Abstract
Although denitrification or nitrate respiration has been found among a few eukaryotes, its phylogenetic relationship with the bacterial system remains unclear because orthologous genes involved in the bacterial denitrification system were not identified in these eukaryotes. In this study, we isolated a gene from the denitrifying fungus Fusarium oxysporum that is homologous to the bacterial nirK gene responsible for encoding copper-containing nitrite reductase (NirK). Characterization of the gene and its recombinant protein showed that the fungal nirK gene is the first eukaryotic ortholog of the bacterial counterpart involved in denitrification. Additionally, recent genome analyses have revealed the occurrence of nirK homologs in many fungi and protozoa, although the denitrifying activity of these eukaryotes has never been examined. These eukaryotic homolog genes, together with the fungal nirK gene of F. oxysporum, are grouped in the same branch of the phylogenetic tree as the nirK genes of bacteria, archaea, and eukaryotes, implying that eukaryotic nirK and its homologs evolved from a single ancestor (possibly the protomitochondrion). These results show that the fungal denitrifying system has the same origin as its bacterial counterpart.
Denitrification plays an important role in the global nitrogen cycle and reduces nitrate (NO3−) and/or nitrite (NO2−) to a gaseous form of nitrogen, generally to dinitrogen (N2) or nitrous oxide (N2O) (27). It typically follows four reduction stages, NO3− → NO2− → NO → N2O → N2, each of which is catalyzed by a specific reductase: dissimilatory NO3− reductase (dNaR), dissimilatory NO2− reductase (dNiR), nitric oxide (NO) reductase (NoR), and N2O reductase, respectively. These enzymes receive electrons from a respiratory chain functioning as a “terminal reductase.” Thus, denitrification exhibits a physiological significance in its ability to anaerobically respire through the processes of nitrate respiration, nitrite respiration, and so forth. Denitrification was previously thought to be a characteristic of bacteria; however, similar reactions have been found to occur in a few eukaryotes and archaea (6, 27). Eukaryotic nitrate respiration was first found in protozoa that reside in an anaerobic freshwater habitat (8). The organism particularly reduces NO3− to NO2− in a single step, a process which recovers dNaR activity in the mitochondrial fraction but does not result in denitrification. Eukaryotic denitrification was first found to occur among fungi (19, 20), which generally form N2O from NO3− or NO2−. Recently, eukaryotic denitrification was also found in a benthic foraminifer that forms N2 from NO3− (18). The fungal denitrification system localizes in the mitochondria and couples to the mitochondrial electron transport chain to produce ATP (12, 21), thus exhibiting properties similar to those of the bacterial systems in its ability to respire anaerobically. Moreover, the mechanism of anaerobic respiration in the “aerobic” organelle of eukaryotes (mitochondrion) evokes interest regarding the origin and evolution of the mitochondrion.
The main components of the fungal denitrifying system, the dNaR, dNiR, and NoR proteins, were either completely or partially purified from Fusarium oxysporum. Fungal NoR of the cytochrome P450 (P450) type, referred to as P450nor (CYP55) (11, 16), is a distinct species of bacterial cytochrome cb-type NoR. By contrast, the previously isolated fungal dNiR protein is a copper-containing type (NirK) that closely resembles its bacterial counterpart (13). Furthermore, dNaR activity partially purified from the mitochondrial membrane fraction showed that fungal dNaR possibly resembles its bacterial counterpart, NarGHI (12, 23). Therefore, while a portion of the fungal system appears to resemble its bacterial counterpart, the phylogenetic relationship between the fungal and bacterial denitrification systems remained unclear because the genes of the fungal components (dNaR and dNiR) have not been sequenced.
Recent genome analyses have revealed the presence of nirK homolog genes in many eukaryotes (fungi and protozoa), a finding consistent with our previous findings on the isolation of the fungal NirK protein (13). Therefore, whether these eukaryotes containing the nirK homolog gene exhibit denitrification activity and whether the denitrifying fungus F. oxysporum really contains a nirK gene deserve a great deal of attention. To address this issue, we used the suppression subtractive hybridization (SSH) technique (7) and succeeded in isolating the nirK gene from the denitrifying fungus F. oxysporum.
MATERIALS AND METHODS
Organism, media, and culture conditions.
F. oxysporum MT-811 (JCM11502) was used throughout this work, in which fungal denitrification was first confirmed (20). The medium used for fungal growth was glycerol peptone (GP) medium, as previously reported (13). The preculture was prepared by culturing F. oxysporum aerobically in GP medium in the absence of nitrate or nitrite (nondenitrifying conditions) at 30°C for 3 days. A portion (30 ml) of the preculture was transferred into 120 ml of fresh GP medium and further incubated at 30°C at 150 rpm in a 500-ml Erlenmeyer flask that was sealed with a rubber stopper (initially aerobic conditions), which is appropriate for fungal denitrification (20). Denitrifying cells were grown in the presence of 20 mM NaNO2, and nondenitrifying cells were grown in the absence of nitrite. Evolution of N2O was confirmed to occur only with the denitrifying culture, as previously observed (20).
SSH.
SSH (7) was examined by comparing mRNA preparations (2 μg) extracted from denitrifying and nondenitrifying mycelia (grown with and without nitrite). All of the SSH steps were performed with the PCR-select cDNA subtraction kit (BD Biosciences-Clontech, Palo Alto, CA). SSH-generated libraries typically contain some background clones representing nondifferentially expressed transcripts. To overcome this problem, we used the mirror orientation selection (MOS) method (3, 17). The PCR-amplified cDNA fragments generated by MOS were then ligated into plasmid pT7blue-2 (Novagen, San Diego, CA).
Differential screening of the subtracted cDNA library by array dot blotting.
cDNA clones were randomly picked from the collection of forward subtracted cDNA plasmid libraries prepared as described above, and their cDNA inserts were amplified by colony PCR with nested primer 2Rs (17). The PCR product (0.5 μl) was blotted in duplicate onto a Hybond N+ membrane (GE Healthcare, Little Chalfont, United Kingdom). The membrane was denatured, neutralized, and then fixed according to the DIG (digoxigenin) application manual (Roche, Mannheim, Germany). Probes were produced with the DIG probe synthesis kit (Roche, Mannheim, Germany). About 100 ng of the forward and reverse subtracted PCR-amplified cDNA fragments generated by MOS was used as the template, and nested primer 2Rs was used as the primer for probe synthesis. The membrane was hybridized at 68°C for 16 h with DIG-labeled probes. After hybridization, membranes were washed twice in a primary wash solution (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]) at room temperature for 5 min and then twice in a secondary wash solution (0.5× SSC, 0.1% SDS) at 68°C for 15 min. Membranes were then detected with the DIG nucleic acid detection kit (Roche, Mannheim, Germany). The duplicate membranes were then hybridized with either forward or reverse subtracted DIG-labeled probes for identical blots. The reverse subtracted cDNA probe is a negative control because this population is theoretically enriched for genes expressed only under NO2−-limited culture conditions (driver cDNA population). For the cDNA array dot blot screenings, see Fig. S1 in the supplemental material. Sequencing of differentially expressed clones was performed with the M13 forward and reverse primers and the CEQ 2000XL DNA analysis system (Beckman Coulter, Fullerton, CA). Analysis of sequence data and sequence similarity searches were performed by using the BlastX program of the National Center for Biotechnology Information (NCBI) (1).
Expression profile analysis by Northern blotting.
To exclude false clones and obtain quantitative expression levels, the clones obtained by the SSH technique were analyzed by Northern blot analysis. A total of 5 μg of RNA was separated by electrophoresis on a denaturing 1% agarose gel, followed by blotting onto a Hybond N+ membrane (GE Healthcare, Little Chalfont, United Kingdom). The membrane was baked at 70°C for 2 h and stored at −20°C until use. The DIG-labeled probes were generated by PCR with the respective cloned fragments as templates and gene-specific primers (see Table S1 in the supplemental material). Hybridization and detection were performed as described above.
Identification of the full-length cDNA of F. oxysporum dNiR (FoNiR) and the genomic structure.
Oligonucleotide-capped full-length cDNAs were prepared with the GenRacer kit (Invitrogen, Carlsbad, CA). Total RNA (2 μg) from the denitrifying cells (grown in the presence of nitrite) was used as the starting material. Rapid amplification of cDNA 5′ ends (5′ RACE) and 3′ RACE reactions were performed with the Easy-A high-fidelity PCR cloning enzyme and a gene-specific primer paired with the 5′ or 3′ NP primer (see Table S1 in the supplemental material) and with oligonucleotide-capped, full-length cDNAs as the template. RACE amplification products were sequenced. Fungal genomic DNA was prepared with the Genomic DNA Purification kit (Promega, Madison, WI). FoNiR genomic DNA was cloned by PCR with a gene-specific primer (see Table S1 in the supplemental material) and then sequenced. The exon-intron boundary of the FoNiR gene was determined by aligning the sequence of the cloned genomic DNA and the sequence of the cDNA. Analyses of sequence data and sequence homology were performed with the Bl2seq program of the NCBI (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi).
dNiR activity assay.
dNiR activity was assayed as in a previous study (23), with the slight modification described below. The standard reaction mixture (final volume of 3.0 ml in a 15-ml test tube) contained 2 mM sodium nitrite, 0.1 mM methyl viologen, and 0.05 μg of recombinant FoNiR (rFoNiR) in 50 mM morpholineethanesulfonic acid-NaOH buffer (pH 6.5). After the reaction, residual nitrite was determined with the Griess reagent [0.02% (vol/vol) N-(1-naphthyl)ethylenediamine and 1% (vol/vol) sulfanilic acid in 25% (vol/vol) HCl]. One unit of dNiR activity was defined as the amount that reduces 1 μmol of NO2−/min.
Other determinations.
SDS-PAGE (polyacrylamide gel electrophoresis) was performed with a 10% gel for checking the purity and estimating the Mrs of purified proteins. Protein concentration was determined with a protein assay reagent (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard. The Mr of rFoNiR was also determined by gel filtration with a Superdex 200 HR 10/60 column (GE Healthcare, Little Chalfont, United Kingdom) equilibrated with 20 mM Tris-HCl buffer (pH 7.5) containing 150 mM NaCl. The standard proteins used were thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), bovine serum albumin (66 kDa), and carbonic anhydrase (29 kDa) (Sigma, St. Louis, MO). Amino-terminal sequence analysis was performed with the protein sequencer Procise 491HT (Applied Biosystems, Foster City, CA). UV-visible light absorption spectra were recorded at room temperature with a V-550 spectrophotometer (Jasco, Tokyo, Japan). Electron paramagnetic resonance (EPR) spectra were measured with a JES-FA100 ESR spectrophotometer (JEOL, Tokyo, Japan) at 20 K. Atomic absorption analysis was performed with an SPS1200VR plasma spectrometer (Seiko, Tokyo, Japan).
For additional details, see Materials and Methods in the supplemental material.
Nucleotide sequence accession number.
The DNA and deduced amino acid sequences of the full-length FoNiR protein have been deposited in GenBank under accession number EF600898.
RESULTS AND DISCUSSION
Isolation of the genes of F. oxysporum differentially expressed under denitrifying conditions.
We first isolated the genes that are specifically expressed under denitrifying conditions (initially aerobic, in the presence of NO2−) by the SSH technique. The patterns of hybridization signal intensity appeared to differ between the duplicate array membranes that were, respectively, hybridized with the forward (see Fig. S1A in the supplemental material) and reverse (see Fig. S1B in the supplemental material) subtracted probes, indicating the success of the SSH procedure in identifying the differentially expressed clones. The cDNA inserts of 40 clones were partially sequenced. A search in the National Center for Biotechnology Information nonredundant database with the BlastX algorithm (1) revealed that 36 clones had at least one significant (E value, <10e−7) hit in the database and comprised 12 distinct mRNA species (Table 1).
TABLE 1.
Characteristics of the cDNA fragments in the subtracted library that evolved from the genes of F. oxysporum differentially expressed by the addition of NO2−
| Clone | No. of clones | Size (bp) | Putative homologa | Organism | E value | Putative function |
|---|---|---|---|---|---|---|
| B16 | 6 | 586 | Flavohemoglobin (NO deoxygenase) | Fusarium oxysporum | 7e−99 | Nitrogen metabolism |
| D12 | 4 | 492 | Flavohemoglobin (NO deoxygenase) | Fusarium oxysporum | 2e−88 | Nitrogen metabolism |
| E8 | 6 | 313 | Nitrite reductase I | Ajellomyces capsulatus | 7e−41 | Nitrogen metabolism (denitrification) |
| E7 | 3 | 483 | Nitrite reductase I | Ajellomyces capsulatus | 4e−52 | Nitrogen metabolism (denitrification) |
| G4 | 5 | 529 | Hypothetical protein FG00663.1 | Gibberella zeae PH-1 | 1e−59 | Unknown |
| F4 | 4 | 656 | Hypothetical protein FG00663.1 | Gibberella zeae PH-1 | 2e−28 | Unknown |
| N12 | 2 | 457 | NAD-dependent formate dehydrogenase | Gibberella zeae PH-1 | 1e−31 | Glyoxylate metabolism |
| G18 | 2 | 458 | Alcohol dehydrogenase I | Metarhizium anisopliae | 5e−66 | Gluconeogenesis |
| H2 | 2 | 338 | No significant matches | Unknown | ||
| G11 | 1 | 413 | P450nor (NO reductase) | Fusarium oxysporum | 1e−58 | Nitrogen metabolism (denitrification) |
| L3 | 1 | 469 | Zinc finger transcription factor | Pichia stipitis | 7e−10 | Transcription |
| K11 | 1 | 455 | γ-Amino-n-butyrate permease | Emericella nidulans | 4e−42 | Transporter |
| A10 | 1 | 552 | Indoleamine 2,3-dioxygenase family protein | Neosartorya fischeri | 1e−52 | Tryptophan metabolism |
| K4 | 1 | 422 | No significant matches | Unknown | ||
| N15 | 1 | 271 | No significant matches | Unknown |
Putative homologs are based on BlastX searches of the nonredundant GenBank database.
Of the 36 differentially expressed sequences, many clones showed redundancy. Ten clones, including B16 and D12, were derived from the flavohemoglobin (Fhb) gene (GenBank accession no. BAA33011); nine, including E7 and E8, were derived from a homolog of nitrite reductase of Ajellomyces capsulatus (GenBank accession no. AY816318); and nine were derived from hypothetical protein FG00663.1 of Gibberella fujikuroi. These results suggest that the genes are highly expressed under denitrifying conditions. It is important to note that clones E7 and E8 are also homologs of the NirK proteins of bacteria (5). We previously demonstrated that the denitrification process of F. oxysporum is performed by a set of three main enzymes: dNaR, dNiR, and NoR (P450nor) (12, 16, 20). Here, we isolated the genes for dNiR (nirK) and P450nor (CYP55) as differentially expressed genes (Table 1), although no clone was isolated, which was homologous to any known dNaR registered in the public databases. It is possible that dNaR is induced by NO3− but not by NO2−. The high proportion of dNiR homolog clones (9 out of 40 clones) also supports the view that the gene product is the dNiR protein involved in fungal denitrification. On the other hand, only one clone (G11) could be identified as P450nor (CYP55) of F. oxysporum (GenBank accession no. BAA 03390), suggesting that P450nor is, to a lesser extent, constitutively expressed even under nondenitrifying conditions.
Clone G18 showed similarity to alcohol dehydrogenase, and clone N12 was identified as an NAD-dependent formate dehydrogenase. Isolation of these genes is consistent with our previous observation that ethanol and formate, the substrates of alcohol dehydrogenase and formate dehydrogenase, act as the electron donors for anaerobic energy metabolism (ammonia fermentation or denitrification) (23, 26). Of the other clones isolated here, G4, F4, L3, K11, and A10 were homologs for hypothetical protein FG00663.1, a transcription factor, γ-amino-n-butyrate permease, and indoleamine 2,3-dioxygenase-like protein, respectively, all of whose relationships with denitrification are not clear. The remaining clones, H2, K4, and N15, did not match any known sequences in the public databases; however, further studies are required to elucidate the functions of these genes.
Confirmation of differential expression by Northern analysis.
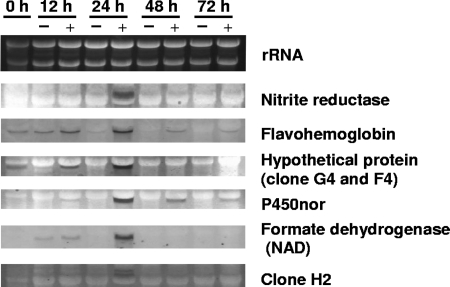
In the SSH experiments described above (Table 1), fungal cells were first grown aerobically (preculture) and then transferred to “initially aerobic” culture conditions (discussed in Materials and Methods) to induce denitrifying activity. In the second culture, the O2 in the headspace air of the sealed flask was consumed within the first 20 h, after which denitrification started, as previously reported (20). This implied that energy metabolism changes from aerobic respiration to denitrification (nitrate/nitrite respiration) during the first day of culture. Next, we initiated a Northern blot analysis to check whether the selected genes, including the nirK homolog, are specifically expressed in denitrification (Fig. 1). The results show that all of the genes tested are expressed most strongly at the 24-h growth stage, but only upon supplementation with NO2−. The sampling time (24 h) corresponds to the stage just when denitrification is initiated (confirmed by N2O formation; data not shown). Therefore, initiation of denitrification is accompanied by synchronous expression of the genes, providing unambiguous evidence that these genes are involved in denitrification. The present results (Table 1 and Fig. 1) also suggest that denitrification is supported by many genes (and their products) in addition to the main components (dNaR, dNiR, and NoR), including proteins with unknown functions.
FIG. 1.
Expression of the putative genes in F. oxysporum responsible for denitrification. The differential expression of genes isolated under denitrifying conditions (Table 1) was examined by the Northern blotting technique. Each lane contained 5 μg of total RNA from fungal cells incubated for 0, 12, 24, 48, and 72 h, respectively, in GP medium with (+) and without (−) NO2−. Each blot was hybridized with the indicated DIG-labeled cDNA probe. Equal loads of 28S and 18S rRNAs are shown in each lane.
Full-length cDNA and genomic structure of the dNiR homolog gene and comparison of the deduced amino acid sequence with that of bacterial NirK proteins.
The DNA sequence of the dNiR homolog gene for FoNiR was determined, and the full-length cDNA was obtained by 5′ and 3′ RACE. The full-length cDNA was 1,838 bp long, comprising a coding region of 1,491 bp, a 53-bp 5′ untranslated region, and a 294-bp 3′ untranslated region. The FoNiR gene consisted of two exons with a 47-bp intron between them.
The FoNiR gene encoded a predicted protein comprising 496 amino acid residues with a calculated Mr of 53,705. For a comparison of the deduced amino acid sequence with that of bacterial NirK proteins, see Fig. S2 in the supplemental material. The alignment shows that the C-terminal domain of FoNiR (from Ile179 to Ser496) exhibits overall sequence similarities to the bacterial NirK proteins (dNiR domain), while that of the N-terminal region (from Met1 to Ala178) contains an extension with few sequence similarities to bacterial NirK proteins.
Sequence analysis (MITOPROT, http://ihg.gsf.de/ihg/mitoprot.html; HMMTOP, http://www.enzim.hu/hmmtop/html/submit.html) showed that the first 65 amino acid residues of the N-terminal domain constitute a mitochondrial targeting signal, followed by a transmembrane sequence and a stem region (see Fig. S2 in the supplemental material). These results provide additional support to the view that fungal dNiR is a mitochondrial membrane protein, as previously suggested by enzyme fraction studies (13, 14). Consequently, the primary sequence identities between the FoNiR and bacterial NirK proteins was higher (up to 50%) (Fig. 2) when only the dNiR domain of FoNiR was used for comparison. The alignment revealed that the ligands of the type 1 and 2 Cu centers are completely conserved in FoNiR. They are His-253, Cys-294, His-302, and Met-307 for the type 1 Cu center and His-258, His-293, and His-456 for type 2 (see Fig. S2 in the supplemental material). Moreover, the Asp and His residues, which are important for enzyme catalysis (4, 10) by forming a hydrogen bond network on the type 2 Cu center, are conserved in FoNiR (Asp-256 and His-405).
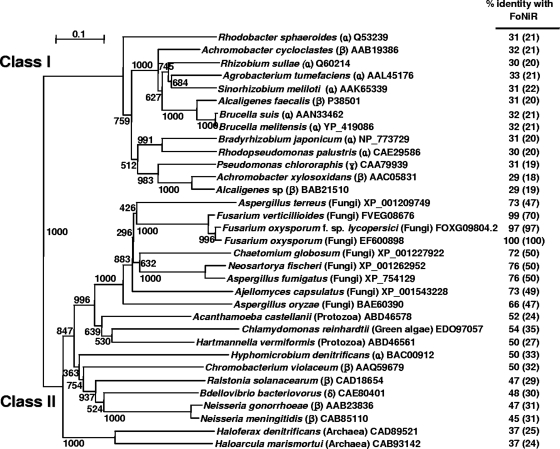
FIG. 2.
Phylogenetic tree of NirK proteins. The construction of an unrooted phylogenetic tree is inferred from the alignment of the complete amino acid sequences based on the ClustalX program. Numbers adjoining the branches indicate bootstrap values (from 1,000 replicates). The scale bar of evolutionary distance indicates the mean number of amino acid substitutions per sequence position. Each NirK protein is shown by its origin (organism), and the classification of each organism (subclass of proteobacteria, archaea, or eukaryotes [fungi, protozoa, and green algae]) is shown in parentheses. The percent identity of NirK from F. oxysporum with respect to the amino acid sequences of selected pairs (dNiR domain) and to the whole protein length (in parentheses) are indicated numerically. The protozoal homolog (Acanthamoeba castellanii and Hartmannella vermiformis) sequences are partial. Accession numbers are given for sequences in the GenPep databank, except for F. verticillioides and F. oxysporum f. sp. lycopersici (Fungal Genetics Stock Center).
Bacterial NirK proteins are divided into two major groups, classes I and II (5). FoNiR was classified as class II (Fig. 2). The NirK proteins in class II are more divergent than are those in class I; e.g., AniA from Neisseria is an exceptional membrane protein among bacterial NirK proteins (5), HdNiR from Hyphomicrobium denitrificans contains an additional cupredoxin domain in its N terminus (24), and both contain extensions in their N termini (see Fig. S2 in the supplemental material). Another characteristic feature of the class II NirK proteins is that they have two short deletions in the linker loop (LL) and tower loop sequences (see box in Fig. S2 in the supplemental material). This is also seen in FoNiR (see Fig. S2 boxed). The LL is located on the bottom of each monomer and creates a protrusion in the AfNiR and AxNiR structures (5). Moreover, the LL serves as a barrier for membrane interaction and thus shortens the deletion in FoNiR (six-residue deletion), as in AniA, thereby favoring interaction with the membrane surface.
Phylogenetic relationship of eukaryotic NirK homologs with prokaryotic NirK proteins.
In recent years, the occurrence of nirK homolog genes has been revealed in many eukaryotic genomes (fungi, protozoa, and green algae) although their physiological functions have never been examined. We constructed a phylogenetic relationship between NirK proteins and NirK homologs of prokaryotes and eukaryotes based on the deduced amino acid sequences. As shown in Fig. 2, eukaryotic NirK homologs including FoNiR cluster together. Potential ligands of the type 1 and type 2 Cu centers are completely conserved in the full sequences of these eukaryotic genes (data not shown). Among prokaryotic NirK proteins, class II bacterial NirK proteins such as those of Hyphomicrobium denitrificans (HdNiR), Chromobacterium violaceum, and so on (5) are phylogenetically more similar to eukaryotic NirK and its homologs. The phylogenetic tree shows that classes I and II diverged first, and then the class II NirK proteins further split into archaeal, bacterial, and eukaryotic groups. The eukaryotic branch is further divided into fungi and protozoa. The random distribution of NirK proteins among proteobacteria suggests that horizontal transfer of nirK genes frequently occurred between these bacteria. By contrast, the eukaryotic NirK homologs were systematically distributed in the same branch, suggesting that they emerged from the same ancestor (Fig. 2).
Expression and purification of rFoNiR in E. coli.
A portion of the FoNiR gene, in which the mitochondrial targeting signal and transmembrane sequences were truncated, was expressed in the heterologous host E. coli HMS174. The truncated cDNA thus encodes the dNiR and stem region domains (from Glu112 to Ser496) with an estimated Mr of 46,919 (containing the His tag region). rFoNiR was purified from the soluble fraction of the cells and exhibits a single band on SDS-PAGE (see Fig. S3B in the supplemental material). The Mr of rFoNiR is estimated to be about 60,000 from SDS-PAGE and greater than 900,000 from a nondenaturing gel filtration measurement (data not shown). This suggests that rFoNiR is aggregated, while the deviation between the values of a monomer calculated and estimated from SDS-PAGE is unclear. Therefore, we determined the N-terminal amino acid sequence of the main band and obtained the sequences GSSHHHHHHSS for the protein as purified and GSHMRGPQPV for the thrombin-cleaved protein. The sequence RGPQPV is completely identical to that of the N terminus of FoNiR introduced into pET28b/FoNiR, indicating that it is rFoNiR.
Characterization of purified rFoNiR and comparison with native FoNiR (nFoNiR).
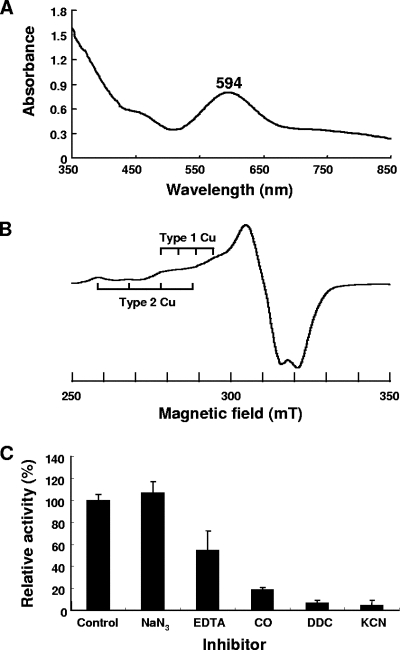
We previously purified and characterized dNiR of F. oxysporum (the same strain used in this study) (13). Here, we compared the properties of purified rFoNiR and nFoNiR (the data for nFoNiR are from a previous paper [13]). Purified rFoNiR exhibits a blue color spectrum showing maximum absorbance at 594 nm, as shown in Fig. 3A, which is almost identical to that of nFoNiR, whose main peak is at 595 nm. The EPR spectrum (shown in Fig. 3B) is also characteristic of NirK proteins that contain both type 1 and type 2 copper centers. The approximate AII and gII values were estimated to be 5.4 mT and 2.25, respectively, for hyperfine splitting originated from type 1 copper and 10 mT and 2.34, respectively, for type 2 copper. These values are similar to those obtained with nFoNiR: 6.82 mT and 2.22 for type 1 copper and 10 mT and 2.32 for type 2 copper. rFoNiR exhibits dNiR activity with optima at pH values below 6.5 (data not shown), which is consistent with the results obtained for nFoNiR (13). Its specific activity (303 U/mg protein; pH 6.5, 35°C) is also comparable to that of nFoNiR (447 U/mg; pH 7.0, 30°C). The results obtained for inhibitors (Fig. 3C) are similar to those obtained for nFoNiR and other NirK proteins and indicate that rFoNiR and nFoNiR resemble each other spectrally and enzymatically.
FIG. 3.
Absorption and EPR spectra of rFoNiR and effects of inhibitors on dNiR activity. (A) Absorption spectrum of purified rFoNiR, showing an absorption maximum at 594 nm, in 20 mM Tris-HCl buffer (pH 7.5) containing 0.15 M NaCl, 0.1 mM CuSO4 · 5H2O, and 17.4 mg/ml protein. (B) EPR spectrum of rFoNiR at 20 K. Purified rFoNiR (26.0 mg/ml) was dissolved in 20 mM Tris-HCl buffer (pH 8.0) containing 0.1 mM CuSO4 · 5H2O. The conditions were as follows: microwave frequency, 8.99 GHz; microwave power, 0.99 mW; modulation amplitude, 1.0 mT; sweep time, 30 s; time constant, 0.03 s. (C) Effects of inhibitors. rFoNiR was incubated with each inhibitor (1 mM, except CO) at 35°C for 20 min prior to the enzyme assay. For examination of CO, helium in the gas phase of the assay vessel was replaced with CO. The control contained no inhibitor. DDC, diethyldithiocarbamate.
On the other hand, the increase in absorbance with a decrease in wavelength (UV region) (Fig. 3A), which is not observed with nFoNiR, suggests that rFoNiR is aggregated. This is consistent with the result obtained by gel filtration mentioned above, which yielded a large Mr value for native rFoNiR (above 900,000). Furthermore, the extinction coefficient of the absorbance at 594 nm is estimated to be 2.16 mM−1 cm−1, which is less than half of that for nFoNiR (5.38 mM−1 cm−1), suggesting that only a portion of the rFoNiR protein contains the type 1 Cu center. It therefore appears that the rFoNiR preparation is a mixture of proteins that were completely and incompletely folded in the heterologous expression system (E. coli cells), resulting in aggregation.
Native fungal dNiR activity of F. oxysporum is recovered in both the soluble and large particle fractions when the cells are disrupted, and nFoNiR was isolated from the soluble fraction. The Mr value of soluble nFoNiR estimated by SDS-PAGE (41,800 per monomer) is almost identical to that of rFoNiR comprising the dNiR and stem region domains (41,722; from Glu112 to Ser496 without the His tag domain). These results suggest that a portion of FoNiR is solubilized from the membrane during the cell disruption process, resulting in the formation of soluble nFoNiR that has lost the membrane-bound domain.
These results do not contradict the assumption that nFoNiR and rFoNiR are the products of the same gene (nirK). We can therefore conclude that the nirK gene homolog isolated in this study from F. oxysporum encodes the NirK protein involved in fungal denitrification and thus is the first eukaryotic ortholog of bacterial nirK genes.
Mitochondrial anaerobic respiration.
It is generally accepted that the mitochondrion of eukaryotes evolved as the result of endosymbiosis of an alphaproteobacterium with an anaerobic host (25) and that the mitochondria of all eukaryotes have a common origin (9). The endosymbiont that gave rise to the mitochondrion was previously believed to be an obligate aerobe. However, the discovery of anaerobic respiration (including fungal denitrification) in mitochondria (22) suggests that the endosymbiont was not an obligate aerobe but a facultative anaerobe and that most eukaryotes discarded their anaerobic respiration ability during the process of adaptation to aerobic environments (15).
In contrast to the orthologous relationship between the eukaryotic and prokaryotic nirK genes, the P450-type NoR (P450nor) protein has not been found among prokaryotes. However, many fungi that contain nirK homologs also contain P450nor homologs, suggesting that this type of denitrification system (comprising NirK and P450nor) is ubiquitous among fungi. Therefore, it appears that the mitochondrial denitrifying system replaced the original NoR protein with P450nor, whose gene was initially obtained from bacteria by means of horizontal gene transfer. The prototype P450 gene would have encoded a usual monooxygenase, whereas fungi would have modulated the gene so as to give NoR activity.
There is another distinct dNiR species, cytochrome cd1-type dNiR (NirS), which is also widely distributed among denitrifying bacteria (27). Denitrifying bacteria usually contain either NirK or NirS, but not both. No correlation seems to exist between the phylogeny of bacteria and the type of dNiR (NirK or NirS). For example, both types are distributed equally among alphaproteobacteria; H. denitrificans and Bradyrhizobium have NirK, whereas Paracoccus denitrificans contains NirS. By contrast, the distribution of eukaryotic nirK homologs is systematic (Fig. 2). Further, only NirK homologs (and not NirS) have been found in eukaryotic genomes. These results and facts indicate that eukaryotic nirK homologs evolved from a single ancestor and diverged with each host that harbored the gene.
The results described above suggest that eukaryotic nirK homologs originated from the protomitochondrion (the endosymbiont that gave rise to the mitochondrion), which harbored NirK-type (but not NirS-type) dNiR. Although eukaryotic nirK homologs are localized in the nuclear genome, a mitochondrial targeting signal can be found in most of them. Some eukaryotes, in particular those harboring both NirK and P450nor, like F. oxysporum, would have conserved the denitrification system derived from the protomitochondrion, although it was modified (adoption of P450nor). It is also possible that some eukaryotic NirK proteins have modulated its function, like NirK of Rhizobium sullae, which has acquired the ability to reduce selenite (2).
Supplementary Material
Acknowledgments
We are grateful to Shinnichiro Suzuki of the Osaka University Department of Chemistry for his timely discussion. We also thank Etsuro Yoshimura and Mitsuru Abo of the University of Tokyo Graduate School of Agricultural and Life Sciences for their helpful discussion and support of the atomic absorption analysis.
This work was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to H.S.; no. 20248009) and The Research and Development Program for New Bio-industry Initiatives.
Footnotes
Published ahead of print on 6 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basaglia, M., A. Toffanin, E. Baldan, M. Bottegal, J. P. Shapleigh, and S. Casella. 2007. Selenite-reducing capacity of the copper-containing nitrite reductase of Rhizobium sullae. FEMS Microbiol. Lett. 269:124-130. [DOI] [PubMed] [Google Scholar]
- 3.Boengler, K., F. Pipp, W. Schaper, and E. Deindl. 2003. Rapid identification of differentially expressed genes by combination of SSH and MOS. Lab. Investig. 83:759-761. [DOI] [PubMed] [Google Scholar]
- 4.Boulanger, M. J., M. Kukimoto, M. Nishiyama, S. Horinouchi, and M. E. Murphy. 2000. Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase. J. Biol. Chem. 275:23957-23964. [DOI] [PubMed] [Google Scholar]
- 5.Boulanger, M. J., and M. E. Murphy. 2002. Crystal structure of the soluble domain of the major anaerobically induced outer membrane protein (AniA) from pathogenic Neisseria: a new class of copper-containing nitrite reductases. J. Mol. Biol. 315:1111-1127. [DOI] [PubMed] [Google Scholar]
- 6.Cabello, P., M. D. Roldan, and C. Moreno-Vivian. 2004. Nitrate reduction and the nitrogen cycle in archaea. Microbiology 150:3527-3546. [DOI] [PubMed] [Google Scholar]
- 7.Diatchenko, L., Y. F. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlay, B. J., A. S. W. Span, and J. M. P. Harman. 1983. Nitrate respiration in primitive eukaryotes. Nature 303:333-336. [Google Scholar]
- 9.Gray, M. W., G. Burger, and B. F. Lang. 1999. Mitochondrial evolution. Science 283:1476-1481. [DOI] [PubMed] [Google Scholar]
- 10.Kataoka, K., H. Furusawa, K. Takagi, K. Yamaguchi, and S. Suzuki. 2000. Functional analysis of conserved aspartate and histidine residues located around the type 2 copper site of copper-containing nitrite reductase. J. Biochem. (Tokyo) 127:345-350. [DOI] [PubMed] [Google Scholar]
- 11.Kizawa, H., D. Tomura, M. Oda, A. Fukamizu, T. Hoshino, O. Gotoh, T. Yasui, and H. Shoun. 1991. Nucleotide sequence of the unique nitrate/nitrite-inducible cytochrome P-450 cDNA from Fusarium oxysporum. J. Biol. Chem. 266:10632-10637. [PubMed] [Google Scholar]
- 12.Kobayashi, M., Y. Matsuo, A. Takimoto, S. Suzuki, F. Maruo, and H. Shoun. 1996. Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J. Biol. Chem. 271:16263-16267. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi, M., and H. Shoun. 1995. The copper-containing dissimilatory nitrite reductase involved in the denitrifying system of the fungus Fusarium oxysporum. J. Biol. Chem. 270:4146-4151. [DOI] [PubMed] [Google Scholar]
- 14.Kubota, Y., N. Takaya, and H. Shoun. 1999. Membrane-associated, dissimilatory nitrite reductase of the denitrifying fungus Cylindrocarpon tonkinense. Arch. Microbiol. 171:210-213. [Google Scholar]
- 15.Martin, W., M. Hoffmeister, C. Rotte, and K. Henze. 2001. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol. Chem. 382:1521-1539. [DOI] [PubMed] [Google Scholar]
- 16.Nakahara, K., T. Tanimoto, K. Hatano, K. Usuda, and H. Shoun. 1993. Cytochrome P-450 55A1 (P-450dNIR) acts as nitric oxide reductase employing NADH as the direct electron donor. J. Biol. Chem. 268:8350-8355. [PubMed] [Google Scholar]
- 17.Rebrikov, D. V., O. V. Britanova, N. G. Gurskaya, K. A. Lukyanov, V. S. Tarabykin, and S. A. Lukyanov. 2000. Mirror orientation selection (MOS): a method for eliminating false positive clones from libraries generated by suppression subtractive hybridization. Nucleic Acids Res. 28:E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Risgaard-Petersen, N., A. M. Langezaal, S. Ingvardsen, M. C. Schmid, M. S. M. Jetten, H. J. M. Op den Camp, J. W. M. Derksen, E. Pina-Ochoa, S. P. Eriksson, L. P. Nielsen, N. P. Revsbech, T. Cedhagen, and G. J. van der Zwaan. 2006. Evidence for complete denitrification in a benthic foraminifer. Nature 443:93-96. [DOI] [PubMed] [Google Scholar]
- 19.Shoun, H., D. H. Kim, H. Uchiyama, and J. Sugiyama. 1992. Denitrification by fungi. FEMS Microbiol. Lett. 73:277-281. [DOI] [PubMed] [Google Scholar]
- 20.Shoun, H., and T. Tanimoto. 1991. Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction. J. Biol. Chem. 266:11078-11082. [PubMed] [Google Scholar]
- 21.Takaya, N., S. Kuwazaki, Y. Adachi, S. Suzuki, T. Kikuchi, H. Nakamura, Y. Shiro, and H. Shoun. 2003. Hybrid respiration in the denitrifying mitochondria of Fusarium oxysporum. J. Biochem. (Tokyo) 133:461-465. [DOI] [PubMed] [Google Scholar]
- 22.Tielens, A. G., C. Rotte, J. J. van Hellemond, and W. Martin. 2002. Mitochondria as we don't know them. Trends Biochem. Sci. 27:564-572. [DOI] [PubMed] [Google Scholar]
- 23.Uchimura, H., H. Enjoji, T. Seki, A. Taguchi, N. Takaya, and H. Shoun. 2002. Nitrate reductase-formate dehydrogenase couple involved in the fungal denitrification by Fusarium oxysporum. J. Biochem. (Tokyo) 131:579-586. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi, K., K. Kataoka, M. Kobayashi, K. Itoh, A. Fukui, and S. Suzuki. 2004. Characterization of two type 1 Cu sites of Hyphomicrobium denitrificans nitrite reductase: a new class of copper-containing nitrite reductases. Biochemistry 43:14180-14188. [DOI] [PubMed] [Google Scholar]
- 25.Yang, D., Y. Oyaizu, H. Oyaizu, G. J. Olsen, and C. R. Woese. 1985. Mitochondrial origins. Proc. Natl. Acad. Sci. USA 82:4443-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou, Z., N. Takaya, A. Nakamura, M. Yamaguchi, K. Takeo, and H. Shoun. 2002. Ammonia fermentation, a novel anoxic metabolism of nitrate by fungi. J. Biol. Chem. 277:1892-1896. [DOI] [PubMed] [Google Scholar]
- 27.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.