Abstract
Pyrrolobenzodiazepines, a class of natural products produced by actinomycetes, are sequence selective DNA alkylating compounds with significant antitumor properties. Among the pyrrolo[1,4]benzodiazepines (PBDs) sibiromycin, one of two identified glycosylated PBDs, displays the highest affinity for DNA and the most potent antitumor properties. Despite the promising antitumor properties clinical trials of sibiromycin were precluded by the cardiotoxicity effect in animals attributed to the presence of the C-9 hydroxyl group. As a first step toward the development of sibiromycin analogs, we have cloned and localized the sibiromycin gene cluster to a 32.7-kb contiguous DNA region. Cluster boundaries tentatively assigned by comparative genomics were verified by gene replacement experiments. The sibiromycin gene cluster consisting of 26 open reading frames reveals a “modular” strategy in which the synthesis of the anthranilic and dihydropyrrole moieties is completed before assembly by the nonribosomal peptide synthetase enzymes. In addition, the gene cluster identified includes open reading frames encoding enzymes involved in sibirosamine biosynthesis, as well as regulatory and resistance proteins. Gene replacement and chemical complementation studies are reported to support the proposed biosynthetic pathway.
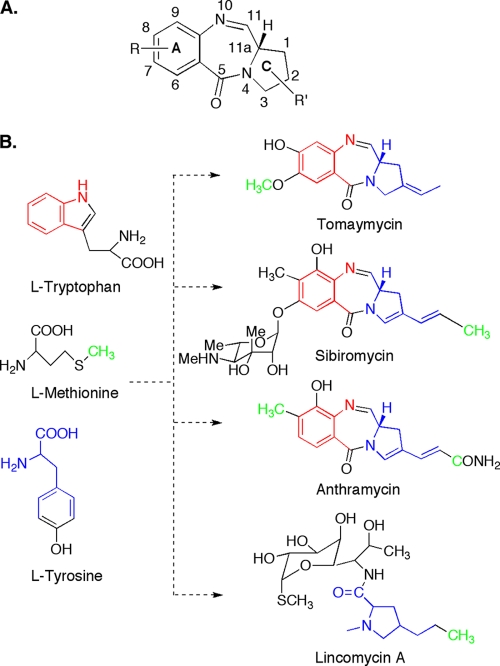
Pyrrolo[1,4]benzodiazepines (PBDs) are a class of natural products found in actinomycetes (Fig. 1) and defined by a common pyrrolo[1,4]benzodiazepine ring system (41). They are sequence-selective DNA alkylating agents with significant antitumor properties (21). Once in the minor groove of DNA an aminal bond is formed between the electrophilic C-11 of a PBD and the exocyclic N-2 of a guanine base in a double-stranded DNA (20). Formation of the PBD-DNA complex causes very little distortion of the double-helical structure of DNA (20), and as such this complex is less readily repaired by DNA repair proteins compared to DNA adducts with other alkylating agents (4), significantly contributing to the potency of PBDs. Successful syntheses of PBD analogs have been reported, but synthetic procedures for the more chemically diverse PBDs are laborious and have modest yields (1, 44). In addition, a chemical synthesis for glycosylated PBDs has not yet been accomplished. Structure-activity relationship studies on the synthetically and naturally produced PBDs showed that the C-9 hydroxylation present in anthramycin is the source of the cardiotoxic properties of this compound (Fig. 1) (3, 17, 26, 38). These studies also showed that O glycosylation at C7 significantly enhanced DNA-binding affinity (Fig. 1) (17). The only known glycosylated PBDs are sibiromycin and sibanomicin produced by Streptosporangium sibiricum and Micromonospora sp., respectively, both containing a sibirosamine moiety (16, 35). Only the producer of sibiromycin is commercially available. A loose correlation between DNA binding affinity and cytotoxicity has been shown with naturally and synthetically produced PBDs (42). Sibiromycin has the highest DNA binding affinity and cytotoxicity with 50% inhibitory concentrations varying from 4 to 1.7 pM in leukemia, plasmacytoma, and ovarian cancer cell lines (42). Despite its potency, further testing of sibiromycin is precluded due to the presence of C-9 hydroxyl group responsible for the cardiotoxic properties. In order to generate analogs of glycosylated PBDs by combinatorial biosynthesis and to exploit their potency, we chose to characterize the sibiromycin gene cluster.
FIG. 1.
(A) Pyrrolobenzodiazepine common ring system. (B) Metabolic precursors and chemical structures of sibiromycin, anthramycin, tomaymycin, and lincomycin A.
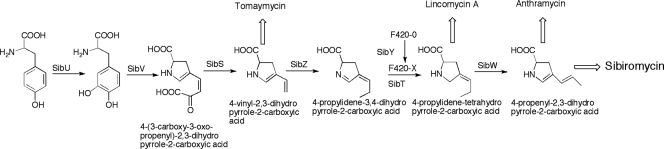
The metabolic precursors of the pyrrolobenzodiazepine ring of three PBDs (anthramycin, sibiromycin, and tomaymycin) were identified by feeding experiments to be l-tryptophan via the kynurenine pathway for the anthranilate moiety and l-tyrosine for the hydropyrrole moiety (11), suggesting a common biosynthetic pathway for these moieties in PBDs. The tyrosine-to-hydropyrrole transformation has been also identified by feeding studies in the biosynthesis of lincomycin, a lincosamide antibiotic (2) (Fig. 1B). Despite the sequencing of the biosynthetic gene clusters of anthramycin (10) and lincomycin (37), limited functional assignment of open reading frames (ORFs) and elucidation of the biosynthetic pathways were reported partly due to the presence of several gene products with no significant similarities to functionally characterized enzymes. We reasoned that we could take advantage of the identification of the sibiromycin gene cluster not only to try to lay the groundwork for the production of analogs of sibiromycin by combinatorial biosynthesis but also to establish the biosynthetic pathways of the anthranilate and the hydropyrrole moieties by a comparative analysis of the PBDs and lincomycin gene clusters. To help in this analysis, we have also utilized the gene cluster of another PBD, tomaymycin, whose characterization is reported in the accompanying study (24a). The comparative analysis takes advantage of the presence of similarity and differences at the anthranilate and hydropyrrole moieties among these natural products (Fig. 1). For example, both anthramycin and sibiromycin contain C-8 methyl and C-9 hydroxyl substituents not present in tomaymycin. However, tomaymycin shares with sibiromycin a C-7 hydroxyl substituent. Therefore, homologous proteins involved in C-9 hydroxylation are expected to be present in the anthramycin and sibiromycin gene cluster but absent in the tomaymycin gene cluster. We applied a similar approach for the biosynthesis of the hydropyrrole moiety using also the lincomycin gene cluster.
In the present study, we describe the cloning and sequencing of the sibiromycin gene cluster, the first biosynthetic gene cluster for a glycosylated PBD. Gene replacement experiments were used to confirm that the identified gene cluster was involved in sibiromycin biosynthesis, to define the boundaries of the sibiromycin gene cluster, and to elucidate the biosynthesis of the anthranilate moiety. Using the comparative approach, we were able not only to elucidate the sibiromycin biosynthetic pathway with a certain degree of confidence but also to assign ORFs in the anthramycin gene cluster contributing to the determination of the anthramycin biosynthetic pathway. The proposed biosynthetic pathway for the anthranilic moiety was supported by gene replacement and chemical complementation studies. The data reported here provide the basis for future studies on the enzymes involved in the biochemistry present in these pathways and for combinatorial biosynthetic experiments for the production of glycosylated PBDs.
MATERIALS AND METHODS
Reagents, bacterial strains, plasmids, and cosmids.
l-Kynurenine and l-3-hydroxykynurenine were purchased from Sigma, while 3-hydroxyanthranilic acid was from Tyger Scientific, Inc. 3-Hydroxy-4-methylanthranilic acid was obtained by catalytic hydrogenation of 2-nitro-3-hydroxy-4-methylbenzoic acid (Aldrich). The nuclear magnetic resonance (NMR) values were as follows: for 1H NMR (400 MHz, CD3OD), δ 2.22 (s, 3H), δ 6.35 (d, J = 8 Hz, 1H), and δ 7.30 (d, J = 8 Hz, 1H); and for 13C NMR (125 MHz, CD3OD), δ 171.4, δ 142.5, δ 142.3, δ 129.5, δ 123.4, δ 118.0, δ 110.5, and δ 16.1. All other chemicals, biochemicals, and molecular biology reagents used were purchased from common commercial sources. Bacterial strains, plasmids, and cosmids used are listed in Table S1 in the supplemental material. Escherichia coli ET12567 (27) and plasmids pIJ790 and pIJ773 were a generous gift from B. Gust (University of Tubingen). Growth and maintenance of S. sibiricum (DSM 44039) were performed as described by Hurley et al. (14) using mycelial plugs. The plugs were prepared routinely every 2 months due to the previously observed loss of antibiotic production (14) by inoculating the sibiromycin medium (3% [wt/vol] corn starch, 1.5% [wt/vol] Bacto soytone, 0.4% [wt/vol] NaCl, 0.5% [wt/vol] CaCO3) with previously prepared plugs accounting for 2% of the total volume. Flasks were shaken at 250 rpm and at 30°C for 84 h. Optimal aeration was obtained by using flasks with springs and by filling a maximum one-fifth of the flask volume with medium. Mycelial plugs of 2 ml of the grown-up culture of S. sibiricum were frozen at −80°C.
DNA isolation and manipulation.
The CTAB (cetyltrimethylammonium bromide) standard method (19) was used for the isolation of genomic DNA from S. sibiricum. DNA was isolated from E. coli using QIAprep spin miniprep kits (Qiagen) and manipulated according to standard methods (40). PCR amplification was carried out on an MJ Research PTC-200 thermal cycler using either Taq DNA polymerase or cloned Pfu DNA polymerase (Stratagene). The methods described in the DIG System user's guide (Roche Molecular Biochemicals) were used for primer labeling of probes, blotting, hybridization, and colorimetric detection.
Genomic library construction and screening.
Genomic DNA from S. sibiricum was partially digested with Sau3AI, dephosphorylated, and ligated into SuperCos 1 (Stratagene), packaged using Gigapack III XL packing extract (Stratagene), and transduced into E. coli XL1-Blue MR (Stratagene) according to the manufacturer's protocols. The degenerate primers Glu1 (5′-CSGGSGSSGCSGGSTTCATSGG-3′) and Glu2 (5′-GGGWRCTGGYRSGGSCCGTAGTTG-3′) (6) were used to amplify a DNA fragment of 541 bp. This dTDP-glucose 4,6-dehydratase gene probe was confirmed by sequencing and used for library screening. The positive clones identified were further screened with a second probe (724 bp), the nonribosomal peptide synthetase (NRPS) gene probe, obtained by PCR with degenerate primers (5′-GCSTACSYSATSTACACSTCSGG-3′ and 5′-SASGTCVCCSGTSCGGTAS-3′) and confirmed by sequencing. One of the positive clones identified, pSuperSib1, was chosen for sequencing and found to contain only a portion of the sibiromycin gene cluster. A second clone harboring the missing section of the sibiromycin gene cluster was identified by screening of the original S. sibiricum library with the NRPS probe and with probe S2 (455 bp) amplified with primers (5′-CGGGACGGTGTTGTTCG-3′ and 5′-GTGGTCAGCTACCTGATG-3′). The clone pSuperSib2, which did not hybridize with the NRPS probe but hybridized with the S2 probe, was chosen for sequencing.
DNA sequencing and analysis.
Sequencing of the shotgun libraries of pSuperSib1 and pSuperSib2 and assembly with Sequencer (Gene Codes Corp.) were performed by the DNA Sequencing Core (University of Michigan, seqcorebrcf.med.umich.edu/). PCR-based techniques and primer walking on plasmids were used to fill the gaps and to elucidate misassembled regions. Sequencing of plasmids was performed by the DNA Sequencing Core (University of Michigan) or by the Sequencing Facility from the University of Maryland (www.umbi.org/cbr/core-facilities/dna-sequencing/facility.php). ORF and sequence homology analyses were performed by using SoftBerry (Softberry, Inc.) and GeneMark (exon.gatech.edu/GeneMark/) and by using BLAST programs (blast.ncbi.nlm.nih.gov/blast.cgi), respectively.
Production, isolation, and analysis of sibiromycin.
A 2% inoculum of mycelial plugs of wild-type and mutant S. sibiricum strains was added to flasks with springs filled up to one-fifth of the total volume with the sibiromycin medium described above. Flasks were shaken at 250 rpm and 30°C for 60 h. Sibiromycin was isolated by using a modified protocol described by Hurley et al. (14). Production growths were extracted three times with equal volumes of dichloromethane. The combined organic layers were extracted three times with equal volumes of extraction buffer (0.1 M citric acid, 0.2 M Na2HPO4 [pH 4.0]). The combined aqueous layers were adjusted to pH 7.8 and extracted three times with equal volumes of chloroform. The combined organic layers were concentrated to dryness and dissolved in a volume of methanol corresponding to 1% of the production medium's volume. The methanolic solutions were stored at −20°C. The 7-deoxyaglycone of sibiromycin produced by the ΔsibG strain was isolated by extraction three times with equal volumes of ethyl acetate. The combined organic layers were concentrated to dryness and dissolved in a volume of methanol corresponding to 0.01% of the production medium's volume and stored as described above.
Analysis of the isolated sibiromycin by thin-layer chromatography (ethyl acetate-methanol, 4:1) and UV absorbance showed a characteristic Rf of 0.17 and λmax at 226 and 310 nm (12), respectively. The antibiotic properties were tested on LB agar plate with Bacillus subtilis (NRRL 354) incubated at 37°C for 12 h. Sibiromycin production was further confirmed by high-pressure liquid chromatography-electrospray ionization (HPLC-ESI) analysis on a Zorbax Elipse XDB-C8 column (4.6 by 150 mm; Agilent Technologies) preequilibrated in 90% solvent A (0.1% trifluoroacetic acid in distilled H2O) and 10% solvent B (methanol) at a flow rate of 1 ml/min. A linear gradient method (hold for 2 min in solvent A at 90%, 10 to 70% solvent B for 18 min) was applied to the column. The various compounds were detected at 230 and 310 nm by using an Agilent 1100 HPLC system and by ESI-mass spectrometry in the positive-ion mode using a JEOL AccuTOF-CS mass spectrometer. PBDs exist in aqueous solution in equilibrium between the imine and carbinolamine forms. Because neither form is ideal for long-term storage, PBDs are usually stored in methanol as the carbinolamine methyl ether. Depending on the conditions of the analytical technique used, one or more forms can be detected. Therefore, sibiromycin eluted at 15.6, 16.1, and 17.1 min from the dichloromethane and ethyl acetate extractions, yielding a characteristic [M + H]+ at m/z = 458.2 for the imine form, m/z = 476.3 for the carbinolamine form, and m/z = for the 490.3 for the carbinolamine methyl ether form, consistent with the molecular formulas C24H31N3O6 (calculated 457.2), C24H33N3O7 (calculated 475.2), and C25H35N3O7 (calculated 489.3), respectively. Fragmentation at the anomeric bond induced by increasing the orifice voltage resulted in the disappearance of the sibiromycin m/z peaks and in the appearance of the following peaks at 285.2, 303.2, and 317.2 m/z in addition to the 174.1 m/z assigned to the aglycone and sibirosamine fragments, respectively, in the ESI mass spectra.
Gene inactivation by gene replacement.
Single gene inactivation experiments were carried out in E. coli using the protocol based in REDIRECT technology (8). An apramycin resistance [aac(3)IV] cassette containing oriT was amplified from pIJ773 with forward and reverse primers containing 39 nucleotide extensions homologous to regions immediately upstream and downstream to the gene to be disrupted. The primers used for each inactivation experiment are listed in Table S2 in the supplemental material. E. coli Gene Hogs cells with λ-RED plasmid pIJ790 and pSuperSib1 or pSuperSib2 were transformed with the PCR product obtained by electroporation. The isolated modified pSuperSib1 or pSuperSib2 were transformed into E. coli ET12567 containing pUZ8002 and then used for intergenetic conjugation in S. sibiricum. Successful exconjugants were selected by three rounds of screening for kanamycin sensitivity and apramycin resistance. Insertional inactivation of the target gene was confirmed by Southern blot analysis (see Fig. S1 in the supplemental material) and PCR using primers complementary to regions outside the target gene. The primers used to confirm gene inactivation experiments are listed in Table S2 in the supplemental material. At least two parallel fermentations of each mutant strain were performed concomitantly to two fermentations of a positive control consisting of wild-type S. sibiricum conjugated with empty cosmid modified by gene replacement of bla gene with the apramycin cassette. Sibiromycin production was tested as described above.
Chemical complementation.
Chemical complementation experiments with ΔsibC strain were performed by adding the corresponding compound for a final concentration of 1 mM after 24 h of the start of the fermentation. Duplicate fermentations were cultured concomitantly to the positive control previously described and to the mutant strain grown in the absence of the corresponding compound. Sibiromycin production was tested as described above.
Nucleotide sequence accession number.
The sequence of the sibiromycin gene cluster has been deposited in GenBank under accession number FJ768674.
RESULTS AND DISCUSSION
Cloning, sequencing, and assignment of the sibiromycin gene cluster from S. sibiricum ATCC 29053.
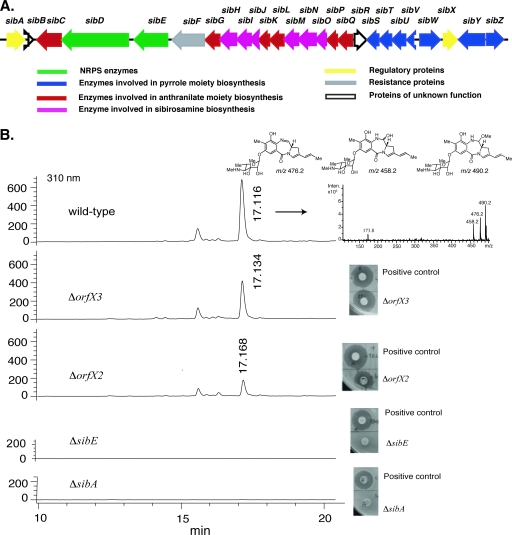
dTDP-glucose 4,6-dehydratase catalyzes the conversion of dTDP-glucose into dTDP-4-keto-6-deoxyglucose, the metabolic precursor of several 6-deoxyhexose moieties of glycosylated natural products (39). Since sibirosamine, the glycosyl moiety of sibiromycin, is a 6-deoxyhexose, the presence of dTDP-glucose 4,6-dehydratase in the gene cluster was hypothesized. Degenerate primers designed based on conserved regions in dTDP-glucose 4,6-dehydratases from actinomycetes (6) were used to clone by touchdown PCR a DNA fragment of 541 bp that, after sequencing, reveals part of a putative dTDP-glucose 4,6-dehydratase. This DNA fragment was then used to screen a cosmid library of S. sibiricum resulting in several positive clones from Southern hybridization. These positive clones were further screened using degenerate primers for the A3/A7 conserved regions of the adenylation domain of NRPSs (29). The amplified DNA fragment encoded an amino acid sequence similar to the A3/A7 region of the NRPS ORF22 in the anthramycin gene cluster (10). One of the positive clones, pSuperSib1, was sequenced by a shotgun approach and subsequent primer walking to cover the gaps. ORF analysis showed the presence at the downstream boundary of five ORFs coding for proteins, including two NRPSs, with similarity to proteins contained in the anthramycin gene cluster (10). An ORF encoding a dTDP-glucose 4,6-dehydratase was identified in a gene cluster coding for sugar biosynthetic enzymes difficult to assign to sibirosamine biosynthesis. The S. sibiricum cosmid library was rescreened using probe S2 to identify a clone, pSuperSib2, which harbors the remaining part of the sibiromycin gene cluster. The two overlapping cosmids contain a continuous region of 32.7 kb with 26 ORFs identified and tentatively assigned to the sibiromycin gene cluster by bioinformatics analysis (Fig. 2A and Table 1). The identification of two genes, sibD and sibE, encoding for NRPSs highly similar to and with the same domain organization of the NRPSs in the anthramycin gene cluster (10) supports the assignment of the sibiromycin gene cluster. Furthermore, the loss of sibiromycin production by inactivation of sibE (Fig. 2) is consistent with a role of this gene in sibiromycin biosynthesis.
FIG. 2.
(A) Genetic organization of the sibiromycin gene cluster. The proposed functions of individual ORFs are summarized in Table 1. (B) HPLC-ESI and bioassay analyses of the secondary metabolites produced by wild-type, ΔsibA, ΔsibE, ΔorfX2, and ΔorfX3 strains.
TABLE 1.
Deduced functions of ORFs in the sibiromycin biosynthetic gene cluster
| Gene | Protein size (aa)a | Putative function | NCBI accession no. of protein homologb,c | % Identity/ similarity |
|---|---|---|---|---|
| sibA | 299 | Putative regulator | BAC79018 | 31/43 |
| sibB | 94 | None | ABW71856 (ORF25) | 33/46 |
| sibC | 477 | Kynurenine 3-monoxygenase | XP_001514157 | 31/48 |
| ABW7854 (ORF23) | 44/58 | |||
| sibD | 1,504 | Nonribosomal peptide synthetase | ABW71853 (ORF22) | 41/53 |
| TomB | 42/54 | |||
| sibE | 597 | Nonribosomal peptide synthetase | ABW71852 (ORF21) | 47/58 |
| TomA | 40/53 | |||
| sibF | 771 | UvrA-drug resistance pump | YP_001822519 | 55/70 |
| ABW71839 (ORF8) | 61/75 | |||
| TomM | 61/75 | |||
| sibG | 352 | NADH-dependent flavin oxidoreductase | YP_001823080 | 20/25 |
| TomO | 40/49 | |||
| sibH | 393 | Glycosyltransferase | ABO28818 | 48/57 |
| sibI | 332 | dTDP-glucose synthase | AAS79450 | 57/69 |
| sibJ | 582 | dTDP-4-keto-6-deoxyglucose 3,5-epimerase | BAC55217 | 48/61 |
| sibK | 822 | Esterase/aryl formamidase | YP_001676677 | 38/53 |
| ABW71851 (ORF20) | 45/54 | |||
| sibL | 345 | C-Methyltransferase | AAL33761 | 38/53 |
| ABW71850 (ORF19) | 45/54 | |||
| sibM | 417 | Sugar C-methyltransferase | AAF01816 | 54/66 |
| sibN | 390 | dTDP-4-keto-6-deoxyglucose transaminase | ZP_02842724 | 51/68 |
| sibO | 246 | N-Methyltransferase | CAA63163 | 38/49 |
| sibP | 262 | Tryptophan 2,3-dioxygenase | YP_001812683 | 37/57 |
| ABW71848 (ORF17) | 58/68 | |||
| sibQ | 394 | Kynurenine hydrolase | CAJ89348 | 50/60 |
| ABW71847 (ORF16) | 45/57 | |||
| sibR | 251 | Unknown | AAK59995 | 33/39 |
| sibS | 300 | Unknown | CAA55771 (LmbX) | 41/52 |
| AX657735 (ORF15) | 32/43 | |||
| TomK | 43/52 | |||
| sibT | 293 | F420-dependent reductase | ABX00622 (LmbY) | 54/67 |
| ABW71845 (ORF14) | 57/70 | |||
| TomJ | 53/64 | |||
| sibU | 318 | Tyrosine hydroxylase | ABX00599 (LmbB2) | 39/49 |
| ABW71844 (ORF13) | 36/46 | |||
| TomI | 39/48 | |||
| sibV | 152 | l-DOPA 2,3-dioxygenase | CAA55747 (LmbB1) | 56/67 |
| ABW71843 (ORF12) | 45/59 | |||
| TomH | 51/58 | |||
| sibW | 492 | FAD-dependent oxidoreductase | ABW71838 (ORF7) | 41/48 |
| sibX | 391 | Transcriptional regulator | YP_001823517 | 21/33 |
| sibY | 598 | γ-Glutamyltransferase | ABX00597 (LmbA) | 57/68 |
| ABW71837 (ORF6) | 56/67 | |||
| TomL | 52/62 | |||
| sibZ | 337 | Methyltransferase | ABX00619 (LmbW) | 56/65 |
| ABW71836 (ORF5) | 60/74 |
aa, amino acids.
The proteins encoded by the lincomycin A and anthramycin gene clusters are indicated in parentheses.
See the accompanying study (24a) for proteins encoded by the tomaymycin gene cluster.
Determination of the boundaries of the sibiromycin gene cluster.
Gene replacement experiments using REDIRECT technology were carried out to assign the gene cluster boundaries (8). The ORFs upstream to sibA, orfX1 and orfX2, encode proteins with high similarity with S-adenosylmethionine synthetase and transmembrane efflux protein (83 and 50% identity, respectively), respectively. Although gene inactivation of orfX1 and orfX2 did not abolish sibiromycin production, gene inactivation of sibA resulted in a complete loss of sibiromycin production, confirming the start of the gene cluster at this gene (Fig. 2). sibZ was assigned to the sibiromycin gene cluster due to the high similarities of its gene product with proteins encoded in the anthramycin (10) and lincomycin A (37) gene clusters (Table 1). Downstream to sibZ, orfX3 encodes a putative polysaccharide deacetylase. Inactivation by gene replacement of orfX3 did not affect sibiromycin production confirming that orfX3 does not belong to the sibiromycin gene cluster (Fig. 2). Therefore, the sibiromycin gene cluster was assigned to start at sibA and end at sibZ (Fig. 2 and Table 1).
Biosynthesis of the 4-methyl-3,5-hydroxyanthranilic acid moiety.
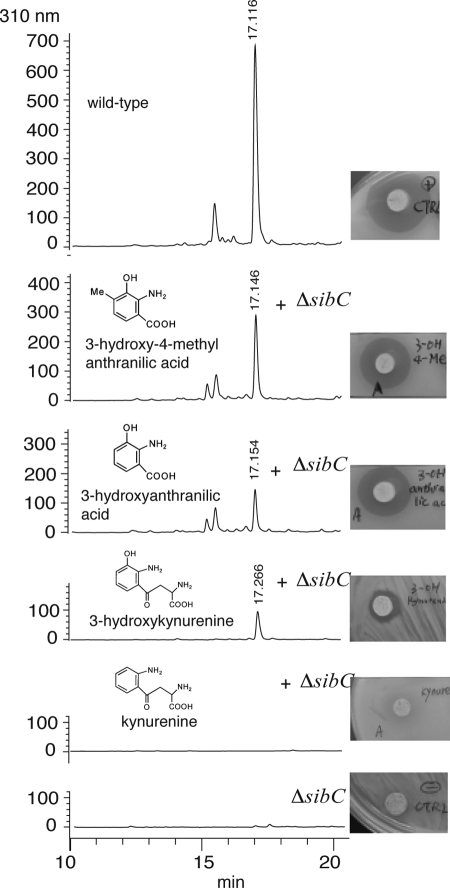
Feeding experiments showed that l-tryptophan is incorporated via the kynurenine pathway in the anthranilate moiety of sibiromycin and that l-3-hydroxykynurenine is a likely intermediate (13). In primary metabolism, de novo biosynthesis of NAD+ from l-tryptophan via l-3-hydroxykynurenine and 3-hydroxyanthranilic acid occurs in eukaryotes (28) and some prokaryotes (22). l-Tryptophan via the kynurenine pathway is also the metabolic precursor of natural products such as quinolobactin, daptomycin, and actinomycin (18, 30, 32). Four genes (sibC, sibK, sibP, and sibQ) of the sibiromycin gene cluster encode proteins highly similar to kynurenine 3-monoxygenase, aryl formamidase, tryptophan-2,3-dioxygenase and kynurenine hydrolase, respectively, known enzymes of the kynurenine pathway responsible to generate 3-hydroxyanthranilic acid (Table 1). The anthranilate moiety, absent in lincomycin A, is differently substituted in sibiromycin, tomaymycin and anthramycin. The anthranilate moiety of sibiromycin is C methylated at C-8 and hydroxylated at C-9 as in anthramycin and is hydroxylated at C-7 as in tomaymycin. Hydroxylation at C-9 is likely to occur prior to the diazepine ring formation due to the similarity of SibC to kynurenine-3-monoxygenases (Table 1). Inactivation of sibC resulted in a mutant strain incapable of producing sibiromycin, suggesting that at least one enzyme catalyzing a reaction downstream in the pathway is specific for a 3-hydroxykynurenine or 3-hydroxyanthranilic acid derivatives or that the kynurenine is shunted in other pathways (Fig. 3 and 4). Although no production was observed by feeding l-kynurenine to this mutant strain, sibiromycin production was rescued by feeding 3-hydroxy-l-kynurenine, 3-hydroxyanthranilic, and 3-hydroxy-4-methylanthranilic acids (Fig. 3). These results not only confirm the functional assignment of SibC as a kynurenine-3-monoxygenase but also support the intermediacy of 3-hydroxy-l-kynurenine, 3-hydroxyanthranilic, and 3-hydroxy-4-methylanthranilic acids.
FIG. 3.
HPLC-ESI and bioassay analyses of the secondary metabolites produced by wild-type and ΔsibC strains, and by chemical complementation of ΔsibC strains with l-kynurenine, 3-hydroxy-l-kynurenine, 3-hydroxyanthranilic acid, and 3-hydroxy-4-methylanthranilic acid.
FIG. 4.
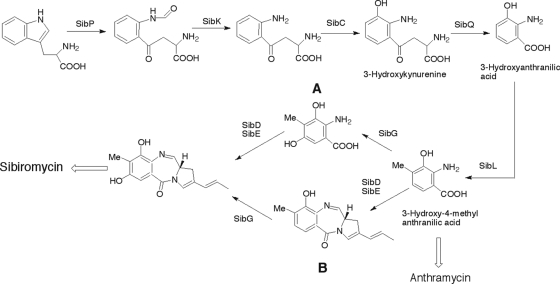
Proposed pathway for the biosynthesis of the 3,5-hydroxy-4-methylanthranilic acid moiety in sibiromycin, which is the suggested substrate for the NRPS enzymes catalyzing diazepine ring formation. Pathway A is favored. The anthramycin biosynthesis is proposed to diverge at the formation of 3-hydroxy-4-methylanthranilic acid.
The inactivation of sibC and feeding experiments reported above allow revisiting the biosynthesis of the 3-hydroxy-4-methylanthranilate moiety of anthramycin. The intermediacy of 3-hydroxy-4-methylanthranilic acid supported by radiotracer and competition experiments (11) was questioned based on the failure of 3-hydroxy-4-methylanthranilic acid to complement anthramycin production in a Δorf23 strain of Streptomyces refuineus (10). However, if one considers the known impermeability of S. refuineus toward this compound (13), the failure to rescue anthramycin production is not inconsistent with the intermediacy of 3-hydroxy-4-methylanthranilic acid. Thus, based on the chemical complementation results in ΔsibC strain and on the comparative analysis we propose that 3-hydroxy-4-methylanthranilic acid is an intermediate obtained by a common biosynthetic step in the anthramycin and sibiromycin biosyntheses (Fig. 4). The product of orf24 from the anthramycin gene cluster was proposed to be involved in the biosynthesis of the anthranilate moiety of anthramycin based on inactivation and feeding experiments (10). Hu et al. speculated that ORF24 could be involved in the formation of 3-hydroxy-l-kynurenine and l-kynurenine (10). The absence of a gene encoding a homologous protein in the sibiromycin gene cluster raises questions regarding this assignment. Thus, the precise role of this protein in the anthramycin biosynthesis remains unclear.
The genes encoding the proteins catalyzing the C-7 hydroxylation and C-8 methylation in sibiromycin were identified by comparative analysis of the anthramycin (10), tomaymycin (24a), lincomycin A (37), and sibiromycin gene clusters. Of the four putative S-adenosylmethionine-dependent methyltransferases in the sibiromycin gene cluster, two are similar to sugar methyltransferases (see below), and two (SibL and SibZ) are similar to proteins in the anthramycin biosynthetic pathway. Of these latter two, only SibZ is similar to a protein in the lincomycin A biosynthetic pathway and, therefore, likely to be involved in the biosynthesis of the dihydropyrrole moiety (see below). By elimination SibL was implicated in the C-8 methylation of the anthranilate moiety. Methylation on an aromatic ring catalyzed by S-adenosylmethionine-dependent enzymes occurs in ortho to a phenolic oxygen (34). Thus, C-8 methylation step occurs either after C-7 or C-9 hydroxylation step. The intermediacy of the 3-hydroxy-4-methylanthranilic acid shown (Fig. 3) above strongly supports the biosynthetic pathway proposed in Fig. 4 in which C-8 methylation occurs prior to C-7 hydroxylation and after C-9 hydroxylation. Using the comparative analysis, SibG was proposed to catalyze the hydroxylation at C-7 in sibiromycin. Tomaymycin contains the same C-7 hydroxyl substitution at the anthranilate moiety while anthramycin does not. Thus, a homolog of SibG was found only in the tomaymycin gene cluster (TomO; see the accompanying study [24a]) (Table 1).
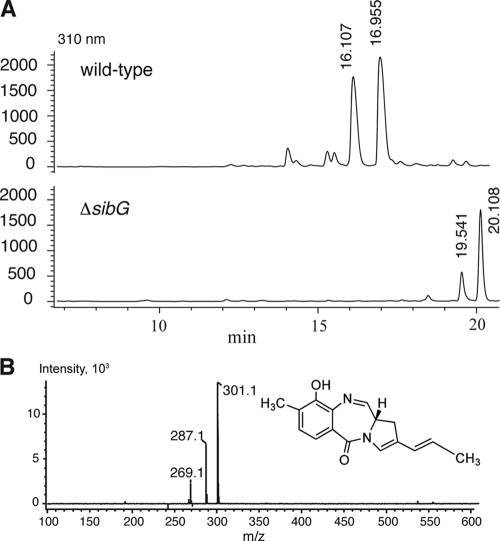
The ΔsibG mutant strain of S. sibiricum was obtained to test our hypothesis that SibG catalyzes C-7 hydroxylation. The absence of the C-7 hydroxyl group would hamper glycosylation of the PBD core ring, and the 7-deoxyaglycone of sibiromycin will be obtained. Extraction of the production medium of ΔsibG with ethyl acetate yielded to a compound eluting at 19.5 and 20.1 min with [M+H]+ at m/z = 269.1 for the imine form, m/z = 287.1 for the carbinolamine form, and m/z = 301.1 for the carbinolamine methyl form (Fig. 5) consistent with the molecular formulas C16H16N2O2 (calculated, 268.1), C16H18N2O3 (calculated, 286.1), and C17H20N2O3 (calculated, 300.1), respectively. This molecular formula could correspond to either the 7-deoxy or 9-deoxyaglycone of sibiromycin. We propose that this compound is the 7-deoxyaglycone of sibiromycin because the presence of the C-7 hydroxyl group in the 9-deoxyaglycone would likely yield to a glycosylated compound, and it would not stop at the aglycone. No glycosylated analog was identified in either dichloromethane or ethyl acetate extractions. The accumulation of the 7-deoxyaglycone of sibiromycin in the production medium of ΔsibG strain confirms the assignment of SibG, but it does not allow us to distinguish whether this enzyme acts on the 3-hydroxy-4-ethylanthranilic acid or on the PBD ring (Fig. 4). However, chemical complementation experiments on a mutant strain of Streptomyces achromogenes with tomO inactivated (see the accompanying study [24a]) supports the assignment of TomO and of the homolog protein, SibG, as the enzymes catalyzing hydroxylation at C-5 of an anthranilate derivative in the tomaymycin and sibiromycin biosyntheses. Taken together, these data support a biosynthetic pathway of sibiromycin in which all of the hydroxylation and methylation substitutions at the anthranilate moiety occur prior to diazepine ring formation as described in Fig. 4, pathway A.
FIG. 5.
(A) HPLC analyses of the secondary metabolites produced by wild-type and ΔsibG strains. Isolation of the metabolites was carried out by ethyl acetate extractions. (B) ESI spectrum of the HPLC peak eluting at 20.1 min and the chemical structure of the 7-deoxyaglycone of sibiromycin.
Biosynthesis of the 4-propenyl-2,3-dihydropyrrole-2-carboxylic acid moiety.
Labeling experiments have identified l-tyrosine as the biosynthetic precursor for the hydropyrrole moieties of PBDs, as well as of lincomycin A (2, 11). The functional assignment of the biosynthetic enzymes involved in this transformation with the exception of the first two steps catalyzed by a tyrosine hydroxylase and an l-DOPA 2,3-dioxygenase (31, 33) remains undetermined in the PBDs and lincomycin A biosyntheses (10, 37). We propose a common biosynthetic route for the formation of 4-propylidene-tetrahydropyrrole-2-carboxylic acid in the PBDs and lincomycin biosyntheses, the last intermediate in common in the anthramycin, sibiromycin, and lincomycin A biosyntheses (Fig. 6). The six enzymes (SibS, SibT, SibU, SibV, SibY, and SibZ) involved in this multistep conversion were assigned based on the comparative analysis of the anthramycin, sibiromycin, and lincomycin A gene clusters and on the accumulation of the 4-propylidene-3,4-dihydropyrrole-2-carboxylic acid in an F420-deficient Streptomyces lincolnensis strain (23). The first two steps are catalyzed by SibU and SibV, highly similar to LmbB2 and LmbB1, respectively, whose tyrosine hydroxylase and l-DOPA 2,3-dioxygenase activities have been previously shown in vitro (Table 1) (31). A BLAST search of protein databases fails to identify proteins similar to SibS and to its homologous proteins in the PBDs and lincomycin A biosyntheses. The presence of homologous proteins in all PBDs and lincomycin gene clusters strongly supports a role for SibS in the formation of 4-vinyl-2,3-dihydropyrrole-2-carboxylic acid. We propose that SibS catalyzes the unusual C-C hydrolysis of 4-(3-carboxy-3-oxo-propenyl)-2,3-dihydropyrrole-2-carboxylic acid similarly to BphD, a C-C bond hydrolase (9). The terminal methyl group of the propylidene and propyl side chains of anthramycin, sibiromycin, and lincomycin A derives from methionine (2, 11). Genes (sibZ, orf5, and lmbW) encoding homologous methyltransferases are present in the three gene clusters. The last step in the biosynthesis of 4-propylidene-3,4-dihydropyrrole-2-carboxylic acid is the F420-dependent reduction to 4-propylidene-tetrahydropyrrole-2-carboxylic acid. F420 is a flavinlike cofactor substituted with a side chain with different numbers of glutamate residues. F420-dependent enzymes catalyze different redox reactions especially those involved in methanogenesis (24). F420 cofactors are synthesized from F420-0, a form of the cofactor depleted of glutamyl side chains. SibY closely related to γ-glutamyltransferases is proposed to catalyze the formation of an undetermined F420 cofactor from F420-0. SibT containing conserved sequence motifs of F420-dependent reductases is proposed to catalyze this reductive formation of 4-propylidene-tetrahydropyrrole-2-carboxylic acid, the branching point between the sibiromycin and anthramycin biosyntheses and the lincomycin biosynthesis.
FIG. 6.
Proposed pathway for the biosynthesis of the 4-propenyl-2,3-dihydropyrrole-2-carboxylic acid moiety in sibiromycin, which is the suggested substrate for the NRPS enzymes catalyzing diazepine ring formation. The branching points for the anthramycin, tomaymycin, and lincomycin A biosyntheses are indicated.
The biosynthesis of lincomycin requires an additional reduction of 4-propylidene-tetrahydropyrrole-2-carboxylic acid to yield the propyl side chain (Fig. 6). Instead, the biosyntheses of anthramycin and sibiromycin diverge with the oxidation of 4-propylidine-tetrahydropyrrole-2-carboxylic acid to 4-propenyl-2,3-dihydropyrrole-2-carboxylic acid. Therefore, only the anthramycin and sibiromycin gene clusters must encode an enzyme catalyzing this oxidation (Fig. 6). The putative FAD-dependent oxidoreductase SibW presumably catalyzes the oxidation of the tetrahydropyrrole. The sibiromycin biosynthesis of the dihydropyrrole moiety terminates at the formation of the 4-propenyl-2,3-dihydropyrrole-2-carboxylic acid. However, the last steps in the biosynthesis of the dihydropyrrole moiety of anthramycin are oxidation and amidation of the propenyl side chain of this intermediate. Enzymes involved in these steps would only be encoded in the anthramycin gene cluster and not in the sibiromycin gene cluster. Therefore, by a process of elimination, the formation of the acrylamide side chain of anthramycin is likely catalyzed by ORF4, ORF3, and ORF2 involved in the oxidation of the terminal methyl group to a carboxylic acid and, as previously established (10), by ORF1, responsible for the amidation reaction. It remains to be determined whether these modifications occur prior to or after diazepine ring formation.
Biosynthesis of the sibirosamine moiety.
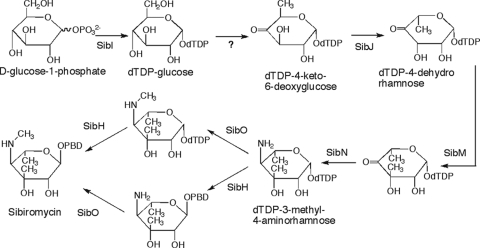
Sibirosamine is a rare aminosugar present only in the glycosylated PBDs, sibiromycin (36) and sibanomicin (16). Many glycosylated natural products owe their biological activities in part or completely to the sugar modification of the aglycone (46). The presence of the sibirosamine moiety enhances significantly the DNA binding affinity and antitumor properties of sibiromycin compared to anthramycin and tomaymycin (42). Five genes (sibI, sibJ, sibM, sibN, and sibO) encode enzymes responsible for the biosynthesis of sibirosamine based on homology search. With the exception of sibO, all of these genes encode proteins bearing high similarity to biochemically characterized enzymes (Fig. 7 and Table 1). As expected, the first step in the biosynthesis of sibirosamine is the formation of dTDP-glucose catalyzed by SibI highly similar to dTDP-glucose synthetase (Table 1). Since sibirosamine is a 6-deoxyhexose, the presence of a dTDP-glucose 4,6-dehydratase was expected. Surprisingly, the sibiromycin gene cluster does not contain a gene encoding this enzyme. However, endogenous dTDP-glucose dehydratase in S. sibiricum, such as the one identified in pSuperSib1 ∼9 kb from the start of the gene cluster, could make up for this activity. SibJ catalyzes the epimerization of C-5 and C-3 based on the similarity (41% identity) with RmlC, a dTDP-4-keto-6-deoxyglucose 3,5-epimerase in rhamnose biosynthesis (7). Methylation at C-3 by SibM, highly similar (48% identity) to the C-methyltransferase NovU of the novobiocin biosynthesis (43), and subsequent reductive amination of C-4 by SibN, a homolog (45% identity) of dTDP-4-keto-6-deoxyhexose transaminase WecE (15), yields dTDP-3-methyl-4-aminorhamnose (Fig. 7). SibO with sequence similarity to a putative N-methyltransferase in Streptomyces anulatus is proposed to catalyze N methylation at the 4-amino group (Table 1). Whether this reaction occurs before or after loading of the sugar moiety to the PBD aglycone is unclear. Glycosylation of the PBD aglycone is likely catalyzed by SibH. This protein bears similarity (43% identity) to JadS, an O-glycosyltransferase in the jadomycin B biosynthesis (45). JadS is an inverting glycosyltransferase, meaning that glycosyl transfer occurs with concomitant inversion of the stereochemistry at the anomeric carbon (5). Thus, we propose that SibH transfers a β-dTDP-aminodeoxyhexose on the PBD aglycone forming sibiromycin, an α-glycoside (35) (Fig. 7).
FIG. 7.
Proposed pathways for the biosynthesis of the sibirosamine moiety with different sequences of the N methylation and glycosyl transfer reactions.
Genes encoding resistance, regulation, and unknown function.
SibA and SibX bear sequence similarity to transcription regulators (Table 1). SibA is also a homolog of ORF25 encoded by the anthramycin gene cluster, suggesting a shared regulatory mechanism for the biosyntheses of anthramycin and sibiromycin. Resistance to PBDs in the native producers of sibiromycin, anthramycin, and tomaymycin is achieved by at least one common export mechanism carried out by the homologous SibF, ORF8, and TomM (see the accompanying study [24a] and Table 1). These enzymes are highly similar to DrrC, a biochemically characterized resistance protein in the daunorubicin gene cluster (25). There are two additional ORFs (sibB and sibR) in the sibiromycin gene cluster that encode proteins of unknown functions. A homology search resulted in no hits in the protein databases for SibB and a weak similarity of SibR to proteins of unknown function. Any of these proteins bears no similarity to proteins of unknown function encoded by the anthramycin gene cluster (ORF11 and ORF18). The absence of a homolog in the anthramycin and tomaymycin gene clusters led us to conclude that they are probably not essential for the biosyntheses of the anthranilate moiety, of the diazepine ring, and of 4-propenyl-2,3-dihydropyrrole-2-carboxylic acid moiety. However, their roles remain unclear.
Conclusions.
In conclusion, the sibiromycin gene cluster presented here revealed a similar “modular” biosynthetic strategy previously identified in the anthramycin gene cluster. Limitations in the assignment of the ORFs of the anthramycin gene cluster were overcome by the identification of the sibiromycin gene cluster. By considering chemical differences and similarities in anthramycin, sibiromycin, and tomaymycin (see the accompanying study [24a]), we were able to assign with a certain degree of confidence the gene encoding the dihydropyrrole moiety. Using gene replacement and chemical complementation experiments, we were able to verify the biosynthetic pathway of the anthranilate moiety proposed by comparative gene analysis in the sibiromycin biosynthesis and to conciliate contradictory data previously reported for the anthramycin biosynthesis (10, 13). The availability of the first gene cluster for a glycosylated PBD lays the foundation for engineering of the biosynthetic machinery for the production of a glycosylated PBD.
Supplementary Material
Acknowledgments
We are grateful to Bruno Duro for his contribution to the screening of the S. sibiricum genomic DNA library and to B. Gust (University of Tubingen) for the E. coli ET12567 cells and plasmids pIJ790, pIJ773, and pUWL201.
This study was supported by the Marlene and Stewart Greenebaum Cancer Center's American Cancer Society Institutional Research Grant (01-4-30170), by the National Institutes of Health (GM084473), and by startup funds from the University of Maryland, College Park.
Footnotes
Published ahead of print on 6 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Antonow, D., N. Cooper, P. W. Howard, and D. E. Thurston. 2007. Parallel synthesis of a novel C2-aryl pyrrolo[2,1-c][1,4]benzodiazepine (PBD) library. J. Comb. Chem. 9:437-445. [DOI] [PubMed] [Google Scholar]
- 2.Brahme, N. M., J. E. Gonzalez, J. P. Rolls, E. J. Hessler, S. Mizsak, and L. H. Hurley. 1984. Biosynthesis of the lincomycins. 1. Studies using stable isotopes on the biosynthesis of the propyl-l-hygric and ethyl-l-hygric acid moieties of lincomycin-A and lincomycin-B. J. Am. Chem. Soc. 106:7873-7878. [Google Scholar]
- 3.Cargill, C., E. Bachmann, and G. Zbinden. 1974. Effects of daunomycin and anthramycin on electrocardiogram and mitochondrial metabolism of the rat heart. J. Natl. Cancer Inst. 53:481-486. [DOI] [PubMed] [Google Scholar]
- 4.Clingen, P. H., I. U. De Silva, P. J. McHugh, F. J. Ghadessy, M. J. Tilby, D. E. Thurston, and J. A. Hartley. 2005. The XPF-ERCC1 endonuclease and homologous recombination contribute to the repair of minor groove DNA interstrand crosslinks in mammalian cells produced by the pyrrolo[2,1-c][1,4]benzodiazepine dimer SJG-136. Nucleic Acids Res. 33:3283-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307-317. [DOI] [PubMed] [Google Scholar]
- 6.Decker, H., S. Gaisser, S. Pelzer, P. Schneider, L. Westrich, W. Wohlleben, and A. Bechthold. 1996. A general approach for cloning and characterizing dNDP-glucose dehydratase genes from actinomycetes. FEMS Microbiol. Lett. 141:195-201. [DOI] [PubMed] [Google Scholar]
- 7.Graninger, M., B. Nidetzky, D. E. Heinrichs, C. Whitfield, and P. Messner. 1999. Characterization of dTDP-4-dehydrorhamnose 3,5-epimerase and dTDP-4-dehydrorhamnose reductase, required for dTDP-l-rhamnose biosynthesis in Salmonella enterica serovar Typhimurium LT2. J. Biol. Chem. 274:25069-25077. [DOI] [PubMed] [Google Scholar]
- 8.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horsman, G. P., S. Bhowmik, S. Y. K. Seah, P. Kumar, J. T. Bolin, and L. D. Eltis. 2007. The tautomeric half-reaction of BphD, a C-C bond hydrolase: kinetic and structural evidence supporting a key role for histidine 265 of the catalytic triad. J. Biol. Chem. 282:19894-19904. [DOI] [PubMed] [Google Scholar]
- 10.Hu, Y., V. Phelan, I. Ntai, C. M. Farnet, E. Zazopoulos, and B. O. Bachmann. 2007. Benzodiazepine biosynthesis in Streptomyces refuineus. Chem. Biol. 14:691-701. [DOI] [PubMed] [Google Scholar]
- 11.Hurley, L. H. 1980. Elucidation and formulation of novel biosynthetic pathways leading to the pyrrolo[1,4]benzodiazepine antibiotics anthramycin, tomaymycin, and sibiromycin. Acc. Chem. Res. 13:263-269. [Google Scholar]
- 12.Hurley, L. H. 1977. Pyrrolo(1,4)benzodiazepine antitumor antibiotics: comparative aspects of anthramycin, tomaymycin, and sibiromycin. J. Antibiot. 30:349-370. [DOI] [PubMed] [Google Scholar]
- 13.Hurley, L. H., and C. Gairola. 1979. Pyrrolo (1,4) benzodiazepine antitumor antibiotics: biosynthetic studies on the conversion of tryptophan to the anthranilic acid moieties of sibiromycin and tomaymycin. Antimicrob. Agents Chemother. 15:42-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurley, L. H., W. L. Lasswell, R. K. Malhotra, and N. V. Das. 1979. Pyrrolo[1,4]benzodiazepine antibiotics: biosynthesis of the anti-tumor antibiotic sibiromycin by Streptosporangium sibiricum. Biochemistry 18:4225-4229. [DOI] [PubMed] [Google Scholar]
- 15.Hwang, B. Y., H. J. Lee, Y. H. Yang, H. S. Joo, and B. G. Kim. 2004. Characterization and investigation of substrate specificity of the sugar aminotransferase WecE from Escherichia coli K-12. Chem. Biol. 11:915-925. [DOI] [PubMed] [Google Scholar]
- 16.Itoh, J., H. O. Watabe, S. Ishii, S. Gomi, M. Nagasawa, H. Yamamoto, T. Shomura, M. Sezaki, and S. Kondo. 1988. Sibanomicin, a new pyrrolo[1,4]-benzodiazepine antitumor antibiotic produced by a Micromonospora sp. J. Antibiot. 41:1281-1284. [DOI] [PubMed] [Google Scholar]
- 17.Kamal, A., M. V. Rao, N. Laxman, G. Ramesh, and G. S. Reddy. 2002. Recent developments in the design, synthesis and structure-activity relationship studies of pyrrolo[2,1-c][1,4]benzodiazepines as DNA-interactive antitumour antibiotics. Curr. Med. Chem. Anticancer Agents 2:215-254. [DOI] [PubMed] [Google Scholar]
- 18.Keller, U. 1984. Acyl pentapeptide lactone synthesis in actinomycin-producing streptomycetes by feeding with structural analogs of 4-methyl-3-hydroxyanthranilic acid. J. Biol. Chem. 259:8226-8231. [PubMed] [Google Scholar]
- 19.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics, 2nd ed. John Innes Foundation, Norwich, United Kingdom.
- 20.Kopka, M. L., D. S. Goodsell, I. Baikalov, K. Grzeskowiak, D. Cascio, and R. E. Dickerson. 1994. Crystal structure of a covalent DNA drug adduct: anthramycin bound to C-C-a-a-C-G-T-T-G-G and a molecular explanation of specificity. Biochemistry 33:13593-13610. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, R., and J. W. Lown. 2003. Recent developments in novel pyrrolo[2,1-c][1,4]benzodiazepine conjugates: synthesis and biological evaluation. Mini Rev. Med. Chem. 3:323-339. [DOI] [PubMed] [Google Scholar]
- 22.Kurnasov, O., V. Goral, K. Colabroy, S. Gerdes, S. Anantha, A. Osterman, and T. P. Begley. 2003. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 10:1195-1204. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, M. S., D. A. Yurek, J. H. Coats, S. T. Chung, and G. P. Li. 1992. Isolation and identification of 3-propylidene-delta 1-pyrroline-5-carboxylic acid, a biosynthetic precursor of lincomycin. J. Antibiot. 45:1773-1777. [DOI] [PubMed] [Google Scholar]
- 24.Li, H., H. Xu, D. E. Graham, and R. H. White. 2003. Glutathione synthetase homologs encode alpha-l-glutamate ligases for methanogenic coenzyme F420 and tetrahydrosarcinapterin biosyntheses. Proc. Natl. Acad. Sci. USA 100:9785-9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Li, W., S. C. Chou, A. Khullar, and B. Gerratana. 2009. Cloning and characterization of the biosynthetic gene cluster for tomaymycin, an SJG-136 monomeric analog. Appl. Environ. Microbiol. 75:2958-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomovskaya, N., S. K. Hong, S. U. Kim, L. Fonstein, K. Furuya, and R. C. Hutchinson. 1996. The Streptomyces peucetius drrC gene encodes a UvrA-like protein involved in daunorubicin resistance and production. J. Bacteriol. 178:3238-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lubawy, W. C., R. A. Dallam, and L. H. Hurley. 1980. Protection against anthramycin-induced toxicity in mice by coenzyme Q10. J. Natl. Cancer Inst. 64:105-109. [PubMed] [Google Scholar]
- 27.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 28.Magni, G., A. Amici, M. Emanuelli, N. Raffaelli, and S. Ruggieri. 1999. Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 73:135-182. [DOI] [PubMed] [Google Scholar]
- 29.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2674. [DOI] [PubMed] [Google Scholar]
- 30.Matthijs, S., C. Baysse, N. Koedam, K. A. Tehrani, L. Verheyden, H. Budzikiewicz, M. Schafer, B. Hoorelbeke, J. M. Meyer, H. De Greve, and P. Cornelis. 2004. The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol. Microbiol. 52:371-384. [DOI] [PubMed] [Google Scholar]
- 31.Neusser, D., H. Schmidt, J. Spizek, J. Novotna, U. Peschke, S. Kaschabeck, P. Tichy, and W. Piepersberg. 1998. The genes lmbB1 and lmbB2 of Streptomyces lincolnensis encode enzymes involved in the conversion of l-tyrosine to propylproline during the biosynthesis of the antibiotic lincomycin A. Arch. Microbiol. 169:322-332. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen, K. T., D. Kau, J. Q. Gu, P. Brian, S. K. Wrigley, R. H. Baltz, and V. Miao. 2006. A glutamic acid 3-methyltransferase encoded by an accessory gene locus important for daptomycin biosynthesis in Streptomyces roseosporus. Mol. Microbiol. 61:1294-1307. [DOI] [PubMed] [Google Scholar]
- 33.Novotna, J., A. Honzatko, P. Bednar, J. Kopecky, J. Janata, and J. Spizek. 2004. l-3,4-Dihydroxyphenyl alanine-extradiol cleavage is followed by intramolecular cyclization in lincomycin biosynthesis. Eur. J. Biochem. 271:3678-3683. [DOI] [PubMed] [Google Scholar]
- 34.Pacholec, M., J. Tao, and C. T. Walsh. 2005. CouO and NovO: C-methyltransferases for tailoring the aminocoumarin scaffold in coumermycin and novobiocin antibiotic biosynthesis. Biochemistry 44:14969-14976. [DOI] [PubMed] [Google Scholar]
- 35.Parker, K. A., and R. E. Babine. 1982. Revision of assignment of structure to the pyrrolodiazepinone anti-tumor antibiotic sibiromycin. J. Am. Chem. Soc. 104:7330-7331. [Google Scholar]
- 36.Parker, K. A., and R. E. Babine. 1982. Revision of the assignment of relative stereochemistry in sibirosamine: synthesis of methyl N-tosyl alpha-d-sibirosaminopyranoside. Tetrahedron Lett. 23:1763-1766. [Google Scholar]
- 37.Peschke, U., H. Schmidt, H. Z. Zhang, and W. Piepersberg. 1995. Molecular characterization of the lincomycin production gene cluster of Streptomyces lincolnensis 78-11. Mol. Microbiol. 16:1137-1156. [DOI] [PubMed] [Google Scholar]
- 38.Petrusek, R. L., G. L. Anderson, T. F. Garner, Q. L. Fannin, D. J. Kaplan, S. G. Zimmer, and L. H. Hurley. 1981. Pyrrolo[1,4]benzodiazepine antibiotics: proposed structures and characteristics of the in vitro deoxyribonucleic-acid adducts of anthramycin, tomaymycin, sibiromycin, and neothramycin-A and neothramycin-B. Biochemistry 20:1111-1119. [DOI] [PubMed] [Google Scholar]
- 39.Salas, J. A., and C. Mendez. 2005. Biosynthesis pathways for deoxysugars in antibiotic-producing actinomycetes: isolation, characterization and generation of novel glycosylated derivatives. J. Mol. Microbiol. Biotechnol. 9:77-85. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Thurston, D. E., and D. S. Bose. 1994. Synthesis of DNA-interactive pyrrolo[2,1-C][1,4]benzodiazepines. Chem. Rev. 94:433-465. [DOI] [PubMed] [Google Scholar]
- 42.Thurston, D. E., D. S. Bose, P. W. Howard, T. C. Jenkins, A. Leoni, P. G. Baraldi, A. Guiotto, B. Cacciari, L. R. Kelland, M. P. Foloppe, and S. Rault. 1999. Effect of A-ring modifications on the DNA-binding behavior and cytotoxicity of pyrrolo[2,1-c][1,4]benzodiazepines. J. Med. Chem. 42:1951-1964. [DOI] [PubMed] [Google Scholar]
- 43.Thuy, T. T., H. C. Lee, C. G. Kim, L. Heide, and J. K. Sohng. 2005. Functional characterizations of novWUS involved in novobiocin biosynthesis from Streptomyces spheroides. Arch. Biochem. Biophys. 436:161-167. [DOI] [PubMed] [Google Scholar]
- 44.Tiberghien, A. C., D. Hagan, P. W. Howard, and D. E. Thurston. 2004. Application of the Stille coupling reaction to the synthesis of C2-substituted endo-exo unsaturated pyrrolo[2,1-c][1,4]benzodiazepines (PBDs). Bioorg. Med. Chem. Lett. 14:5041-5044. [DOI] [PubMed] [Google Scholar]
- 45.Wang, L., R. L. White, and L. C. Vining. 2002. Biosynthesis of the dideoxysugar component of jadomycin B: genes in the jad cluster of Streptomyces venezuelae ISP5230 for l-digitoxose assembly and transfer to the angucycline aglycone. Microbiology 148:1091-1103. [DOI] [PubMed] [Google Scholar]
- 46.Weymouth-Wilson, A. C. 1997. The role of carbohydrates in biologically active natural products. Nat. Prod. Rep. 14:99-110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.