Abstract
This paper describes a molecular-based method which is able to discriminate between viable and inactivated Bacillus subtilis spores by utilizing the DNA-intercalating dye propidium monoazide. The approach should be valuable in our attempt to employ molecular methods to streamline the evaluation of process validation using bacterial endospores.
There is a continued need for the development and application of rapid methods for the detection and enumeration of bacterial endospores, especially as investigators seek to evaluate the efficacy of emerging food-processing technologies. For such thermal-process validation studies, surrogates of Clostridium botulinum, including Bacillus subtilis and Clostridium sporogenes, are commonly used. Currently, standard plating methods remain the “gold standard” for enumeration of survivors of these processes. Molecular amplification approaches, such as quantitative real-time PCR (qPCR), have shown promise but have not been applied to the enumeration of surviving spores because (i) release of nucleic acid from spores is difficult and (ii) the detection of DNA does not necessarily equate with the presence of viable spores.
Recently, the DNA-intercalating agents ethidium monoazide and propidium monoazide (PMA) have been used in conjunction with qPCR for the selective detection of live cells of food-borne pathogens (2, 3, 5, 6, 9). These compounds selectively penetrate the membranes of dead cells and form stable DNA monoadducts upon photolysis, resulting in DNA which cannot be amplified by PCR (3). To our knowledge, this technique has not yet been applied to bacterial spores. The purpose of this study was to demonstrate that DNA-intercalating agents could be used in conjunction with qPCR for the selective enumeration of viable, but not inactivated, spores of B. subtilis.
All media were supplied by Difco (Franklin Lakes, NJ), and chemicals were obtained from Sigma-Aldrich (St. Louis, MO). Bacillus subtilis ATCC 35021 (American Type Culture Collection, Manassas, VA) was grown overnight in 10 ml of brain heart infusion broth at 37°C. Five-hundred-microliter aliquots of the vegetative cells were spread onto 150- by 15-mm petri dishes containing sporulation agar comprised of 13 g/liter nutrient broth, 15 g/liter agar, 0.51 g/liter MgSO4·7H2O, 0.97 g/liter KCl, 0.2 g/liter CaCl2·2H2O, 3 mg/liter MnSO4·H20, and 0.5 mg/liter FeSO4·7H2O. The plates were incubated aerobically at 37°C for 3 to 5 days until more than 95% of cells had sporulated, as determined by phase-contrast microscopy. Spores were harvested in cold, sterile distilled water (dH2O) and washed repeatedly (5 to 10 times). Before final resuspension, the spores were treated with 80 U/ml DNase (Sigma-Aldrich, St. Louis, MO) at 37°C for 90 min (to degrade residual DNA), with subsequent DNase inactivation by heating at 65°C for 10 min. The final populations were approximately 107 spores/ml, and crops were stored at 4°C until use. Using the capillary tube method (8), the D121°C value (decimal reduction time, i.e., time at 121°C required to kill 90% of the spores) and the Z value (temperature required to change the D value by 1 log) for this spore crop were calculated to be 0.23 min and 7.66°C (data not shown). This is comparable to the D121°C and Z values of 0.21 min and 10°C, respectively, for C. botulinum (1).
A number of different DNA isolation methods were attempted, including using a Masterpure gram-positive DNA purification kit and a Soilmaster DNA extraction kit (both from Epicentre Biotechnologies, Madison, WI), a Qiagen DNeasy plant mini kit (Valencia, CA), and bead beating. Only one (bead beating) resulted in efficient DNA extraction, as measured by dilution series qPCR, while at the same time preserving the intercalation effect of the PMA. Specifically, DNA isolation was done using 500 μl of 106-μm, acid-washed glass beads (Sigma Aldrich, St. Louis, MO) with beating for 1 min at maximum speed in a Biospec mini bead beater (Biospec Products, Inc., Bartlesville, OK). Thereafter, samples were placed on ice and centrifuged at 18,500 × g for 5 min to remove the glass beads and cellular debris. Approximately 300 μl of supernatant was removed, and the DNA precipitated by using 1/10 volume of 3 M sodium acetate, 75 μg/ml Glycoblue (Ambion, Austin, TX), and 1 volume isopropanol at −20°C for 15 min, followed by centrifugation. The resulting DNA pellet was washed with 70% ethanol, dried, and resuspended in 10 μl of ultrapure water.
The qPCR method used the previously reported degenerate primers Bacillus F1 (5′-ATYATGYTVACRGCVTTYGGBCARGAAGA) and Bacillus R1 (5′-TAKCCTTTWATRTGIGCDGGIACRCCGATTTC) which target the B. subtilis spoOA gene (7). A TaqMan probe (5′-FAMCAGCAGCCAGCCTGAACCAAAGAABHQ1-3′) was designed by using Beacon Designer 7 software (Premier Biosoft, Palo Alto, CA). PCR mixes included 1× Jumpstart PCR buffer (Sigma-Aldrich, St. Louis, MO), 4 mM MgCl2, 600 nM Bacillus F1, 600 nM Bacillus R1, 200 nM Bacillus subtilis probe, 5 mM deoxynucleoside triphosphate blend (Applied Biosystems, Foster City, CA), 1.25 U Jumpstart Taq polymerase (Sigma-Aldrich, St. Louis, MO), 2% dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO), and 2.5 μl DNA in a final volume of 25 μl. The reactions were run on a SmartCycler real-time PCR machine (Cepheid, Sunnyvale, CA) with 1 cycle at 95°C for 120 s followed by 40 cycles of 95°C for 15 s, 55°C for 20 s, and 72°C for 20 s. The qPCR assay was able to detect as few as 10 spores per reaction, with log-linear detection in the range of 101 to 106 spores/ml (data not shown). A standard curve was generated by plotting the threshold cycle (CT) values of the known samples against the calculated spore concentration (spores/ml).
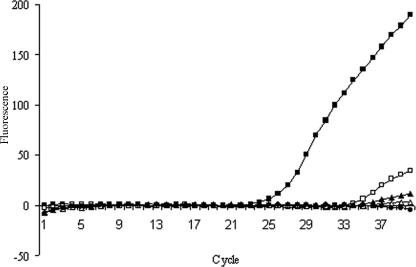
Preliminary experiments established that PMA but not ethidium monoazide (Biotium, Inc., Hayward, CA) was able to enter inactivated but not live spores (data not shown). To establish the amount of PMA required to inhibit PCR amplification of DNA derived from inactivated spores, increasing concentrations (0 to 25 μM) were tested on spore suspensions which had been inactivated by being autoclaved (121°C for 20 min) twice in sequence. As can be seen in Fig. 1, the CT values were completely eliminated at concentrations of ≥10 μM PMA; the 10 μM concentration was also shown not to inhibit qPCR (data not shown). Based on these data, a final protocol was developed. Specifically, a 500-μl aliquot of spore suspension was treated with 10 μM PMA, followed by incubation in the dark for 50 min at room temperature. The tubes were then placed on ice with the lids open and exposed to a Regent 500 W halogen lamp (Cooper Lighting, Peachtree City, GA) at a distance of 12 cm for 3 min.
FIG. 1.
The effect of PMA concentration on qPCR results for B. subtilis spores (107 spores/ml) pretreated with PMA prior to DNA extraction and qPCR. ▪, 0 μM PMA; □, 5 μM PMA; ▴, 10 μM PMA; ▵, 25 μM PMA; •, no-template control.
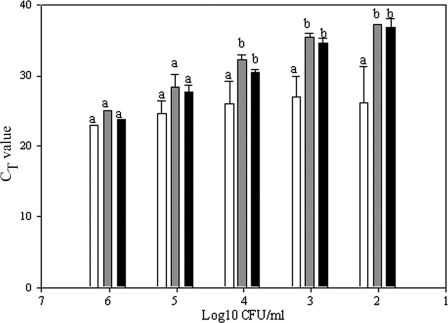
To determine if this protocol would be able to distinguish live from inactivated spores in a mixed suspension, spore suspensions were serially diluted in dH2O or in a solution of inactivated (twice autoclaved) spores, followed by qPCR with and without prior PMA treatment. There were no statistically significant differences between CT values obtained for the water-diluted or inactivated-spore-diluted samples treated with PMA. However, statistically significant differences in CT values (i.e., lower) were observed when comparing these values to those obtained for the viable spores diluted in inactivated-spore suspension without prior PMA treatment (Fig. 2). Regression lines were plotted (Sigma Plot 9.0; Systat Software, Inc., Chicago, IL) for the results of the three treatments as a function of viable-spore concentration; mean slope values ± standard deviations were 3.42 ± 0.17, 3.32 ± 0.74, and 0.73 ± 0.45 for water-diluted viable spores and viable spores diluted in heat-inactivated-spore suspension with and without PMA treatment, respectively. There was no statistically significant difference in slopes when comparing the results for the PMA-treated spores suspended in inactivated-spore suspension and the water-diluted spores.
FIG. 2.
CT values obtained for viable spores diluted in either dH2O (black bars) or a background of 107 inactivated spores and subjected to PMA treatment (gray bars) or a direct DNA extraction without PMA pretreatment (white bars). Results are means ± standard deviations of the results of three replicate experiments. The letters indicate statistically significant differences when comparing data within each spore concentration (P ≤ 0.05).
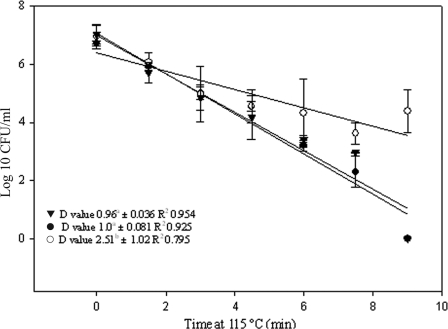
To establish whether the approach could be used to enumerate viable spores surviving thermal inactivation, 1.2 ml of diluted spore crop (approximately 106 spores/ml) was added to aluminum capillary tubes, followed by heating at 115°C in an oil bath (Haake L/D8 Fisions, Waltham, MA) (4). Tubes were removed at 1.5-min intervals over a period of 9 min, placed on ice for 5 min, and then sterilized by immersion in 3% hypochlorite. Three different aliquots of each treatment were subjected to the following: (i) plating for enumeration; (ii) immediate extraction for DNA isolation, followed by qPCR; and (iii) treatment with PMA followed by DNA extraction and qPCR. Preliminary data revealed that under these conditions, it was necessary to pretreat the heated spores with 10 mM dithiothreitol at 65°C for 15 min to facilitate selective entry of PMA into inactivated spores (data not shown). For the PMA-treated samples, the spore concentrations calculated by qPCR approximated those obtained by cultural enumeration for all time points examined (Fig. 3). This was not the case for the samples that were not treated with PMA, in which case qPCR overestimated the viable spore population by ≥4 log10 when counts were <105 spores/ml. When D115°C values were calculated, using linear regression, and compared, there was no statistically significant difference between the results of PMA-qPCR and cultural enumeration, but statistically significant differences were observed when comparing these D values to those obtained for the samples subjected to qPCR without prior PMA treatment.
FIG. 3.
Thermal inactivation of B. subtilis at 115°C as evaluated by cultural enumeration (•), PMA-qPCR (▾), and qPCR without prior PMA treatment (○). The qPCR results were compared to the standard curve to calculate approximate log10 CFU/ml. Results are means ± standard deviations of the results of three replicate experiments. The D115°C values followed by different superscript letters are statistically significantly different (P < 0.05).
Perhaps as expected, we demonstrated that spore populations surviving thermal inactivation are overestimated when using qPCR for quantification. DNA-intercalating agents have recently been used in conjunction with qPCR for the selective detection of live vegetative bacterial cells, including many of the food-borne pathogens (2, 3, 5, 6, 9), but such methods have not yet been applied to endospore quantification. We report the successful use of a combined PMA-qPCR method to estimate surviving spore populations after heat inactivation and which provided D values which were not statistically significantly different from those obtained by cultural enumeration. In the future, we intend to use the approach for validation of delivered thermal treatments on the pilot scale. The method may also be appropriate for validation of advanced thermal (ohmic, radiofrequency, and microwave) and nonthermal (ultrahigh pressure, radiation, pulsed electric field, etc.) sterilization technologies. Another potential area of application is in the validation of sterilization treatments of sterile zones in aseptic fillers and/or sterilization of packaging materials used in aseptic packaging. We are also in the process of modifying the method so that it can be applied to other thermal-process indicators, including C. sporogenes, an organism which is notably unpleasant to culture and difficult to enumerate. Altogether, the method described here takes us one step closer to being able to employ molecular methods for the discrimination of viable from nonviable endospores and, hence, their selective enumeration, as an alternative to cultural methods in the validation of food-processing efficacy.
Acknowledgments
We acknowledge the Center for Aseptic Processing and Packaging (CAPPS) for partial funding of this research.
The manuscript is designated FSR09-04 in the journal series of the Department of Food, Bioprocessing, and Nutrition Sciences, North Carolina State University.
The use of trade names in this publication does not imply endorsement by the North Carolina Agricultural Research Service or criticism of similar ones not mentioned.
Footnotes
Published ahead of print on 6 March 2009.
REFERENCES
- 1.Holdsworth, S. D. 1997. Thermal processing of packaged foods. Chapman and Hall, London, United Kingdom.
- 2.Nocker, A., and A. K. Camper. 2006. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl. Environ. Microbiol. 72:1997-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nogva, H. K., S. M. Drømtorp, H. Nissen, and K. Rudi. 2003. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. BioTechniques 4:804-813. [DOI] [PubMed] [Google Scholar]
- 4.Odlaug, T. E., and I. J. Pflug. 1977. Thermal destruction of Clostridium botulinum spores suspended in tomato juice in aluminum thermal death time tubes. Appl. Environ. Microbiol. 34:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudi, K., B. Moen, S. M. Drømtorp, and A. L. Holck. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudi, K., K. Naterstad, S. M. Drømtorp, and H. Holo. 2005. Detection of viable and dead Listeria monocytogenes on Gouda-like cheeses by real-time PCR. Lett. Appl. Microbiol. 40:301-306. [DOI] [PubMed] [Google Scholar]
- 7.Rueckert, A., R. S. Ronimus, and H. W. Morgan. 2006. Development of a real-time PCR assay targeting the sporulation gene, spo0A, for the enumeration of thermophilic bacilli in milk powder. Food Microbiol. 23:220-230. [DOI] [PubMed] [Google Scholar]
- 8.Stern, J. A., and B. E. Proctor. 1954. A micro-method and apparatus for the multiple determination of rates of destruction of bacteria and bacterial spores subjected to heat. Food Technol. 8:139-143. [Google Scholar]
- 9.Wang, S., and R. E. Levin. 2006. Discrimination of viable Vibrio vulnificus cells from dead cells in real-time PCR. J. Microbiol. Methods. 64:1-8. [DOI] [PubMed] [Google Scholar]