Abstract
Anecdotes, both historical and recent, recount the curing of skin infections, including diaper rash, by using red soils from the Hashemite Kingdom of Jordan. Following inoculation of red soils isolated from geographically separate areas of Jordan, Micrococcus luteus and Staphylococcus aureus were rapidly killed. Over the 3-week incubation period, the number of specific types of antibiotic-producing bacteria increased, and high antimicrobial activity (MIC, ∼10 μg/ml) was observed in methanol extracts of the inoculated red soils. Antibiotic-producing microorganisms whose numbers increased during incubation included actinomycetes, Lysobacter spp., and Bacillus spp. The actinomycetes produced actinomycin C2 and actinomycin C3. No myxobacteria or lytic bacteriophages with activity against either M. luteus or S. aureus were detected in either soil before or after inoculation and incubation. Although protozoa and amoebae were detected in the soils, the numbers were low and did not increase over the incubation period. These results suggest that the antibiotic activity of Jordan's red soils is due to the proliferation of antibiotic-producing bacteria.
There is a growing recognition of the pressing need for new antimicrobial agents for the treatment of infectious diseases (11, 38). As just one cogent example, new antibiotics are in high demand for the treatment of Staphylococcus aureus infections (25), particularly due to the emergence of methicillin-resistant S. aureus in communities and hospitals (25, 41). In addition, providing effective and affordable antibiotics to people in epidemic-prone developing countries remains a major challenge (37).
Historically, natural products have played a key role in the discovery and development of many antibiotics (34). In particular, soil-based actinomycetes have been the source of countless drugs, such as streptomycin, actinomycin, erythromycin, and vancomycin, to name only a few (18). One approach to the discovery of new antimicrobial agents from natural sources has been to use folklore or historical records to guide the collection of samples (20).
Through our ongoing studies of the biodiversity of the Hashemite Kingdom of Jordan (Jordan) (2-7, 35, 36), we were intrigued by anecdotes of the antibiotic-like properties of red soil, used historically for treating skin infections and diaper rash and still in use in some communities as an inexpensive alternative to pharmaceutical products. Within Jordan, there are four major biogeographic regions (8, 21), and red soils are most commonly found in the Mediterranean region of the northwestern portion of Jordan, near cities such as Irbid, Ramtha, and Ajloun. An area away from housing, preferably not touched by feet and thus considered clean, is chosen, and soil below the surface is collected, as the surface is considered contaminated or not clean. After the infected area of skin is washed and dried, the sieved soil is applied daily as either a powder or paste until the infection subsides. The basis for the antimicrobial activity of red soils is not known.
Antimicrobial activity of soils against inoculated microorganisms has been attributed to abiotic or biotic factors. Abiotic activity has been shown to be responsible for the antimicrobial activities of clay minerals used in the treatment of a mycobacterial skin infection known as Buruli ulcer (28). Soil texture was found to influence the survival of Pseudomonas fluorescens and Bacillus subtilis in soil (40), while soil temperature and pH and the presence of roots affected the leaching of a genetically modified strain of P. fluorescens in soil (29). Biotic factors, including predation and antimicrobial-producing or lytic microorganisms, were suggested as mechanisms for killing microorganisms introduced into soils (1, 14, 31, 32). Prior inoculation of soil with one strain of P. fluorescens reduced the ability of a second P. fluorescens strain to colonize (19). A phenazine pigment produced by a P. fluorescens strain was shown to be responsible for biological control of a root disease of wheat caused by Gaeumannomyces graminis var. tritici (39), and it has been shown that filaments of the biocontrol fungus Trichoderma grow toward fungal pathogens and release antibiotics and lytic enzymes (9). With that background as a guide, we undertook an investigation to identify the basis for the antimicrobial activity of Jordan's red soils.
MATERIALS AND METHODS
Sample collection.
Red soil samples were collected from two separate sites, and the geographic relationships of these and other soil samples collected by our team in Jordan in 2004 are described and mapped in a recent article (36). Sample A460-9-3 was collected from an agricultural field on the grounds of the National Center for Agricultural Research and Technology Transfer, north of Amman at 32°04′42"N and 35°50′32"E (elevation, 633 m). Sample A460-14-2 was collected from a farm in the northeastern portion of Jordan near the village of Hoffa Alwestyia at 32°34′07"N and 35°42′57"E (elevation, 345 m). As a control, non-red soil sample A460-2-2 was collected from a citrus orchard in the northern portion of the Jordan River Valley at 32°37′23"N and 35°35′55"E (elevation, 219 m).
Growth and enumeration of Micrococcus luteus and Staphylococcus aureus.
M. luteus is a normal inhabitant of soil (14) and can be easily identified and enumerated because of its yellow colony pigmentation and resistance to 7.5% (wt/vol) NaCl. Although S. aureus is not a normal inhabitant of soil, it is a member of the human skin microbiota (23) and is involved in skin infections (25). M. luteus and S. aureus cells were grown in 10 ml one-fourth-strength tryptic soy broth (TSB; BBL Microbiology Systems, Cockeysville, MD) in a 125-ml cotton-stoppered flask with aeration (120 rpm) at 30°C for 3 days. Cells were collected by centrifugation (5,000 × g for 10 min) and washed twice with an equal volume of sterile tap water and suspended in 10 ml of sterile tap water (approximately 2 × 108 CFU/ml). Colonies of M. luteus were enumerated on one-fourth-strength TSB agar containing 5% (wt/vol) NaCl (TSB plus salt). Colonies of S. aureus were enumerated on mannitol salt agar (beef extract, 1 g; proteose peptone no. 3, 10 g; NaCl, 75 g; mannitol, 10 g; agar, 15 g; and phenol red, 0.025 g, in 1 liter of distilled water).
Measurement of M. luteus and S. aureus killing by red soils.
Samples (5 g each) of red soils A460-9-3 and A460-14-2 and control agricultural soil A460-2-2 were added to sterile 50-ml screw-cap centrifuge tubes. In this manner, soil moisture content and headspace could be maintained throughout the duration of the experiment. One set of soils were not inoculated and employed as negative controls for antibiotic measurement. M. luteus (0.5 ml) and S. aureus (0.5 ml) were added to the soil samples and mixed thoroughly (approximately 2 × 107 CFU of M. luteus and S. aureus per gram of soil). Immediately, a 0.5-g sample was removed aseptically, weighed, transferred to a sterile 50-ml centrifuge tube, suspended in 5 ml of one-fourth-strength TSB, vortexed at the highest setting for 60 s, and shaken at 1 reciprocation per second for 30 min at room temperature. The resulting suspension was diluted in one-fourth-strength TSB, and 0.1 ml of the undiluted and diluted soil suspensions was spread (in duplicate) on TSB plus salt (M. luteus) and mannitol salt agar (S. aureus). Inoculated soil suspensions were incubated at room temperature, and a 0.5-g soil sample was removed and processed to enumerate M. luteus and S. aureus as described above after 1, 2, and 3 weeks of incubation. The results are reported as mean CFU per gram (± standard deviation) of inoculated soil samples from duplicate, independent experiments. Identity of M. luteus (yellow-pigmented colonies on one-fourth-strength TSB salt agar) and S. aureus (orange-pigmented colonies surrounded by a yellow halo on mannitol salt agar) colonies was confirmed by subculturing and cultural, biochemical, and enzymatic tests.
Measurement of MICs of methanol extracts of soils.
Samples (0.5 g) of each uninoculated soil (negative control) and inoculated and uninoculated soils after 3 weeks of incubation were suspended in 5 ml of methanol, vortexed at the highest setting for 60 s, and shaken at 1 reciprocation per second for 30 min at room temperature. The soil was pelleted by centrifugation (5,000 × g for 10 min), and the supernatant methanol solution was collected without collecting any soil. The methanol extract was evaporated to dryness overnight in a beaker at room temperature, and the residual extract was dissolved in 5 ml of dimethyl sulfoxide (DMSO). The dry weight of the DMSO solution was measured by drying 2 ml of it in a tared aluminum pan overnight at 105°C. The MICs of the DMSO solutions were measured by broth microdilution in 96-well plates against M. luteus, S. aureus, Mycobacterium smegmatis, Saccharomyces cerevisiae, and Aspergillus niger. The target microorganisms were grown and prepared for MIC measurement as described previously (12). The results are reported as the minimal concentration of dry weight that completely inhibits growth of the target microorganism. To serve as quality assurance standards, MICs for known antibiotics were included (Table 1).
TABLE 1.
Antimicrobial activity of methanol extracts in red soils prior to and after inoculation with M. luteus and S. aureus
| Sample(s) | MIC (fold increase)a
|
|||||
|---|---|---|---|---|---|---|
| M. luteus | S. aureus | M. smegmatis | S. cerevisiae | A. niger | P. acnes | |
| Red soil A460-9-3 | ||||||
| Before inoculation | >0.55 | >0.55 | >0.55 | >0.55 | >0.55 | >0.35 |
| 3 Weeks later | 0.003 (183) | 0.006 (92) | 0.040 (14) | 0.23 (2) | 0.007 (79) | 0.050 (7) |
| Red soil A460-14-2 | ||||||
| Before inoculation | >0.35 | >0.35 | >0.35 | >0.35 | >0.35 | >0.35 |
| 3 Weeks later | 0.004 (88) | 0.011 (32) | 0.011 (32) | 0.150 (2) | 0.011 (32) | 0.050 (7) |
| Positive controlsb | 0.0001 | 0.0008 | 0.0002 | 0.025 | 0.025 | ND |
Values are reported in mg/ml (n-fold increase). ND, not determined.
Positive control data (from left to right) are for ampicillin, ampicillin, kanamycin, amphotericin B, and amphotericin B. These are typical average values, as reported previously (30).
Enumeration, isolation, and identification of antimicrobial-producing microorganisms in inoculated red soils.
Antimicrobial-producing colonies from the inoculated soils were enumerated on lawns of target microorganisms. Cultures (0.1 ml) of M. luteus, S. aureus, Candida albicans, and A. niger (spore suspension) grown as described previously (12) were spread on 1/10-strength brain heart infusion (BHI) broth agar (BBL Microbiology Systems, Cockeysville, MD). Immediately, 0.1 ml of the undiluted and diluted soil suspensions (in duplicate) was spread onto plates, and the plates were allowed to air dry and incubated at 30°C for 2 to 5 days. In addition, 0.1 ml of the undiluted (supernatant) and diluted suspensions was spread on copper agar (i.e., 4.63 g BHI broth [1/10 strength; Difco, Detroit, MI], 0.04 g CuCl2 [anhydrous], 7.5 g agar, and 500 ml distilled water]), and the medium was incubated at 30°C (16). Empirically, colonies appearing on copper agar are likely to be antibiotic producers (15, 16). Colonies of bacteria appearing on the copper agar or colonies surrounded by zones of inhibition on the lawns were picked, and isolated colonies were used to inoculate 2 ml of one-fourth-strength TSB and were incubated for 3 days at 30°C. The antimicrobial-producing strains were identified by colony and microscopic morphology (i.e., actinomycetes and Bacillus) and profiles of cellular fatty acids (MIDI Labs, Newark, DE). To determine the range of antimicrobial activity of the isolates, 10 μl of a cell-free filtrate of the 2-ml cultures was spotted on lawns of the target microorganisms (see above). Clearing of the lawn was taken as evidence of antimicrobial activity (Table 1). To determine the susceptibility of Propionibacterium acnes strain ATCC 11827 to the isolates, 100 μl of each cell-free filtrate or of a methanol extract of the inoculated soils was added to 1.9 ml of reinforced clostridial medium (Oxoid, United Kingdom). Each was then inoculated with 100 μl of a P. acnes culture grown at 37°C for 3 days under anaerobic conditions (GasPak Plus; Becton Dickinson, BBL, Sparks, MD) and, after 3 days, scored visually for a reduction in turbidity (Table 1) by comparison to a control.
Detection of enzymatic activity.
Protease (17), lipase (33), and alkaline phosphatase (42) activities were measured in untreated and boiled cell-free culture filtrates and in organic and aqueous fractions of chloroform-methanol (1:1 dilution) extracts of representative Lysobacter isolates from red soils.
Detection and enumeration of myxobacteria, protozoa, amoebae, and bacteriophage.
Samples (0.5 g) of uninoculated, inoculated, and incubated (3 weeks) soils were suspended in 10 ml sterile tap water and vortexed at the highest speed for 60 s. The suspension was allowed to settle for 1 h, and 1 ml was filtered through a 0.22-μm-pore-size filter. To detect myxobacteria, 0.1-ml samples of the diluted and undiluted unfiltered suspension were spread on CF agar with and without a lawn of Escherichia coli strain C (26). After 1 week of incubation at 30°C, the agar medium was examined under 10-fold magnification for evidence of fruiting bodies. Protozoa and amoebae were detected by mixing 3 ml of molten (45°C), 1/10-strength BHI broth containing 0.7% agar (top agar) with 0.1 ml of the filtrate and 0.1 ml of either M. luteus or S. aureus and by pouring the suspension over the surface of BHI agar. After 1 to 3 days of incubation at 30°C, cleared zones were inspected under 100-fold magnification to determine whether protozoa or amoebae were present. For bacteriophage enumeration, 0.1 ml of the filtrate and 0.1 ml of either M. luteus or S. aureus were added to 3.0 ml of top agar, and the contents were mixed and poured over the surface of BHI agar. After overnight incubation at 30°C, the number of plaques was counted.
Detection of antimicrobial activity of organic extracts of isolates.
Isolates were grown, extracted, and tested for antimicrobial activity using a modification of procedures described previously (13). Briefly, isolates were grown to 500-ml volumes in one-fourth-strength TSB plus salt broth, frozen, and thawed, and the entire culture was freeze-dried. The resultant powder was extracted by stirring it overnight in either methanol or chloroform-methanol (1:1 dilution). The resultant slurry was filtered to remove solids, and the solvent was removed in vacuo. This extract was then partitioned between chlorofom-methanol-H2O (4:1:5 dilution). The organic fraction (lower portion) was removed and dried in vacuo. A measured aliquot of this fraction was dissolved in 1 ml of DMSO, and the antimicrobial activity was measured as described previously (5).
Isolation of actinomycins from isolate 14-2-1.
A culture of isolate 14-2-1 was extracted as described above. The organic fraction, showing antimicrobial activity, was purified further using a Varian ProStar high-pressure liquid chromatography (HPLC) system (Walnut Creek, CA) equipped with ProStar 210 pumps and a 330 photodiode array detector, with data collected and analyzed using Star Chromatography Workstation software (version 5.52) via an ODS-A column (250 by 25 mm, inside diameter of 5 μm; YMC, Wilmington, NC). The CH3CN-H2O gradient solvent system initiated at a dilution of 1:1 and increased to 8:2 linearly over 70 min. Two major compounds (coded 11065-92-1 and 11065-92-2) that were eluted between approximately 40 and 50 min were isolated, and these were identified via the experiments described below as actinomycin C2 and actinomycin C3, respectively. These assignments were deduced via 1H and 13C nuclear magnetic resonance using a Varian Unity Inova 500 instrument with a 5-mm broadband inverse probe with a z-gradient. They were confirmed by high-resolution mass spectrometry (performed on a Finnigan MAT 95Q hybrid-sector instrument [Thermo Finnigan; San Jose, CA]), where compound 11065-92-1 (1.5 mg) yielded a sodiated molecular ion that correlated with actinomycin C2 (1,291.63192 atomic mass units [amu] measured versus 1,291.633346 amu calculated for C63H88N12O16Na) and where compound 11065-92-2 (3.0 mg) yielded a sodiated molecular ion that correlated with actinomycin C3 (1,305.65237 amu measured versus 1,305.648996 amu calculated for C64H90N12O16Na). Finally, these assignments were verified by comparison to actinomycin C reference standards (Axxora, LLC, San Diego, CA). Compound 11065-92-1 was eluted with the same retention time (8.75 min) and same UV spectrum as actinomycin C2, and compound 11065-92-3 was eluted with the same retention time (9.59 min) and same UV spectrum as actinomycin C3; both were measured using the aforementioned HPLC system, with a gradient solvent system of CH3CN-H2O that initiated at a dilution of 60:40 and increased linearly to 80:20 over 10 min on an ODS-A column (150 by 4.6 mm, inside diameter of 5 μm; YMC, Wilmington, NC).
RESULTS
Red soils kill M. luteus and S. aureus.
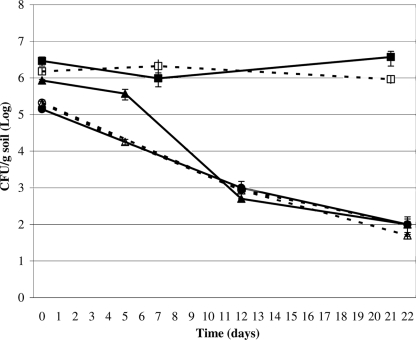
Following the inoculation of M. luteus and S. aureus into the red soils, their numbers fell dramatically by day 12 (Fig. 1). Numbers did not fall in the inoculated agricultural soil (Fig. 1). The values in the figure represent averages of duplicate counts from two experiments with the same soil sample. The surviving numbers of both S. aureus and M. luteus in CFU per gram were significantly below those of corresponding inoculum densities and the agricultural soil after 12 and 22 days of incubation (P of <0.05 by one-tailed Student's t test). Separate or combined inoculation of the two bacteria yielded the same results (data not shown). Autoclaved red soils did not demonstrate any killing (data not shown), illustrating that biotic factors were responsible for killing. One consequence of using small amounts of soil was that 10% of soil was removed for analysis at each sampling point. The benefit was that distribution of the inoculum was more uniform using the small soil volumes, thus reducing variation in the recovery of bacteria. Further, soil texture, moisture content, and headspace were maintained throughout the 3-week incubation period. Experiments with larger amounts of soil led to the formation of aggregates with entirely different soil textures.
FIG. 1.
Killing of M. luteus and S. aureus by red soils A460-9-3 and A460-14-2. Soil A460-2-2 is an agricultural, non-red soil, which was used as a negative control. For all experiments, M. luteus and S. aureus were inoculated into the soil samples, and the number of CFU of the inoculated bacteria per gram of soil was measured at intervals for 22 days. A460-2-2 plus M. luteus, ▪; A460-2-2 plus S. aureus, □; A460-9-3 plus M. luteus, •; A460-9-3 plus S. aureus, ○; A460-14-2 plus M. luteus, ▴; A460-14-2 plus S. aureus, ▵.
Increased antimicrobial activity in red soils following M. luteus and S. aureus inoculation.
Three weeks of incubation for the M. luteus- and S. aureus-inoculated red soils led to the appearance of significantly higher levels of antimicrobial activity of methanol extracts in the inoculated soil (Table 1) (P of <0.05 by Student's t test). The data reported in Table 1 were taken from one of the two experiments in which the results were nearly identical. There was no detectable antimicrobial activity recovered from either of the uninoculated red soils incubated in parallel for 3 weeks (negative control). Soil samples were inoculated with M. luteus and S. aureus, and immediately, or after 3 weeks of incubation at room temperature, methanol extracts were obtained as described. Although there was no detectable antimicrobial activity of the methanol extract in the two red soils at the time of inoculation, there was broad-spectrum antimicrobial activity in the methanol extracts obtained after 3 weeks of incubation (Table 1). The methanol extracts in both inoculated soils also exhibited antibiotic activity against P. acnes (Table 1), a member of the normal microbiota of skin that is implicated in dermal infections (23). Although the MICs for the methanol extracts are moderate (i.e., 0.003 to 0.150 mg/ml) (Table 1) compared to those of pure antibiotics, it is to be understood that the values are for crude extracts, not purified compounds, and antibiotics are subject to adsorption by soils and therefore difficult to extract (24).
Absence of myxobacteria, protozoa, amoebae, or bacteriophages.
Neither myxobacteria-like fruiting bodies nor bacteriophage plaques were detected in red soils examined prior to and following inoculation and 3 weeks of incubation. Protozoa and amoebae were detected at low levels in the uninoculated red soils (≤10/g); however, their numbers did not increase over the course of 3 weeks of incubation.
Increased number of antibiotic-producing bacteria following M. luteus and S. aureus inoculation in red soils.
Coincident with the fall in numbers of M. luteus and S. aureus (Fig. 1) was a moderate increase in the number of colonies producing zones of inhibition against M. luteus and S. aureus (Table 2). Only a few of the increases were significantly higher, as indicated in the footnote to Table 2. There were no differences in the number of colonies producing zones of inhibition in the uninoculated red soils over the course of the incubation period (data not shown). The data in Table 2 were from experiments where S. aureus and M. luteus were separately inoculated into the red soils to demonstrate that the increase in zones of inhibition against both S. aureus and M. luteus occurred when just one was inoculated into the soil. Almost identical results were obtained from experiments where both were inoculated. Inoculation of the red soils with either M. luteus or S. aureus also increased the number of antibiotic-producing microorganisms against C. albicans (3- and 4.5-fold, respectively) and A. niger (3.5- and 13-fold, respectively) for red soil A460-9-3. Although Table 2 lists the total number of colonies surrounded by zones of inhibition in the target microbial lawns, certain types predominated. For example, in red soil A460-9-3 inoculated with M. luteus, the total number of colonies with zones of inhibition was 2.0 × 107 colonies/g, of which 0.3 × 107 colonies/g were Bacillus, 0.2 × 107 colonies/g were actinomycetes, and 1.5 × 107 colonies/g had a transparent slimy or transparent mucoid colony type whose zones of inhibition on lawns of either M. luteus or S. aureus lacked any clear evidence of a colony.
TABLE 2.
Increase in number of antibiotic-producing microorganisms following inoculation of red soils
| Soil sample + inoculum | Incubation period (days) | CFU/g soil (fold increase)a against:
|
|||
|---|---|---|---|---|---|
| M. luteus | S. aureus | C. albicans | A. niger | ||
| A460-9-3 + M. luteus | 0 | 3.2 × 105 | 1.0 × 107 | 9.0 × 104 | 2.0 × 105 |
| 20 | 6.8 × 105 (2.1) | 2.0 × 107 (2.0) | 2.7 × 105 (3.0) | 7.0 × 105 (3.5) | |
| A460-9-3 + S. aureus | 0 | 2.4 × 106 | 1.0 × 106 | 2.0 × 105 | 2.0 × 105 |
| 20 | 9.1 × 107 (38)c | 9.0 × 106 (9.0)c | 9.0 × 105 (4.5) | 2.6 × 106 (13)c | |
| A460-14-2 + M. luteus | 0 | 3.4 × 105 | 3.0 × 105 | NDb | ND |
| 20 | 1.2 × 106 (3.5) | 3.3 × 106 (11)c | ND | ND | |
| A460-14-2 + S. aureus | 0 | 7.7 × 105 | 4.0 × 105 | ND | ND |
| 20 | 1.4 × 106 (1.8) | 1.1 × 106 (2.8) | ND | ND | |
Data from the experiment where M. luteus and S. aureus were inoculated separately into the same soil samples.
ND, not determined.
Significantly higher numbers after incubation (P of <0.05 by Student's t test).
Antimicrobial-producing bacteria from red soils.
Table 3 lists the antibiotic-producing isolates, their identification, spectra of antimicrobial activities for cell-free culture filtrates, and in some cases, the MICs of the organic fraction of individual cultures. Identification of Actinomycetes was based on colony and microscopic morphology, whereas Bacillus and Lysobacter were identified by cellular fatty acid analysis (MIDI Labs, Newark, DE). The Lysobacter isolates had either a transparent mucoid or transparent slimy colony morphology and differed in colony pigmentation (i.e., yellow, orange, red, and brown). Cell-free culture filtrates of Lysobacter isolates 9-3-16 (mucoid, yellow), 9-3-17 (slime, brown), and 14-2-10 (slime, orange-red) all had protease, lipase, and alkaline phosphatase activities that were absent in boiled cell-free culture filtrates (data not shown). Significantly, the boiled cell-free culture filtrates retained antimicrobial activity, and the organic and aqueous fractions of chloroform-methanol extracts of the Lysobacter cultures lacked enzymatic activities. These data demonstrate that the antimicrobial activity of the cell-free culture filtrates or culture extracts was not due to extracellular Lysobacter enzymes.
TABLE 3.
Antimicrobial-producing isolates recovered from two different red soils and the relative activity of cell-free culture filtrates against a panel of microorganisms
| Isolate no. | Identification (genus) | Colony morphology | Antimicrobial activity of cell-free culture filtrates by spot platea
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| M. luteus | S. aureus | E. coli | M. smegmatis | S. cerevisiae | A. niger | P. acnes | |||
| Soil sample A460-9-3 | |||||||||
| 9-3-12 | Lysobacterd | Transparent | − | − | − | + | − | − | NDb |
| 9-3-13 | Lysobacterd | Slime | + (1.85) | + (0.93) | + (0.93) | − | − | − | ND |
| 9-3-14 | Actinomycetesd | + (0.35) | + | − | + (0.05) | + (0.05) | + (0.20) | ND | |
| 9-3-15 | Lysobacterd | Slime | + (0.90) | + (0.11) | − | − | − | − | ND |
| 9-3-16 | Lysobacterd | Mucoid | + (0.09) | + (0.04) | + (0.35) | + (0.35) | + (0.18) | + (0.09) | − |
| 9-3-17 | Lysobacterd | Slime | + (0.19) | + | + | + (0.03) | − | + (0.08) | + |
| 9-3-19 | Lysobacterd | Mucoid | + (0.34) | + (0.17) | − | − | − | − | ND |
| 9-3-20 | Lysobacterd | Slime | + | + | + | + | − | − | ND |
| 9-3-21 | Actinomycetesd | + (0.14) | + (0.14) | + (0.58) | + (0.29) | + | − | ND | |
| 9-3-22 | Lysobacterd | Slime | − (>1.20) | − (>0.60) | − (>0.60) | − (>0.60) | ND | − (>0.60) | ND |
| 9-3-23 | Lysobacterd | Slime | + | + | − | − | + | + | ND |
| 9-3-26 | Bacillusd | Target | + | + | + | + | − | + | + |
| Soil sample A460-14-2 | |||||||||
| 14-2-1 | Actinomycetesc | Yellow | + | + | + | − | − | − | + |
| 14-2-3 | Actinomycetesc | Brown | + | + | + | − | − | − | ND |
| 14-2-6 | Actinomycetesc | − | − | + | − | − | − | ND | |
| 14-2-10 | Lysobacterd | Slime | + (0.40) | + (0.05) | − | − | − | − | − |
| 14-2-13 | Lysobacterd | Slime | + | + | + | − | − | − | ND |
Antimicrobial activity is expressed as clearing (+) or no clearing (−) on a lawn of target microorganism spotted with 10 μl cell-free culture filtrate and as MICs of organic extracts in mg/ml (in parentheses) prepared as described in Materials and Methods.
ND, not determined.
Isolated on copper agar.
Isolated from a zone of inhibition.
Evidence that isolates from red soils produce compounds with antibiotic activity.
One actinomycete, isolate 14-2-1, identified on the basis of colony and microscopic morphology, was examined using a bioactivity-directed fractionation approach for compounds with antibiotic activity (13, 30). Briefly, an organic extract of a 500-ml culture of this isolate grown in one-fourth-strength TSB containing 0.5% (wt/vol) sucrose exhibited MICs of 5 and 1 μg/ml against M. luteus and S. aureus, respectively. Upon further purification using reverse-phase HPLC, two compounds were isolated (11065-92-1 and 11065-92-2), and these were identified as actinomycin C2 and actinomycin C3, respectively. These structures were elucidated using a suite of spectroscopic and spectrometric tools, particularly nuclear magnetic resonance and high-resolution mass spectrometry. Moreover, the chromatographic profiles of the isolated compounds were compared directly with actinomycin C reference standards.
DISCUSSION
The data herein suggest that the killing of inoculated S. aureus and M. luteus by Jordan's red soils was due to the proliferation of antibiotic-producing bacteria and their concomitant production of antimicrobial compounds. The mechanism for induction of antibiotic-producing bacteria and antibiotic production is still unknown and particularly intriguing in soils where there is little diffusion of compounds produced by prey microorganisms. Abiotic factors do not appear to be responsible, because autoclaved red soils failed to kill the inoculated bacteria, unlike the case of the antimicrobial clays (28). Rather than due to protozoan, bacteriophage, or myxobacterial predation, killing by red soils appears to be mediated by a consortium of antibiotic-producing bacteria, namely actinomycetes, Bacillus strains, and Lysobacter strains. That conclusion was supported by evidence that red soil A460-14-2 harbors an actinomycete, which synthesizes at least a pair of antibiotic compounds of the actinomycin structural class. Although it is likely that the enzymatic activities of the Lysobacter in the native red soils contributed to killing (10), members of this genus have been reported also to produce antimicrobial compounds (27). Likewise, the absence of the three extracellular enzyme activities of the Lysobacter strains, coupled with the presence of antimicrobial activity in the boiled cell-free culture filtrates and the organic and aqueous fractions of chloroform-methanol extracts, argues strongly that the antimicrobial activity of the Lysobacter strains was due to production of compounds rather than enzymes that exhibit antimicrobial activity. Studies are ongoing to isolate and elucidate compounds with antibiotic activities from some of these bacterial isolates from red soil.
The magnitude of the killing of S. aureus appears to be similar to that noted by Liang et al. (31), who showed that killing was due to unidentified biotic factors as S. aureus grew in autoclaved soil (31). In another study demonstrating the killing of microorganisms inoculated into soils, it was proposed, but not proven, that predatory bacteria were responsible for killing (14). In a third study, eukaryotic predation was shown to be involved in killing, although antimicrobial-producing or lytic microorganisms could have been involved as well (1). Although there were only modest increases in the number of colonies surrounded by zones of inhibition after incubation of the inoculated red soils (Table 2), their ability to kill would be expected to be magnified by the production of extracellular antimicrobial compounds (Table 1).
These data provide a rationale for the traditional use of Jordan's red soils for the treatment of skin infections, including diaper rash. In the absence of any inherent, abiotic antibiotic activity of red soils, and as S. aureus is a normal inhabitant of human skin microbiota (23), we hypothesize that the application of red soils to an infected area of skin (i.e., inoculation) leads to the proliferation of bacteria that produce antibiotic compounds, killing the infecting skin microbiota. This is similar to the killing of Shigella paradysenteriae by a colicin-producing strain of E. coli following coinfection in the peritoneal cavity of mice (22). Shigella killing coincided with the multiplication of the E. coli strain and an increase in the colicin (antibiotic) concentration (22). Killing by red soils is probably not restricted to the infecting microorganisms, because inoculation of red soils with either M. luteus or S. aureus led to the appearance of broad-spectrum antimicrobial activity (Table 1). This is likely due to the proliferation of a spectrum of microorganisms producing a suite of antimicrobial compounds (Tables 2 and 3).
Acknowledgments
This work was supported by the Fogarty International Center of the National Institutes of Health, with cofunding from the National Institute of Allergy and Infectious Diseases, the National Institute of Drug Abuse, and the National Science Foundation under grant R21 TW006628 for the International Cooperative Biodiversity Groups.
We thank Myra Williams and Vy Troung for expert technical assistance and Tyler Graf for comparing the HPLC profiles of the isolated compounds with those of actinomycin C reference standards. High-resolution mass spectrometry data were acquired by the staff at the Ohio State University Chemistry Mass Spectrometry Facility.
Footnotes
Published ahead of print on 13 March 2009.
REFERENCES
- 1.Acea, M. J., C. R. Moore, and M. Alexander. 1988. Survival and growth of bacteria introduced into soil. Soil Biol. Biochem. 20:509-515. [Google Scholar]
- 2.Alali, F., A. S. Ma'aya'h, A. Alkofahi, A. Qandil, C. Li, J. P. Burgess, M. C. Wani, and N. H. Oberlies. 2006. A new colchicinoid from Colchicum tauri, an unexplored meadow saffron native to Jordan. Nat. Prod. Commun. 1:95-99. [Google Scholar]
- 3.Alali, F., K. Tawaha, T. El-Elimat, R. Qassaymeh, C. Li, J. P. Burgess, Y. Nakanishi, D. J. Kroll, M. C. Wani, and N. H. Oberlies. 2006. Phytochemical studies and cytotoxicity evaluations of Colchicum tunicatum Feinbr and Colchicum hierosolymitanum Feinbr (Colchicaceae): two native Jordanian meadow saffrons. Nat. Prod. Res. 20:558-566. [DOI] [PubMed] [Google Scholar]
- 4.Alali, F., K. Tawaha, T. El-Elimat, M. Syouf, M. El-Fayad, K. Abulaila, S. J. Nielsen, W. D. Wheaton, J. O. Falkinham III, and N. H. Oberlies. 2007. Antioxidant activity and total phenolic content of aqueous and methanolic extracts of Jordanian plants: an ICBG project. Nat. Prod. Res. 21:1121-1131. [DOI] [PubMed] [Google Scholar]
- 5.Alali, F. Q., T. El-Elimat, C. Li, A. Qandil, A. Alkofahi, K. Tawaha, J. P. Burgess, Y. Nakanishi, D. J. Kroll, H. A. Navarro, J. O. Falkinham III, M. C. Wani, and N. H. Oberlies. 2005. New colchicinoids from a native Jordanian meadow saffron, Colchicum brachyphyllum: isolation of the first naturally occurring dextrorotary colchicinoid. J. Nat. Prod. 68:173-178. [DOI] [PubMed] [Google Scholar]
- 6.Alali, F. Q., A. Gharaibeh, A. Ghawanmeh, K. Tawaha, and N. H. Oberlies. 2008. Colchicinoids from Colchicum crocifolium Boiss.: a case study in dereplication strategies for (-)-colchicine and related analogs using LC-MS and LC-PDA techniques. Phytochem. Anal. 19:385-394. [DOI] [PubMed] [Google Scholar]
- 7.Alali, F. Q., Y. R. Tahboub, E. S. Ibrahim, A. M. Qandil, K. Tawaha, J. P. Burgess, A. Sy, Y. Nakanishi, D. J. Kroll, and N. H. Oberlies. 2008. Pyrrolizidine alkaloids from Echium glomeratum (Boraginaceae). Phytochemistry 69:2341-2346. [DOI] [PubMed] [Google Scholar]
- 8.Al-Eisawi, D. M. 1998. Field guide to wild flowers of Jordan and neighbouring countries. Jordan Press Foundation, Al Rai, Amman, Jordan.
- 9.Barea, J. M., M. J. Pozo, R. Azcon, and C. Azcon-Aguilar. 2005. Microbial co-operation in the rhizosphere. J. Exp. Bot. 56:1761-1778. [DOI] [PubMed] [Google Scholar]
- 10.Begunova, E. A., O. A. Stepnaya, I. M. Tsfasman, and I. S. Kulaev. 2004. The effect of the extracellular bacteriolytic enzymes of Lysobacter sp. on gram-negative bacteria. Microbiology 73:267-270. [PubMed] [Google Scholar]
- 11.Bush, K. 2004. Why it is important to continue antibacterial drug discovery. ASM News 70:282-287. [Google Scholar]
- 12.Cain, C. C., A. T. Henry, R. H. Waldo III, L. J. Casida, Jr., and J. O. Falkinham III. 2000. Identification and characteristics of a novel Burkholderia strain with broad-spectrum antimicrobial activity. Appl. Environ. Microbiol. 66:4139-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cain, C. C., D. Lee, R. H. Waldo III, A. T. Henry, L. E. Casida, Jr., M. C. Wani, M. E. Wall, N. H. Oberlies, and J. O. Falkinham III. 2003. Synergistic antimicrobial activity of metabolites produced by a nonobligate bacterial predator. Antimicrob. Agents Chemother. 47:2113-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casida, L. E., Jr. 1980. Bacterial predators of Micrococcus luteus in soil. Appl. Environ. Microbiol. 39:1035-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casida, L. E., Jr. 1988. Nonobligate bacterial predation of bacteria in soil. Microb. Ecol. 15:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Casida, L. E., Jr. 1987. Relation to copper of N-1, a nonobligate bacterial predator. Appl. Environ. Microbiol. 53:1515-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chavira, R., Jr., T. J. Burnett, and J. H. Hageman. 1984. Assaying proteinases with azocoll. Anal. Biochem. 136:446-450. [DOI] [PubMed] [Google Scholar]
- 18.Clardy, J. 2005. Using genomics to deliver natural products from symbiotic bacteria. Genome Biol. 6:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Compeau, G., B. J. Alachi, E. Platsouka, and S. B. Levy. 1988. Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl. Environ. Microbiol. 54:2432-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordell, G. A., N. R. Farnsworth, C. W. W. Beecher, D. D. Soejarto, A. D. Kinghorn, J. M. Pezzuto, M. E. Wall, M. C. Wani, R. R. Cobb, M. J. O'Neil, R. M. Tait, and T. J. R. Harris. 1994. Novel strategies for the discovery of plant-derived anticancer agents, p. 63-83. In F. A. Valeriote, T. H. Corbett, and L. H. Baker (ed.), Anticancer drug discovery and development: natural products and new molecular models. Kluwer Academic Publishers, Boston, MA.
- 21.Feinbrun-Dothan, N. 1986. Flora Palestina. The Israel Academy of Sciences and Humanities, Jerusalem, Israel.
- 22.Friedman, D. R., and S. P. Halbert. 1960. Mixed bacterial infections in relation to antibiotic activities. IV. Shigella-Escherichia coli infections. J. Immunol. 84:11-19. [PubMed] [Google Scholar]
- 23.Gao, Z., C. H. Tseng, Z. Pei, and M. J. Blaser. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA 104:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 25.Hageman, J. C., T. M. Uyeki, J. S. Francis, D. B. Jernigan, J. G. Wheeler, C. B. Bridges, S. J. Barenkamp, D. M. Sievert, A. Srinivasan, M. C. Doherty, L. K. McDougal, G. E. Killgore, U. A. Lopatin, R. Coffman, J. K. MacDonald, S. K. McAllister, G. E. Fosheim, J. B. Patel, and L. C. McDonald. 2006. Severe community-acquired pneumonia due to Staphylococcus aureus, 2003-04 influenza season. Emerg. Infect. Dis. 12:894-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 27.Hashizume, H., M. Igarashi, S. Hattori, M. Hori, M. Hamada, and T. Takeuchi. 2001. Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. I. Taxonomy, isolation and biological activities. J. Antibiot. 54:1054-1059. [DOI] [PubMed] [Google Scholar]
- 28.Haydel, S. E., C. M. Remenih, and L. B. Williams. 2008. Broad-spectrum in vitro antibacterial activities of clay minerals against antibiotic-susceptible and antibiotic-resistant bacterial pathogens. J. Antimicrob. Chemother. 61:353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kemp, J. S., E. Paterson, S. M. Gammack, M. S. Cresser, and K. Killham. 1992. Leaching of genetically modified Pseudomonas fluorescens through organic soils-influence of temperature, soil pH, and roots. Biol. Fertil. Soils 13:218-224. [Google Scholar]
- 30.Li, C., D. Lee, T. N. Graf, S. S. Phifer, Y. Nakanishi, J. P. Burgess, S. Riswan, F. M. Setyowati, A. M. Saribi, D. D. Soejarto, N. R. Farnsworth, J. O. Falkinham III, D. J. Kroll, A. D. Kinghorn, M. C. Wani, and N. H. Oberlies. 2005. A hexacyclic ent-trachylobane diterpenoid possessing an oxetane ring from Mitrephora glabra. Org. Lett. 7:5709-5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, L. N., J. L. Sinclair, L. M. Mallory, and M. Alexander. 1982. Fate in model ecosystems of microbial species of potential use in genetic engineering. Appl. Environ. Microbiol. 44:708-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, K. C., and L. E. Casida. 1983. Survival of myxobacter strain-8 in natural soil in the presence and absence of host-cells. Soil Biol. Biochem. 15:551-555. [Google Scholar]
- 33.Lonon, M. K., D. E. Woods, and D. C. Straus. 1988. Production of lipase by clinical isolates of Pseudomonas cepacia. J. Clin. Microbiol. 26:979-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman, D. J., and G. M. Cragg. 2007. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70:461-477. [DOI] [PubMed] [Google Scholar]
- 35.Oberlies, N. H., C. Li, R. J. McGivney, F. Q. Alali, J. R. Tanner, and J. O. Falkinham III. 2008. Microbial-mediated release of bisphenol A from polycarbonate vessels. Lett. Appl. Microbiol. 46:271-275. [DOI] [PubMed] [Google Scholar]
- 36.Oberlies, N. H., J. I. Rineer, F. Q. Alali, K. Tawaha, J. O. Falkinham III, and W. D. Wheaton. 2009. Mapping of sample collection data: GIS tools for the natural product researcher. Phytochem. Lett. 2:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarkar, P., and I. M. Gould. 2006. Antimicrobial agents are societal drugs: how should this influence prescribing? Drugs 66:893-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shlaes, D. M., S. J. Projan, and J. E. Edwards, Jr. 2004. Antibiotic discovery: state of the state. ASM News 70:275-281. [Google Scholar]
- 39.Thomashow, L. S., and D. M. Weller. 1988. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Elsas, J. D., A. F. Dijkstra, J. M. Govaert, and J. A. van Veen. 1986. Survival of Pseudomonas fluorescens and Bacillus subtilis introduced into 2 soils of different texture in field microplots. FEMS Microbiol. Ecol. 38:151-160. [Google Scholar]
- 41.van Saene, H. K., W. I. Weir, M. A. de la Cal, L. Silvestri, A. J. Petros, and S. P. Barrett. 2004. MRSA—time for a more pragmatic approach? J. Hosp. Infect. 56:170-174. [DOI] [PubMed] [Google Scholar]
- 42.von Tigerstrom, R. G. 1984. Production of two phosphatases by Lysobacter enzymogenes and purification and characterization of the extracellular enzyme. Appl. Environ. Microbiol. 47:693-698. [DOI] [PMC free article] [PubMed] [Google Scholar]