B, 1C, and 1D) show the dominant DGGE bands, all of which display the same melting behavior, that were subjected to sequencing. The bands occurred in all A. fulva profiles and were absent from those from seawater. In total, 27 clone inserts were sequenced and found to be all highly similar, with only 1 to 2% sequence dissimilarity among them. They were assigned, at an 80% confidence level, to the class Gammaproteobacteria using the RDP classifier tool. A comprehensive phylogenetic analysis of these sequences with those of selected Gammaproteobacteria revealed that they represent a distinct gammaproteobacterial lineage of which no cultured representatives have been found thus far (Fig. 3). Unexpectedly, these sequences formed a concise cluster within a larger group of gammaproteobacterial clones with the closest relatives in the order Chromatiales. Interestingly, the 16S rRNA gene with the closest match to these sequences (clone 27H4, accession no. EU183802) (Fig. 3) was retrieved from a Great Barrier Reef sponge species, namely, Rhopaloeides odorabile (66).
FIG. 3.
Evolutionary relationships of 145 gammaproteobacterial 16S rRNA sequences. Twenty-seven sequences retrieved from a dominant “Pseudomonas” band observed to be enriched in A. fulva DGGE profiles (Fig. 2c) were included in phylogenetic inference. The evolutionary history was inferred by using the neighbor-joining method (49). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed by using the Kimura two-parameter method (29) and are in the units of the number of base substitutions per site (note the scale bar). Bootstrap values (1,000 repetitions) greater than 50% are shown on three nodes. There were a total of 413 aligned nucleotide positions in the final data set. The 16S rRNA gene sequences of Burkholderia cepacia (T) ATCC 25416T (Betaproteobacteria) and Streptomyces thermocoprophilus (T) (Actinobacteria) served as outgroups.
Analysis of bacterial clone libraries obtained from A. fulva-derived DNA.
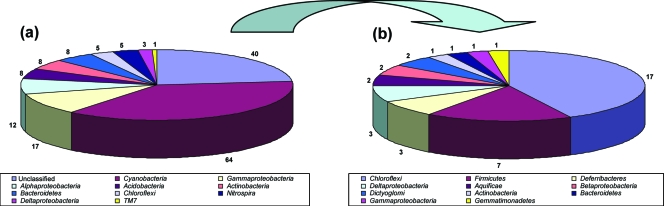
Two 16S rRNA gene clone libraries were generated from A. fulva-derived DNA, representing the two methods of DNA extraction. After sequence quality and chimera checks, totals of 78 and 93 sequences were obtained from the direct and indirect methods, respectively. Sequences were first subjected to library shuffling analysis using LIBSHUFF (53). Although not significant (P < 0.05), the library obtained with the direct DNA extraction method revealed higher diversity at the “species” and “genus” cutoff levels than the one obtained with the indirect method (see Fig. S2 in the supplemental material). Given the nonsignificant difference, we decided to combine both libraries to describe the diversity and composition of the bacterial communities associated with A. fulva. Using the RDP classifier at a confidence threshold of 70%, the majority (i.e., all except 40) of the sequences could be assigned to recognized bacterial phyla, with the Cyanobacteria (37.4%), Chloroflexi (12.8%), Gammaproteobacteria (10.5%), and Alphaproteobacteria (7%) being the most representative library constituents (Fig. 4a). In total, as assessed with the RDP classifier, we found 12 bacterial phyla and one candidate division, i.e., TM7, associated with A. fulva. The 40 sequences that were initially not classifiable at phylum level could later be assigned to the Actinobacteria, Aquificae, Bacteroidetes, Chloroflexi, Deferribacteres, Dictyoglomi, Firmicutes, Gemmatimonadetes, Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria using lower confidence limits (<70%) (Fig. 4b). To assess the closest phylogenetic relatives of these sequences, BLAST-N was used with the nonredundant NCBI database (2). Strikingly, 30 of the 40 sequences were similar to database entries at low levels of similarity (i.e., <97%). Among them, respectively 20, 8, and 2 sequences were affiliated with bacterial 16S rRNA genes found with marine sponges (14, 21, 51, 56), seawater (9, 50, 55), and with a coral reef (48).
FIG. 4.
Phylum distribution of bacterial 16S rRNA gene sequences based on the Classifier tool of the RDP, version 9.0. Bacterial sequences classified at the phylum level with a 70% CI (a) and unclassified sequences at 70% CI and reclassified at ≤69% CI (b) are shown.
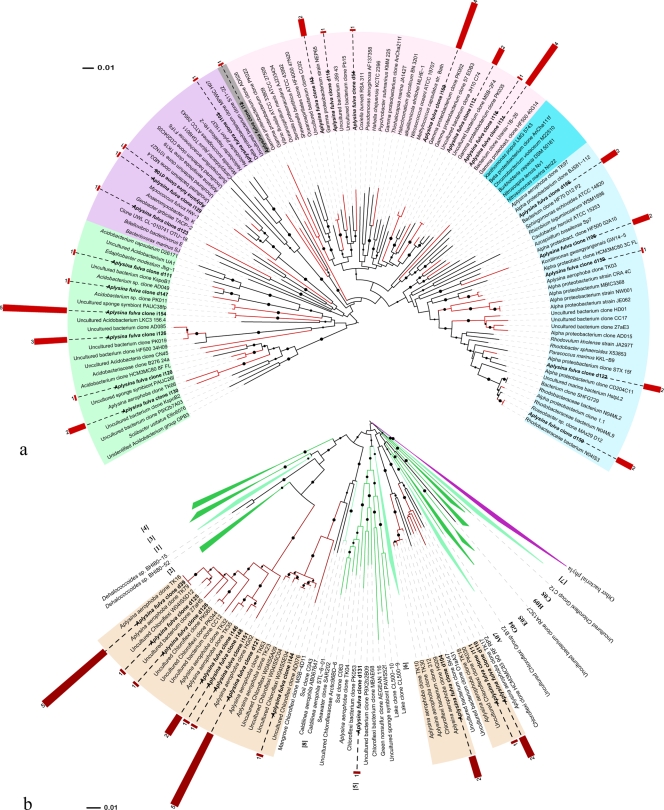
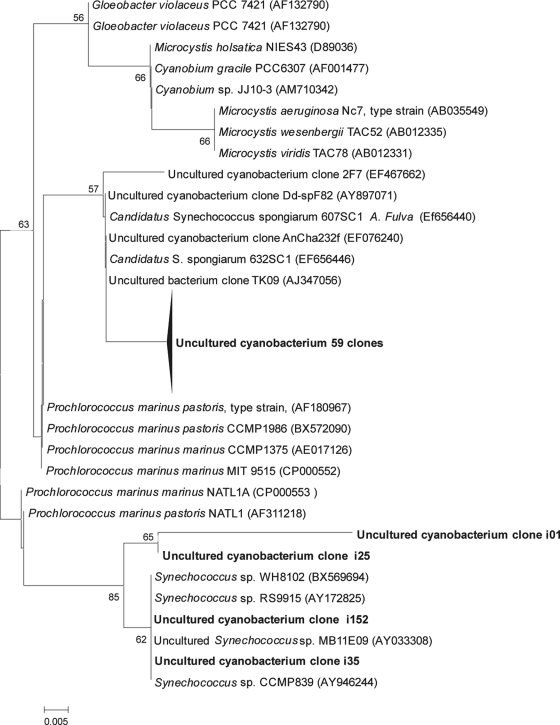
In-depth phylogenetic assessments were performed for the four most abundant phyla and the Acidobacteria (Fig. 5 and 6), the fifth most abundant phylum, along with the Actinobacteria and Bacteroidetes. The analyses were helpful in demonstrating that A. fulva-derived clones are representative of uncultured bacterial populations (Fig. 5 and 6). Clones d122 and d150 in the Alphaproteobacteria clade (Fig. 5a) correspond to OTUs that are an exception to this contention, since they were found in close affiliation with culturable bacteria of the Rhodobacteraceae. Phylogenetic inference also allowed more accurate affiliation of sequences that could not be classified at a 70% confidence interval (CI). This was the case of the OTUs represented by the clones i130 and i126 in the Acidobacteria clade (Fig. 5a), which were, initially, tentatively affiliated with the Beta- and Deltaproteobacteria, respectively, at rather low levels of confidence (35 and 32%). Of note is the extent of diversity found within the Gammaproteobacteria, Acidobacteria (Fig. 5a), and Chloroflexi (Fig. 5b). Strikingly, after extensive phylogenetic assessments, we found as-yet-undescribed, uncultured Chloroflexi phylogenetic clusters that exclusively comprise clones obtained from marine sponges (Fig. 5b). The majority of the cyanobacterial sequences (59 of 63) formed a unique cluster in the tree that contained mainly representatives of the Prochlorales and Chroococcales (Fig. 6). The sequences displayed a high level of conservation, with only 6 of 59 sequences showing slight diversification (data not shown). The closest relatives were 16S rRNA gene clones of uncultured cyanobacteria that resemble the candidate species “Synechococcus spongiarum” (Fig. 6). However, relatedness was still moderate, with levels of similarity of 96 to 97%.
FIG. 5.
Phylogenetic analysis of bacterial 16S rRNA genes retrieved from A. fulva. The trees shown were inferred and drawn as explained in the legend to Fig. 3. Solid circles next to tree branches correspond to bootstrap values higher than 50%. A. fulva-derived clones are indicated in boldface, with their respective tree leaves marked in red. Trees display one representative clone per OTU. A single OTU embraces, here, all of the clone sequences sharing at least 99% similarity. Red bars next to A. fulva clone labels indicate the number of clones belonging to the corresponding OTU. (a) Evolutionary relationships of 125 taxa representative of the Acidobacteria (light green) and the alpha (light blue), beta (blue), gamma (light purple), and delta (purple) classes of Proteobacteria. There were 468 nucleotide positions in the final data set. (b) Phylogeny of the Chloroflexi as delineated by 167 representative taxa. All previously described Chloroflexi subdivisions—as designated by Rappé and Giovanonni (45a) (clades 1 to 8) and Costello and Schmidt (8a) (clades C12, C05, H09, E05, G04, B12, and A07)—are shown and appear, mostly, as collapsed clades. Clades composed exclusively by uncultured (dark green) or containing cultured (light green) representatives are differentiated. Four novel Chloroflexi subdivisions represented solely by sponge-derived uncultured bacteria are highlighted (colored ranges on sequence labels). There were a total of 528 aligned nucleotide positions in the final data set.
FIG. 6.
16S rRNA-based phylogeny of sponge-associated Cyanobacteria. The tree was inferred and drawn as explained in the legend to Fig. 3. Labels in boldface represent A. fulva-derived Cyanobacteria sequences obtained in the present study. The accession numbers of the sequences obtained from the public databank are given in parentheses. There were a total of 612 aligned nucleotide positions in the final data set.
We also used our clone libraries to generate rarefaction curves via DOTUR (52), using 97 and 95% cutoff criteria for grouping OTUs at the “species” and “genus” levels, respectively. In both cases, rarefaction curves did not reach a ‘plateau’ (see Fig. S3 in the supplemental material). The Chao1 indicator of richness revealed that 47.8 and 65%, respectively, of the total “species” and “genera” hypothetically present in A. fulva samples were assessed by the analyses.
DISCUSSION
In this study, the diversity, structure, and composition of the bacterial communities associated with the marine sponge A. fulva were assessed using culture-dependent (plate counting) and -independent (TEM, PCR-DGGE, and clone-and-sequencing) approaches. A. fulva, which belongs to the demosponges, is found on open reefs at the southern Brazilian seashore up to the gulf of Mexico (38). Samples from the mesohyl of A. fulva subjected to TEM revealed high abundances of bacterial cells, encompassing a range of different morphotypes. In addition, enumerations of culturable heterotrophs showed that the bacterial numbers obtained from A. fulva exceeded those observed in seawater by roughly 2 orders of magnitude. Based on both approaches, we consider A. fulva to belong to the HMA sponges (23).
To our knowledge, this is the first study that addresses the diversity and composition of bacterial communities associated with a marine sponge using two different strategies for DNA extraction. In studies in other microbial habitats, such as the soil, riverbed, or marine sediment and the rhizosphere, the use of direct or indirect strategies for the extraction of “whole-community” DNA from environmental samples has been a matter of debate (18, 19). Direct extraction methods commonly provide higher DNA yields (31). Alternatively, DNA extracts of higher purity are obtained through the use of indirect (cell extraction first) approaches (10, 19). Ideally, regardless of the strategy used, DNA representative of the microbial assemblages present in the environmental sample are obtained (65). In the indirect extraction method, maintenance of microbial cell integrity prior to DNA isolation is important. Therefore, in this study, no harsh attempts to immediately inactivate the microbial communities in situ (e.g., freezing) were made, and samples were instead transported to the laboratory on ice baths. We observed a clear effect of DNA extraction method on the DGGE profiles obtained for the total bacterial community (in terms of diversity and ribotype composition) and Actinobacteria (in terms of ribotype composition). In both cases, as determined by constrained ordination analysis, DNA extraction methodology influenced the overall variation to a larger extent than did sampling site. The observed higher diversity of bacterial DGGE ribotypes in indirect DNA extracts could well be explained by a better representation, in this method, of intracellular symbionts as well as of symbionts not easily extracted from the sponge matrix by the direct extraction method. However, LIBSHUFF analysis of the bacterial 16S rRNA gene libraries constructed using both DNA extraction strategies revealed insignificant compositional differences. Similarly, DGGE and clone library analyses also diverged with respect to assessing the bacterial diversity in A. fulva as a function of DNA extraction methodology. Whereas bacterial DGGE fingerprints were indicative of higher diversity obtained with the indirect methodology, the opposite was observed by sequence analysis of bacterial 16S rRNA gene clones. Even though not significant, the number of OTUs found on the basis of the direct DNA extraction method was slightly higher than that obtained when the indirect method was used. The discrepancy between the two types of analysis, observed for both community composition and community diversity, might result from the use of the different primer sets in each of the techniques, as shown in previous investigations of molecular bacterial diversity (4). Our observations highlight the importance of using complementary approaches for a comprehensive investigation of the Bacteria in natural settings.
Regardless of the DNA extraction strategy used, the total bacterial DGGE patterns generated from the sponge samples suggested a strong selection of distinct communities compared to those from seawater samples. This trend was observed at both sampling sites and for both “universal” and group-specific DGGE profiles, a finding that is strongly indicative of a sponge-mediated selective process. Similar effects have been observed for other sponges (57, 58, 60). For instance, the bacterial DGGE profiles from Ircinia felix, Aplysina cauliformis, and Niphates erecta collected at two distinct sites in Key Largo, FL, revealed dissimilar banding patterns compared to seawater samples (69). Similarly, the microbial communities associated with Cymbastela concentrica, collected in the southeastern Australian coast, were found to clearly differ from those of the surrounding seawater (58).
Actinobacteria are notable with respect to their ability to produce diverse natural products, especially cytotoxic compounds, which have been reported to occur in many marine sponge species (6, 28, 56). In turn, metabolically versatile Pseudomonas are known for producing a range of secondary metabolites with potential biotechnological applications, for instance in agriculture or in the pharmaceutical industry (20), but knowledge of the occurrence and activity of Pseudomonas spp. in marine sponges is still rudimentary. Since microbes are believed to play a role in the complex biochemistry of marine sponges (13, 41, 61), supposedly aiding the host organism with its extraordinary defense mechanisms, accessing biotechnologically promising bacterial targets in the sponge environment is a primordial task. Here, we made an attempt to quickly unravel the structure of actinobacterial and Pseudomonas assemblages found in association with A. fulva through PCR-DGGE means. The abundance of these organisms is likely to be low to moderate in A. fulva, since we found no sequence belonging to Pseudomonas and only nine sequences (ca. 5%) belonging to the Actinobacteria within the clone libraries analyzed (n = 171). Indeed, Pseudomonas sequences are barely found in sponge-derived clone libraries. Previous estimates of actinobacterial abundance in sponges—as determined by their proportions in 16S rRNA gene libraries—are contradictory and rank the group both as a minor (i.e., <2% of the obtained sequences) (34, 35) and as a major (i.e., >20%) (37) constituent of sponge-associated bacterial communities. Given that actinobacteria typically have robust, hard-to-lyse cell walls and may also be present in the form of spores, it is not yet clear whether such discrepancies reflect actual changes in bacterial composition in the different sponge species investigated or are affected by the sample processing methodologies implemented in each study. In the present work, the use of group-specific PCR-DGGE permitted analysis of these less-abundant bacterial populations in sponge-associated communities. Revealing and, in some cases, surprising outcomes resulted from such assessments. The actinobacterial and Pseudomonas DGGE profiles revealed moderate diversity compared to that ascertained with “total-community” bacterial primers. Besides the typical water versus sponge differentiation of the profiles, the apparent community structure of the Actinobacteria was influenced by sampling site and DNA extraction method. It appears, therefore, that the efficiency of direct and indirect methods in sampling sponge-associated actinobacteria differs. Moreover, among the nine actinobacterial sequences obtained, eight were most closely affiliated with actinobacterial sequences from sponges, a finding that supports a selective sponge effect on particular actinobacterial types, as also observed by DGGE fingerprinting. We thus posit that a specific mechanistic (interactive) role of these bacteria in the sponge is likely. Such a role might well be in host protection. Further studies are needed to specifically address this issue.
Surprisingly, sequences obtained from the bands excised from the Pseudomonas DGGE profiles were affiliated with uncultured Gammaproteobacteria beyond the Pseudomonadales. Similar observations were obtained by Gomes et al. (17) while investigating mangrove sediment using the same PCR system. The most likely explanation for this is that the primer set used for the first PCR amplification enriches 16S rRNA gene templates from these nonsoil habitats that are different from pseudomonads. These particular environments may contain true Pseudomonas types at rather low levels, and these potentially cross-amplifying types at higher level, thus resulting in reduced-specificity amplification of the abundant templates (17). The PCR-DGGE system used in the present study (36) was clearly suitable for the investigation of Pseudomonas in soil and rhizosphere (8). However, a more adequate tool is needed to more specifically track true pseudomonads in the sponge environment. Interestingly, the sequenced bands appeared in all A. fulva samples, regardless of the DNA extraction method and sampling site, and resembled a unique sequence that was not present in seawater. As observed for some of the actinobacterial ribotypes, these results strengthen the hypothesis that a selective process is exerted by A. fulva, in which the organism detected might have an ecological role, potentially symbiotic. The sequenced bands resembled sequences of members of the Chromatiales. This order contains purple sulfur bacteria that are able to carry out anoxygenic photosynthesis using hydrogen sulfide as the electron donor, and it occurs in marine and freshwater environments (40, 44). Fixation of molecular nitrogen has also been demonstrated in many Chromatiales species.
Clone library analysis revealed that the bacterial community associated with A. fulva was predominantly composed of Cyanobacteria, followed by Chloroflexi and Gamma- and Alphaproteobacteria. Sequences resembling those of the phyla Aquificae, Deferribacteres, Dictyoglomi, and candidate division TM7 are described here for the first time in association with sponges. Particularly remarkable is the uniqueness of many bacterial phylotypes found in our libraries. For instance, we identified 30 sequences that showed <97% identity with publicly available database sequences, most of which were phylogenetically related to sponge-derived sequences retrieved elsewhere. Moreover, of 64 cyanobacterial sequences, 59 formed a unique phylogenetic cluster similar (96 to 97%) to the “Candidatus Synechococcus spongiarum” 16S rRNA gene sequence. This candidate species apparently occurs in a variety of sponge species distributed worldwide (54, 63). Thiel et al. (60) observed that a large proportion of sequences (39%) of an endosome-derived clone library was affiliated with Synechococcus species. In addition, “Candidatus Synechococcus spongiarum” has been reported to be transferred from parental Chondrilla australiensis to the next generation via vertical transmission (62). The high abundance of this organism in A. fulva suggests that vertical transmission also occurs in this sponge. These considerations raise the issue of this species being a “true” endosymbiont. Although Cyanobacteria may exert essential functions in sponges (12), their role and level of putative symbiosis is still to be revealed. In addition, we revealed novel Chloroflexi 16S rRNA gene clusters that are composed solely of uncultured, sponge-derived clones. Although some of the A. fulva clones within these clusters resemble sequences obtained from other sponges elsewhere, some others appear to be unique (e.g., clones d110, i111, and i151 in Fig. 5b). Finally, four of the sequences that fell in the class Alphaproteobacteria are noteworthy, since they are affiliated, at ca. 90% identity, with strain NW001, a true sponge symbiont that occurs in high abundance in the mesohyl of Rhopaloeides odorabile (67). Curiously, in spite of its high dominance among culturable bacteria from this sponge, sequences of strain NW001 could not be detected in sponge-derived DNA (67). Analysis of larvae of the sponge Mycale laxissima revealed that NW001 types, which are also dominant in this sponge, can be transferred between generations through sponge gametes (11). In addition, sequences closely affiliated with that of strain NW001 were retrieved from nine different sponge species, including Aplysina aerophoba, distributed worldwide (11, 22). Webster and Hill (67) postulated that strain NW001 may be implicated in sponge health, given the fact that it was absent in two diseased R. odorabile specimens. However, the mechanisms underlying such a role remain to be elucidated. The recovery of sequences from A. fulva that affiliate with that of strain NW001, though at rather low levels (as seen in Fig. 5a), is indicative of the wide occurrence of a broad and culturable sponge-associated bacterial group that might be important for sponge health.
This study provides initial insights into the diversity, structure, and composition of bacterial communities associated with A. fulva in the south Atlantic. Our data provide sound support for the contention that a highly distinct bacterial community occurs in A. fulva and that most of its members share relatedness at various degrees with other sponge-specific bacterial symbionts discovered worldwide. First, the most abundant sequence type detected in the A. fulva clone libraries resembled that of “Candidatus Synechococcus spongiarum” but was not identical to any other sequence reported to date. Second, novel, thus far “sponge-associated,” Chloroflexi clusters were found, with some clones obtained from A. fulva being unique. Third, sponge-specific gammaproteobacterial sequences related to the Chromatiales were obtained from a dominant band in a “Pseudomonas-specific” DGGE gel. These sequences were unique and hence the underlying, as-yet-uncultured, organism is reported here for the first time. A low level of resemblance of these sequences to a sequence obtained from R. odorabile in Australia was found (66). Fourth, another 30 distinct (below 97% match with database entries) sequences were found, most of them related to sponge-derived bacterial sequences distributed over diverse bacterial phyla. None of these sequences has shown any culturable representative. The present study substantially extends our current knowledge of the diversity of the sponge-associated microbiota. Further analysis of the potential role, biogeography and degree of endemism and species specificity of these bacterial types needs to be undertaken. Metagenomic exploration of these novel genetic reservoirs, along with dedicated efforts to domesticate the relevant members of the A. fulva-associated microbiota, are promising strategies that will shed light on the role of such bacteria in their interaction with the host.
Supplementary Material
Acknowledgments
R.C. was financed by the Soil Biotechnology Foundation, Wageningen, The Netherlands. A.S.R., E.H., and U.L. were supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico and the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro.
We thank Pablo Hardoim for his support in the statistical handling of data.
Footnotes
Published ahead of print on 20 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Althoff, K., C. Schütt, R. Steffen, R. Batel, and W. E. G. Müller. 1998. Evidence for a symbiosis between bacteria of the genus Rhodobacter and the marine sponge Halichondria panicea: harbor also for putatively toxic bacteria? Mar. Biol. 130:529-536. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blunt, J. W., B. R. Copp, M. H. G. Munro, P. T. Northcote, and M. R. Prinsep. 2005. Marine natural products. Nat. Prod. Rep. 22:15-61. [DOI] [PubMed] [Google Scholar]
- 4.Brons, J. K., and J. D. van Elsas. 2008. Analysis of bacterial communities in soil by use of denaturing gradient gel electrophoresis and clone libraries, as influenced by different reverse primers. Appl. Environ. Microbiol. 74:2717-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brusca, R. C., and G. J. Brusca. 2002. Phylum Porifera: the sponges, p. 179-208. In A. D. Sinauer (ed.), Invertebrates. Sinauer Associates, Inc., Cambridge, MA.
- 6.Bull, A. T., and J. E. M. Stach. 2007. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 15:491-499. [DOI] [PubMed] [Google Scholar]
- 7.Costa, R., N. C. M. Gomes, E. Krögerrecklenfort, K. Opelt, G. Berg, and K. Smalla. 2007. Pseudomonas community structure and antagonistic potential in the rhizosphere: insights gained by combining phylogenetic and functional gene-based analyses. Environ. Microbiol. 9:2260-2273. [DOI] [PubMed] [Google Scholar]
- 8.Costa, R., J. F. Salles, G. Berg, and K. Smalla. 2006. Cultivation-independent analysis of Pseudomonas species in soil and in the rhizosphere of field-grown Verticillium dahliae host plants. Environ. Microbiol. 8:2136-2149. [DOI] [PubMed] [Google Scholar]
- 8a.Costello, E. K., and S. K. Schmidt. 2006. Microbial diversity in alpine tundra wet meadow soil: novel Chloroflexi from a cold, water-saturated environment. Environ. Microbiol. 8:1471-1486. [DOI] [PubMed] [Google Scholar]
- 9.DeLong, E. F., C. M. Preston, T. Mincer, V. Rich, S. J. Hallam, N. U. Frigaard, A. Martinez, M. B. Sullivan, R. Edwards, B. R. Brito, S. W. Chisholm, and D. M. Karl. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496-503. [DOI] [PubMed] [Google Scholar]
- 10.Duarte, G. F., A. S. Rosado, L. Seldin, A. C. Keijzer-Wolters, and J. D. van Elsas. 1998. Extraction of rRNA and genomic DNA from soil for studying the diversity of the indigenous bacterial community. J. Microbiol. Methods 32:21-29. [Google Scholar]
- 11.Enticknap, J. J., M. Kelly, O. Peraud, and R. T. Hill. 2006. Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microbiol. 72:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erwin, P. M., and R. W. Thacker. 2007. Incidence and identity of photosynthetic symbionts in Caribbean coral reef sponge assemblages. J. Mar. Biol. Assoc. UK 87:1683-1692. [Google Scholar]
- 13.Fieseler, L., U. Hentschel, L. Grozdanov, A. Schirmer, G. P. Wen, M. Platzer, S. Hrvatin, D. Butzke, K. Zimmermann, and J. Piel. 2007. Widespread occurrence and genomic context of unusually small polyketide synthase genes in microbial consortia associated with marine sponges. Appl. Environ. Microbiol. 73:2144-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fieseler, L., M. Horn, M. Wagner, and U. Hentschel. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich, A. B., I. Fischer, P. Proksch, J. Hacker, and U. Hentschel. 2001. Temporal variation of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 38:105-113. [Google Scholar]
- 16.Friedrich, A. B., H. Merkert, T. Fendert, J. Hacker, P. Proksch, and U. Hentschel. 1999. Microbial diversity in the marine sponge Aplysina cavernicola (formerly Verongia cavernicola) analyzed by fluorescence in situ hybridization (FISH). Mar. Biol. 134:461-470. [Google Scholar]
- 17.Gomes, N. C. M., L. R. Borges, R. Paranhos, F. Pinto, L. C. S. Mendonca-Hagler, and K. Smalla. 2008. Exploring the diversity of bacterial communities in sediments of urban mangrove forests. FEMS Microbiol. Ecol. 66:96-109. [DOI] [PubMed] [Google Scholar]
- 18.Gomes, N. C. M., R. Costa, and K. Smalla. 2004. Simultaneous extraction of DNA and RNA from bulk and rhizosphere soil, p. 59-179. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. L. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 19.Griffiths, R. I., M. Manefield, A. S. Whiteley, and M. J. Bailey. 2004. DNA and RNA extraction from soil, p. 149-158. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. L. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 20.Haas, D., and G. Défago. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307-319. [DOI] [PubMed] [Google Scholar]
- 21.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hentschel, U., M. Schmid, M. Wagner, L. Fieseler, C. Gernert, and J. Hacker. 2001. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 35:305-312. [DOI] [PubMed] [Google Scholar]
- 23.Hentschel, U., K. M. Usher, and M. W. Taylor. 2006. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55:167-177. [DOI] [PubMed] [Google Scholar]
- 24.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heuer, H., J. Wieland, J. Schönfeld, A. Schönwäalder, N. C. M. Gomes, and K. Smalla. 2001. Bacterial community profiling using DGGE and TGGE analysis, p. 177-190. In P. Rouchelle (ed.), Environmental molecular microbiology: protocols and applications. Horizon Scientific Press, Hymondham, United Kingdom.
- 26.Hooper, N. J. A., and R. W. M. van Soest. 2002. System Porifera: a guide to the classification of sponges. Kluwer Academic/Plenum Publishers, New York, NY.
- 27.Kim, T. K., and J. A. Fuerst. 2006. Diversity of polyketide synthase genes from bacteria associated with the marine sponge Pseudoceratina clavata: culture-dependent and culture-independent approaches. Environ. Microbiol. 8:1460-1470. [DOI] [PubMed] [Google Scholar]
- 28.Kim, T. K., M. J. Garson, and J. A. Fuerst. 2005. Marine actinomycetes related to the ‘Salinospora’ group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ. Microbiol. 7:509-518. [DOI] [PubMed] [Google Scholar]
- 29.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide-sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 30.Lafi, F. F., M. J. Garson, and J. A. Fuerst. 2005. Culturable bacterial symbionts isolated from two distinct sponge species (Pseudoceratina clavata and Rhabdastrella globostellata) from the Great Barrier Reef display similar phylogenetic diversity. Microb. Ecol. 50:213-220. [DOI] [PubMed] [Google Scholar]
- 31.Leff, L. G., J. R. Dana, J. V. McArthur, and L. J. Shimkets. 1995. Comparison of methods of DNA extraction from stream sediments. Appl. Environ. Microbiol. 61:1141-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lepš, J., and P. Šmilauer. 1999. Multivariate analysis of ecological data using CANOCO. Cambridge University Press, Cambridge, United Kingdom.
- 33.Li, C.-W., J.-Y. Chen, and T.-E. Hua. 1998. Precambrian sponges with cellular structures. Science 279:879-882. [DOI] [PubMed] [Google Scholar]
- 34.Li, Z. Y., L. M. He, J. Wu, and Q. Jiang. 2006. Bacterial community diversity associated with four marine sponges from the South China Sea based on 16S rDNA-DGGE fingerprinting. J. Exp. Mar. Biol. Ecol. 329:75-85. [Google Scholar]
- 35.Li, Z. Y., and Y. Liu. 2006. Marine sponge Craniella austrialiensis-associated bacterial diversity revelation based on 16S rDNA library and biologically active Actinomycetes screening, phylogenetic analysis. Lett. Appl. Microbiol. 43:410-416. [DOI] [PubMed] [Google Scholar]
- 36.Milling, A., K. Smalla, F. X. Maidl, M. Schloter, and J. C. Munch. 2004. Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil 266:23-39. [Google Scholar]
- 37.Montalvo, N. F., N. M. Mohamed, J. J. Enticknap, and R. T. Hill. 2005. Novel actinobacteria from marine sponges. Antonie van Leeuwenhoek 87:29-36. [DOI] [PubMed] [Google Scholar]
- 38.Muricy, G., and E. Hadju. 2006. Porifera Brasilis: guia de classificação das esponjas marinhas do sudeste do Brasil. Eclesiarte Editora, Rio de Janeiro, Brazil.
- 39.Nuñez, C. V., E. V. R. de Almelda, A. C. Granato, S. O. Marques, K. O. Santos, F. R. Pereira, M. L. Macedo, A. G. Ferreira, E. Hajdu, U. S. Pinheiro, G. Muricy, S. Peixinho, C. J. Freeman, D. F. Gleason, and R. G. S. Berlinck. 2008. Chemical variability within the marine sponge Aplysina fulva. Biochem. Syst. Ecol. 36:283-296. [Google Scholar]
- 40.Ollivier, B., P. Caumette, J. L. Garcia, and R. A. Mah. 1994. Anaerobic bacteria from hypersaline environments. Microbiol. Rev. 58:27-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piel, J. 2004. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 21:519-538. [DOI] [PubMed] [Google Scholar]
- 42.Piel, J., D. Q. Hui, G. P. Wen, D. Butzke, M. Platzer, N. Fusetani, and S. Matsunaga. 2004. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 101:16222-16227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pile, A. J., M. R. Patterson, and J. D. Witman. 1996. In situ grazing on plankton <10 μm by the boreal sponge Mycale lingua. Mar. Ecol. Prog. Ser. 141:95-102. [Google Scholar]
- 44.Proctor, L. M. 1997. Nitrogen-fixing, photosynthetic, anaerobic bacteria associated with pelagic copepods. Aquat. Microb. Ecol. 12:105-113. [Google Scholar]
- 45.Rademaker, J., F. J. Louws, J. Versalovic, and F. J. de Bruijn. 2004. Computer-assisted analysis of molecular fingerprint profiles and database construction, p. 1397-1446. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 45a.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 46.Reiswig, H. M. 1974. Water transport, respiration and energetics of three tropical marine sponges. J. Exp. Mar. Biol. Ecol. 14:231-249. [Google Scholar]
- 47.Ribes, M., R. Coma, and J. M. Gili. 1999. Natural diet and grazing rate of the temperate sponge Dysidea avara (Demospongiae, Dendroceratida) throughout an annual cycle. Mar. Ecol. Prog. Ser. 176:179-190. [Google Scholar]
- 48.Rohwer, F., V. Seguritan, F. Azam, and N. Knowlton. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243:1-10. [Google Scholar]
- 49.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 50.Santelli, C. M., B. N. Orcutt, E. Banning, W. Bach, C. L. Moyer, M. L. Sogin, H. Staudigel, and K. J. Edwards. 2008. Abundance and diversity of microbial life in ocean crust. Nature 453:653-657. [DOI] [PubMed] [Google Scholar]
- 51.Schirmer, A., R. Gadkari, C. D. Reeves, F. Ibrahim, E. F. Delong, and C. R. Hutchinson. 2005. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl. Environ. Microbiol. 71:4840-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steindler, L., D. Huchon, A. Avni, and M. Ilan. 2005. 16S rRNA phylogeny of sponge-associated cyanobacteria. Appl. Environ. Microbiol. 71:4127-4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki, M. T., C. M. Preston, O. Béjà, J. R. de la Torre, G. F. Steward, and E. F. DeLong. 2004. Phylogenetic screening of rRNA gene-containing clones in bacterial artificial chromosome (BAC) libraries from different depths in Monterey Bay. Microb. Ecol. 48:473-488. [DOI] [PubMed] [Google Scholar]
- 56.Taylor, M. W., R. Radax, D. Steger, and M. Wagner. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor, M. W., P. J. Schupp, I. Dahllöf, S. Kjelleberg, and P. D. Steinberg. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6:121-130. [DOI] [PubMed] [Google Scholar]
- 58.Taylor, M. W., P. J. Schupp, R. de Nys, S. Kjelleberg, and P. D. Steinberg. 2005. Biogeography of bacteria associated with the marine sponge Cymbastela concentrica. Environ. Microbiol. 7:419-433. [DOI] [PubMed] [Google Scholar]
- 59.Thiel, V., and J. F. Imhoff. 2003. Phylogenetic identification of bacteria with antimicrobial activities isolated from Mediterranean sponges. Biomol. Eng. 20:421-423. [DOI] [PubMed] [Google Scholar]
- 60.Thiel, V., S. C. Neulinger, T. Staufenberger, R. Schmaljohann, and J. F. Imhoff. 2007. Spatial distribution of sponge-associated bacteria in the Mediterranean sponge Tethya aurantium. FEMS Microbiol. Ecol. 59:47-63. [DOI] [PubMed] [Google Scholar]
- 61.Unson, M. D., N. D. Holland, and D. J. Faulkner. 1994. A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar. Biol. 119:1-11. [Google Scholar]
- 62.Usher, K. M., D. C. Sutton, S. Toze, J. Kuo, and J. Fromont. 2005. Inter-generational transmission of microbial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Mar. Freshw. Res. 56:125-131. [Google Scholar]
- 63.Usher, K. M., S. Toze, J. Fromont, J. Kuo, and D. C. Sutton. 2004. A new species of cyanobacterial symbiont from the marine sponge Chondrilla nucula. Symbiosis 36:183-192. [Google Scholar]
- 64.Vacelet, J., and C. Donadey. 1977. Electron-microscope study of association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30:301-314. [Google Scholar]
- 65.Van Elsas, J. D., and R. Costa. 2007. Molecular assessment of microbial communities in soil: significance for assessing the potential of soil to suppress phytopathogens, p. 498-517. In Z. H. Punja, S. H. de Boer, and H. Sanfacon (ed.), Biotechnology and plant disease. CABI, Wallington, United Kingdom.
- 66.Webster, N. S., R. S. Cobb, and A. P. Negri. 2008. Temperature thresholds for bacterial symbiosis with a sponge. ISME 2:830-842. [DOI] [PubMed] [Google Scholar]
- 67.Webster, N. S., and R. T. Hill. 2001. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an alpha-Proteobacterium. Mar. Biol. 138:843-851. [Google Scholar]
- 68.Webster, N. S., A. P. Negri, M. M. H. G. Munro, and C. N. Battershill. 2004. Diverse microbial communities inhabit Antarctic sponges. Environ. Microbiol. 6:288-300. [DOI] [PubMed] [Google Scholar]
- 69.Weisz, J. B., U. Hentschel, N. Lindquist, and C. S. Martens. 2007. Linking abundance and diversity of sponge-associated microbial communities to metabolic differences in host sponges. Mar. Biol. 152:475-483. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.