Abstract
Vibrio parahaemolyticus is a moderately halophilic bacterium found in estuarine and marine coastal ecosystems worldwide. Although the ability of V. parahaemolyticus to grow and proliferate in fluctuating saline environments is well known, the underlying molecular mechanisms of osmoadaptation are unknown. We performed an in silico analysis of V. parahaemolyticus strain RIMD2210633 for genes homologous to osmotic stress response genes in other bacteria. We uncovered two putative compatible solute synthesis systems (encoded by ectABC and betABI) and six putative compatible solute transporters (encoded by four bcct loci and two proVWX loci). An ectoine synthesis system clustered with a betaine/carnitine/choline transporter and a ProU transporter (encoded by homologues of proVWX from Escherichia coli), and a betaine synthesis system clustered with a ProU transporter (encoded by homologues of proVXW from Pseudomonas syringae). This is at least double the number present in V. cholerae, V. fischeri, or V. vulnificus. Six additional Vibrio species contain both ectABC and betABI, i.e., V. alginolyticus 12G01, V. angustum, V. harveyi BAA-1116, V. splendidus LGP32, Vibrio sp. strain MED222, and Vibrio sp. strain Ex25. V. harveyi HY01 and V. splendidus 12B01 only encoded the betaine system. In addition, V. alginolyticus had a compendium of systems identical to that found in V. parahaemolyticus. Comparative physiological analysis of RIMD2210633 with V. vulnificus YJ016, V. cholerae N16961, and V. fischeri ES114 grown at different salinities and temperatures demonstrated that V. parahaemolyticus had a growth advantage under all of the conditions examined. We demonstrate, by one-dimensional nuclear magnetic resonance analysis, that V. parahaemolyticus is capable of de novo synthesis of ectoine at high salinity whereas a ΔectB knockout strain is not. We constructed a single-knockout mutation in proU1, but no growth defect was noted, indicating transporter system redundancy. We complemented E. coli MKH13, a compatible solute transporter-negative strain, with bcct2 and demonstrated uptake of betaine at high salt concentrations.
Vibrio parahaemolyticus is a moderate halophile prevalent in all of the coastal waters around the world, particularly in the warmer summer months (17). V. parahaemolyticus is found associated with zooplankton and phytoplankton and is present in sea sediment (18-20). V. parahaemolyticus is a pathogen of fish and humans and is the leading cause of seafood-associated bacterial gastroenteritis worldwide. Fish and shellfish, particularly oysters, are implicated as the major vectors for infection (5, 7, 27). Numerous outbreaks of V. parahaemolyticus infection in the Pacific Northwest have resulted in severe economic losses and closures in the seafood industry (27). A number of environmental factors affect the occurrence and distribution of V. parahaemolyticus, such as temperature, salinity, oxygen availability, plankton, and tidal flushing (8-10, 18-20) Because all of the V. parahaemolyticus strains inhabit marine, brackish, and estuarine waters, fluctuations in temporal and persistent salinity pose a constant challenge to the adaptive response of the organism.
In most bacteria, the response to osmotic upshock has two phases (3, 11, 31, 32, 40, 43). The immediate and short-term response to hyperosmotic and high-salinity changes is the accumulation of K+. This is the primary strategy for many extremophiles living in high-salinity environments (37). Because high K+ concentrations are detrimental to most cells, a more long-term strategy to deal with osmotic upshock is required (3, 11, 31, 32, 40, 43). The second strategy, and the one more widely used among halophiles and for salt adaptation in general among bacteria, actinomycetes, algae, fungi, and yeasts, is the synthesis and/or accumulation of organic osmotic solutes (Fig. 1) (3, 11, 31, 32). These are known as compatible solutes or osmolytes since they are amassed in high concentrations without disturbing vital cellular functions (6). Osmolytes include sugars such as trehalose, free amino acids such as proline and glutamate, and their derivatives betaine, glycine betaine, and ectoine, as well as a number of esters and amines (6, 11, 34-36, 40).
FIG. 1.
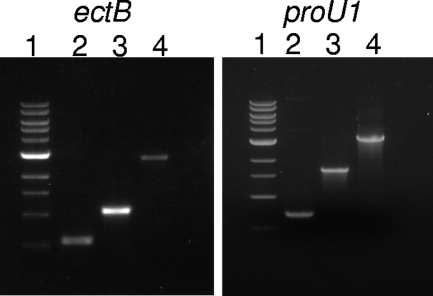
PCR confirmation of truncated alleles and double-crossover events in deletion mutagenesis of the ectB and proU1 genes of V. parahaemolyticus RIMD2210633. ectB: lane 1, 1-kb DNA ladder; lane 2, 533-bp ectAD product generated via SOE PCR; lane 3, 1.04-kb truncated ectB (double crossover); lane 4, 2.73-kb wild type. proU1: lane 1, 1-kb DNA ladder; lane 2, 428 bp; lane 3, 1.64 kb (double crossover); lane 4, 3.18-kb wild type.
The majority of bacteria utilize the trimethylammonium compound glycine betaine (N,N,N-trimethylglycine) as their preferred compatible solute (23, 24, 26, 29, 40, 43). Escherichia coli, which can grow at a maximum NaCl concentration of 0.5 M, can convert choline to betaine by using enzymes encoded by betABI, and choline is transported into the cell by the high-affinity BetT system, as well as by a low-affinity ProU transporter encoded by proVWX (11). One of the most widespread compatible solutes is ectoine (1,4,5,6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid) (23, 24, 26, 29, 40, 43, 44). The pathway for ectoine synthesis has been determined for several moderate halophiles, and in all cases the products of the ectABC genes are required (15, 41, 42). Ectoine was shown to play a role in osmotolerance in V. cholerae; when Pflughoeft et al. exposed a ΔectA mutant strain to high osmolarity, they observed a pronounced growth delay compared to the wild-type strain (33). In E. coli, which lacks an ectoine synthesis system, the ProP (encoded by proP) and ProU transporters were shown to take up a wide variety of osmoprotectants, including ectoine (22). ProU shows a preference for glycine betaine and proline betaine in E. coli and is highly upregulated in high-osmolarity medium (12).
In this study, we first examined the genome of V. parahaemolyticus RIMD2210633 and identified homologues of ectABC and betABI, as well as homologues of four betaine/carnitine/choline transporters (BCCTs) and two ProU compatible solute transporters, triple the number of systems identified in V. cholerae and double the number present in V. vulnificus and V. fischeri. Six additional Vibrio species encode both ectABC and betABI, i.e., V. alginolyticus 12G01, V. angustum, V. harveyi BAA-1116, V. splendidus LGP32, Vibrio sp. strain MED222, and Vibrio sp. strain Ex25. V. alginolyticus 12G01 had the same number and arrangement of compatible solute systems as V. parahaemolyticus. Comparative growth analysis experiments demonstrated that at high salinity and at high or low temperatures, V. parahaemolyticus had a growth advantage over V. cholerae, V. vulnificus, and V. fischeri. We show that the ectABC gene cluster in V. parahaemolyticus is required for de novo ectoine synthesis but that there is functional redundancy due to the large number of compatible solute transporters available.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. V. parahaemolyticus RIMD2210633 was cultured in Luria-Bertani medium (LB) with a final NaCl content of 3% (pH 7.5) and grown at 37°C. E. coli strain DH5α-λpir was grown in LB broth at 37°C. The auxotrophic E. coli DAP strain was supplied with 0.3 mM diaminopimelic acid for growth in LB broth. Chloramphenicol was added at a concentration of 25 μg/ml. The minimal medium used in growth studies was the previously described osmolarity medium (OM) (21, 33). The OM was adjusted by addition of NaCl at 200 mM (1%) for overnight preculture and 500 mM (3%), 1,000 mM (6%), or 1,500 mM (9%) for growth analysis over 24 h.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| V. parahaemolyticus | ||
| RIMD2210633 | O3:K6 clinical isolate | 25 |
| ΔectB | RIMD2210633 ectB mutant (VP1721) | This study |
| ΔproU1 | RIMD2210633 proU1 mutant (VP1726-VP1728) | This study |
| V. vulnificus YJO16 | Clinical isolate | 4 |
| V. cholerae N16961 | O1 El Tor biotype | 13 |
| V. fischeri ES114 | Isolate from Euprymna scolopes | 38 |
| E. coli | ||
| MKH13 | ΔproP2 ΔproU608 ΔputPA101 | 12 |
| DH5α-λpir | λpir φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | |
| DH5α-λpir ΔectB | DH5α/λpir harboring pDS132ΔectB | This study |
| DH5α-λpir ΔproU1 | DH5α/λpir harboring pDS132ΔproU1 | This study |
| β2155 DAP | Donor for bacterial conjugation; thr1004 pro thi strA hsdS lacZΔM15 (F′ lacZΔM15 lacTRQJΔ36 proA+proB+) ΔdapA ErmrpirRP4 (Kmr from SM10) | |
| β2155 DAP-ΔectB | β2155 harboring pDS132ΔectB | This study |
| β2155 DAP-ΔproU1 | β2155 harboring pDS132ΔproU1 | This study |
| Plasmids | ||
| pDS132 | R6K γori mobRP4 sacB Cmr; suicide vector for conjugal transfer and integration | |
| pBBR1MCS | ||
| pDS132ΔectB | pDS132 containing truncated ectABC region | This study |
| pDS132ΔproU1 | pDS132 containing truncated proVWX system | This study |
| pVP1723 | pBBR1MCS containing bcct2 | This study |
Genomic analysis and construction of ΔectB and ΔproU1 deletion mutants.
Homology searches were conducted with the BLAST (basic local alignment search tool) function (1). Genomic DNA was isolated with the Gnome DNA extraction kit in accordance with manufacturer's instructions. Plasmid DNA was isolated with a QIAprep Spin miniprep kit (Qiagen, Hilden, Germany). Splicing by overlap extension (SOE) PCR and allelic exchange were used in the creation of intragenic in-frame mutations in ectB (VP1721) and proU1 (VP1726 to VP1728) in V. parahaemolyticus RIMD2210633, and the sizes of the regions removed from the genes are 1,670 and 2,504 bp, respectively, as described previously (28). Primer pairs SOEAect/SOEBect and SOECect/SOEDect were used to amplify a 246-bp and a 287-bp PCR product, respectively, with V. parahaemolyticus RIMD2210633 as the template (Table 2). The resulting PCR products were purified and ligated together with T4 DNA ligase (New England BioLabs, Beverly, MA). The resulting product was PCR amplified with primer pair SOEAect/SOEDect to generate a 533-bp product named ectAD (Fig. 1, lane 2). The ectAD product was restricted with the XbaI and SacI enzymes (New England BioLabs), as was the suicide vector pDS132, and they were ligated together with T4 DNA ligase (New England BioLabs) and designated pDS132ΔectB. pDS132ΔectB was transformed into E. coli DH5α-λpir via electroporation and then plasmid purified and transformed into auxotrophic E. coli strain β2155 DAP. Transformed cells were designated β2155 DAP-ΔectB. Next, conjugal transfer was performed by cross-streaking each E. coli β2155 DAP-ΔectB donor strain with V. parahaemolyticus RIMD2210633 initially on LB agar with a final salt content of 250 mM (1.5%) NaCl supplemented with 0.3 mM diaminopimelic acid. Growth from these plates was then transferred to LB containing 25 μg/ml chloramphenicol and 200 μg/ml streptomycin. In order to cure exconjugate colonies of the integrated pDS132 plasmid, overnight cultures were serially diluted to 10−9 cells and plated on LB plates containing 10% sucrose. Double-crossover deletion mutant ΔectB isolates were screened for by PCR analysis with SOEFLectF and SOEeFLectR and confirmed by sequencing (Table 1). The ΔectB mutant was designated V. parahaemolyticus strain ΔectB. Deletion mutant ΔproU1, containing a deletion in the ProU1 transporter system, was constructed in the same manner with SOE primer pairs described in Table 2. PCR analysis with primer pair SOEFLproF/SOEFLproR was used to confirm integration of the nonfunctional gene copies in their respective mutant strains (Fig. 1 and Table 2). All of the strains were sequenced at MWG to verify the mutated alleles.
TABLE 2.
Primers used in this study
| Primer | Sequence 5′-3′a | Tab (°C) | %GC | Product size (bp) |
|---|---|---|---|---|
| SOEAect | CTT AGC TTT TAG TCA ACG AGC | 56 | 43 | |
| SOEBect | GAA GGA ACG GAA ACG CAC ATC | 59 | 52 | 246 (SOEABect) |
| SOECect | UGAT GTG CGT TTC CGT TCC TTCU CCG TTA CCT CTG AAT | 47 | 50 | |
| SOEDect | TGA TCA CAT CAG CAC CTT GG | 55 | 48 | 287 (SOECDect) |
| SOEApro | CAT GGA TCC CAT ACT GGA AG | 55 | 50 | |
| SOEBpro | CGT GGC GAG CAG TCA CAT C | 57 | 58 | 409 (SOEABpro) |
| SOECpro | UGAT GTG ACT GCT CGC CAC GCUC GTA TTG GGT AAG TGG | 47 | 60 | |
| SOEDpro | CGT GCG CTT CGA TGA CTG | 53 | 60 | 328 (SOECDpro) |
| SOEFLectF | CTT GCC ATT GAT CGT CAC G | 49 | 53 | |
| SOEFLectR | CAC TGC ATT CTG ACT CAT G | 50 | 47 | 2,733 (wild type) |
| SOEFLproF | CA CTT CTG CAT GAA TGG C | 50 | 53 | |
| SOEFLproR | GCT CGA TAA CTT GGC TCC | 51 | 56 | 4,168 (wild type) |
Overhangs complementary to SOEB primers are underlined.
Ta, annealing temperature.
Growth analysis.
Precultures of either wild-type or mutant bacteria were grown to stationary phase (i.e., 1 × 109 CFU/ml) at 37°C in OM supplemented with 200 mM (1%) NaCl. A 2% inoculum of these overnight cultures was diluted in a 1:100 ratio in 5 ml of fresh OM containing various concentrations of NaCl and organic supplements as indicated. A 200-μl volume of the inoculated medium was then added to a 96-well microtiter plate and incubated at 37°C. All of the assays were done in triplicate, and all of the experiments were performed three times. Optical densities were measured at various time points with a microplate reader. Sigmaplot software was used to construct graphs based on the data obtained. Statistical analysis was carried out with the Student t test. For comparative growth analysis of V. parahaemolyticus RIMD2210633, V. vulnificus YJO16, V. cholerae N16961, and V. fischeri ES114, the bacteria were cultured routinely overnight and diluted in a 1:100 ratio in 5 ml fresh LB containing 3%, 6%, or 9% NaCl. Growth was analyzed over a 24-h period at 42°C, 37°C, 30°C, or 20°C at all of these NaCl concentrations. Graphs were constructed with Sigmaplot software based on the data obtained.
Preparation of cell extracts and nuclear magnetic resonance (NMR) analysis.
The V. parahaemolyticus wild-type and ΔectB mutant strains were incubated with shaking at 37°C in OM supplemented with either 200 mM or 1,000 mM NaCl as previously described (12). Briefly, stationary phase cells were pelleted at 1,000 × g for 10 min and the spent growth medium was removed. Three freeze-thaw cycles were performed on cell pellets to increase cell lysis, and then they were resuspended in 750 μl of ethanol. Debris was removed by centrifugation, and the ethanol extract was transferred to a clean tube. The ethanol was subsequently removed from the extracted material by evaporation under vacuum. The pellet was resuspended in 500 μl of D2O (Aldrich), and insoluble material was removed by centrifugation. The solution was transferred to a 5-mm NMR tube for analysis, which was carried out on a Bruker AVANCE 600 NMR spectrometer operating at a proton frequency of 600.13 MHz (12). A standard proton NMR experiment, with a sweep width of 12,376 Hz and a relaxation delay of 5 s, was used to obtain NMR spectra at 298 K. Sixteen scans were coadded for each spectrum represented here.
Complementation of transporter-deficient E. coli MKH13 with a putative BCCT (VP1723) from V. parahaemolyticus.
V. parahaemolyticus RIMD2210633 genomic DNA was isolated with a GNOME DNA isolation kit (MP Biomedicals) according to the manufacturer's instructions. Primers bcct2F (TCT AGA AAC TTG TGC TTG GTG ATG TG) and bcct2R (GAG CTC ACG GCA CAC TTT CGC ATG), used for PCR amplification of VP1723 from the genomic DNA used as the template, were designed to include 87 bp upstream of the start codon and 47 bp downstream of the stop codon, incorporating an XbaI site on the 5′ end and a SacI site on the 3′ end. The resulting 1,808-bp product was cloned into pCR2.1 with a TOPO-TA cloning kit (Invitrogen), sequenced, and subsequently subcloned into plasmid pBBR1MCS following XbaI/SacI digestion of both the vector and the insert, resulting in the construct pVP1723. Electrocompetent MKH13 was generated according to a standard protocol (39) and transformed with pVP1723, and transformants were selected for chloramphenicol (25 μg/ml) resistance on LB. Cmr MKH13 was subsequently screened for the presence of VP1723 by PCR.
RESULTS AND DISCUSSION
Comparative genome analysis.
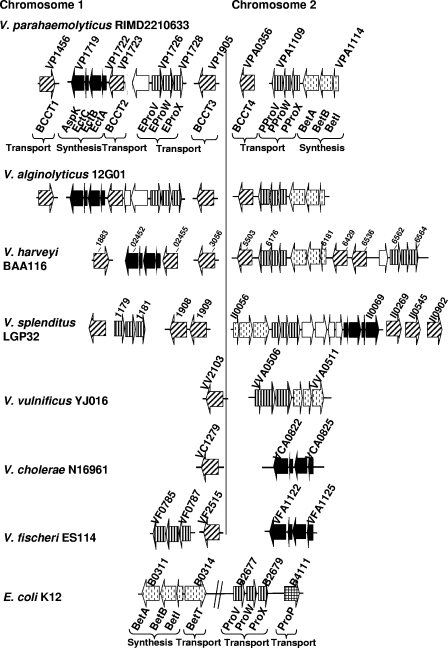
V. parahaemolyticus is a moderately halophilic bacterium that can grow at 0.1 M to 1.5 M NaCl concentrations; however, the underlying molecular mechanisms of osmoadaptation are unknown. We examined the V. parahaemolyticus RIMD2210633 genome sequence for homologues of specialized systems previously shown to play a role in the osmotic stress response in bacteria. In silico genome analysis of RIMD2210633 revealed the presence of two putative compatible solute synthesis systems (ectoine and betaine) and six putative compatible solute transporters (Fig. 2). On chromosome 1, two of the compatible solute transporters, a ProU1 transporter (proVWX) and BCCT2 (bcct), were clustered with the ectoine synthesis system (ectABC). The ectABC genes were homologous to those described in V. cholerae N16961, and the proVWX genes were homologous to those described in E. coli K-12. On chromosome 2, the betaine synthesis system (encoded by betABI) clustered with another ProU2 transporter encoded by proVWX that is homologous to the Pseudomonas syringae sequence (Fig. 2). Three additional BCCTs were identified, open reading frames VP1456, VP1905, and VPA0356, herein referred to as BCCT1, BCCT3, and BCCT4, respectively (Fig. 2). Molecular analysis of 42 V. parahaemolyticus isolates found that the ectABC and betABI gene clusters are present in all of the strains tested (data not shown).
FIG. 2.
Distribution of osmolyte transport and synthesis systems among seven Vibrio species and E. coli K-12. Arrows indicate the direction of transcription. Transporters belonging to the BCCT family are represented by diagonal stripes. Solid black arrows represent putative genes involved in ectoine synthesis. Open arrows indicate hypothetical proteins. Vertical stripes represent genes homologous to that which encodes the ProU (ProVWX) transporter. Dashed lines indicate genes involved in glycine betaine synthesis (betABI). The grid arrow represents the ProP transporter of E. coli K-12.
Bioinformatic analysis of all of the available Vibrio genome sequences in the database revealed an interesting distribution of systems. Among the 36 Vibrionaceae genomes available, representing 15 different species, we found that betABI is present in 12 species, V. alginolyticus 12G01, V. angustum S14, V. campbellii AND4, Vibrio sp. strain Ex25, Vibrio sp. strain MED222, V. harveyi, V. parahaemolyticus, V. splendidus, V. shilonii AK1, V. vulnificus, Photobacterium profundum, and Photobacterium sp., and absent from 3 species, Alivibrio salmonicida, V. cholerae, and V. fischeri. The ectABC cluster is present in 10 species and absent from 5 species, A. salmonicida, V. campbellii, V. shilonii, V. vulnificus, and Photobacterium sp. strain SKA34, and can be absent from some strains within a species. Only A. salmonicida and two V. cholerae strains, V51 and RC385, appear to lack both synthesis systems. We identified seven species that encode both putative osmolyte synthesis systems, V. alginolyticus 12G01, V. angustum, Vibrio sp. strain Ex25, Vibrio sp. strain MED222, V. harveyi BAA-1116, V. splendidus LGP32, and P. profundum 3CTK. V. harveyi HY01, V. splendidus 12B01, P. profundum SS9, and Photobacterium sp. strain Ska34 only encode the betaine system.
In V. alginolyticus 12G01, a close relative of V. parahaemolyticus, the same number and distribution of compatible solute transport and synthesis systems were present (Fig. 2). In V. harveyi BA116, homologues of all of the V. parahaemolyticus systems are present; however, the two ProU transporters are on chromosome 2 and adjacent to one of them is an integrase. In V. splendidus LGP32, three BCCTs and one ProU transporter are present on chromosome 1. On chromosome 2, both ectABC and betABI are present, along with ProU and three BCCTs (Fig. 2). Analysis of the genomes of V. cholerae N16961, V. vulnificus YJ016, and V. fischeri ES114 revealed that V. parahaemolyticus contains at least double the number of osmotolerance systems present in these isolates (Fig. 2). Overall, our data suggest that ectABC is the most variable in its distribution and location on the genome, whereas betABI is always found on chromosome 2 and associated with a ProU transporter, which may be the ancestral state in Vibrio.
Comparative growth analysis of Vibrio species.
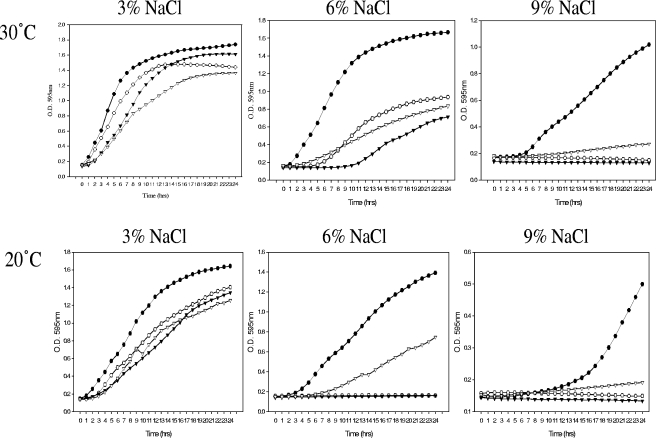
In order to assess whether the carriage of such a large number of systems gives V. parahaemolyticus a growth advantage, we examined the ability of strain RIMD2210633 to grow under increasing NaCl concentrations and at different temperatures compared to V. vulnificus, V. cholerae, and V. fischeri under the same conditions. First, we examined the growth of all of the strains in LB broth containing 3%, 6%, or 9% NaCl at 30°C. V. parahaemolyticus exhibited the best growth of all of the strains at this temperature under all of the salinity conditions examined (Fig. 3). In LB containing 3% NaCl, V. cholerae displayed a 1-h delay in growth while both V. vulnificus and V. fischeri exhibited a 2-h delay in growth. At 6% NaCl, an extended growth delay was observed in all three species; a 4-h lag in growth was observed in V. fischeri, and a 5-h lag was observed in V. vulnificus, while V. cholerae displayed the longest lag in growth at 9 h. V. parahaemolyticus was the only organism capable of growth in LB containing 9% NaCl (Fig. 3). Growth analysis was also performed at 37°C under identical conditions, and similar patterns of growth were observed in all of the strains, with the exception of V. fischeri, which cannot grow at this temperature (data not shown). Growth was examined at 20°C under the same salinity conditions as above, and V. parahaemolyticus again displayed the best growth of all of the species examined at 3% and 6% NaCl (Fig. 3). Similar data were obtained for growth at 42°C in 3% and 6% NaCl, again with the exception of V. fischeri, which cannot grow at this temperature (data not shown). The comparative physiological data presented here highlight the growth advantage V. parahaemolyticus maintains under conditions of elevated salinity at high and low temperatures compared to the other Vibrio species, which contain a limited number of compatible solute systems. Most notable is the pronounced advantage V. parahaemolyticus displays at the highest osmolarity examined. We could speculate from our data that the multisystem osmotolerance strategy available to V. parahaemolyticus allows it to survive in more extreme environments than its close relatives (Fig. 2 and 3). The finding that V. parahaemolyticus also survives better under temperature stress at high salinity suggests that the compatible solute systems may also function to cross-protect against other stresses. This may be particularly important in the human host, where the bacterium must survive the acid, bile, and hydrogen peroxide damage encountered in this niche.
FIG. 3.
Analysis of Vibrio species growth in LB containing 3% (500 mM), 6% (1,000 mM), or 9% (1,500 mM) NaCl at 30°C and 20°C. Solid circles represent V. parahaemolyticus RIMD2210633, open circles represent V. vulnificus YJ016, solid triangles represent V. cholerae N16961, and open triangles represent V. fischeri ES114. O.D., optical density.
Growth analysis of wild-type V. parahaemolyticus and the ΔectB mutant in OM.
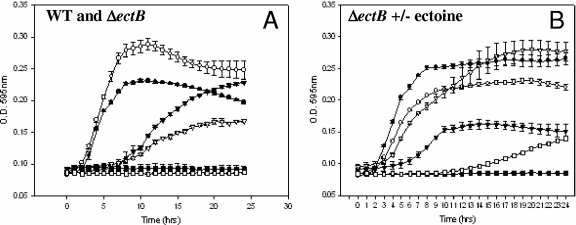
To determine further the role of the compatible solute synthesis systems in osmotolerance in V. parahaemolyticus, we constructed a knockout mutation in the ectB gene by SOE PCR and allelic exchange (Fig. 1). The wild-type RIMD2210633 and ΔectB mutant strains were grown in LB broth containing 500 mM, 1,000 mM, or 1,500 mM, and the two strains showed similar growth patterns (data not shown). This finding suggests that components of LB broth may provide substrates for alternative osmotolerance pathways in V. parahaemolyticus similar to what was noted in V. cholerae (33). Therefore, we used the defined OM, which limits the number of osmotolerance pathways available (33). Our wild-type strain and the ΔectB mutant strain were first grown on OM supplemented with 200 mM NaCl and then transferred to OM supplemented with 500 mM, 1,000 mM, or 1,500 mM NaCl (Fig. 4A). In OM plus 500 mM NaCl, the wild-type and mutant strains showed similar growth patterns; however, in 1,000 mM NaCl, the mutant had an extended lag phase and only reached an optical density of 0.15 and no growth of either strain was found at 1,500 mM NaCl (Fig. 4A). Thus, it appears that the ΔectB mutant has a growth defect at high NaCl concentrations. Next, we examined the growth of the ΔectB mutant in OM supplemented with ectoine at 500 mM, 1,000 mM, or 1,500 mM NaCl (Fig. 4 B). We found that the presence of ectoine in the medium restored the growth of the ΔectB mutant in OM containing 1,000 mM NaCl. In addition, after an 11-h lag phase, growth occurred in 1,500 mM NaCl, suggesting that ectoine is used as a compatible solute under these conditions. In contrast, the ΔectB mutant in OM supplemented with aspartic acid, the precursor for ectoine synthesis, had no effect on growth (data not shown).
FIG. 4.
(A) Growth analysis of wild-type (WT) (solid symbols) and ΔectB mutant (open symbols) V. parahaemolyticus following transfer from OM containing 200 mM (1%) NaCl to OM containing 500 mM (3%) NaCl (circles), 1,000 mM (6%) NaCl (triangles), or 1,500 mM (9%) NaCl (squares). (B) Growth of the ΔectB mutant in OM following transfer from OM containing 200 mM (1%) NaCl to OM containing 500 mM (3%) NaCl (solid circles), 1,000 mM (6%) NaCl (triangles), or 1,500 mM (9%) NaCl (squares) without supplementation (solid symbols) or with supplementation (open symbols) with 500 μM ectoine. O.D., optical density.
Growth in high-osmolarity medium induces ectoine synthesis.
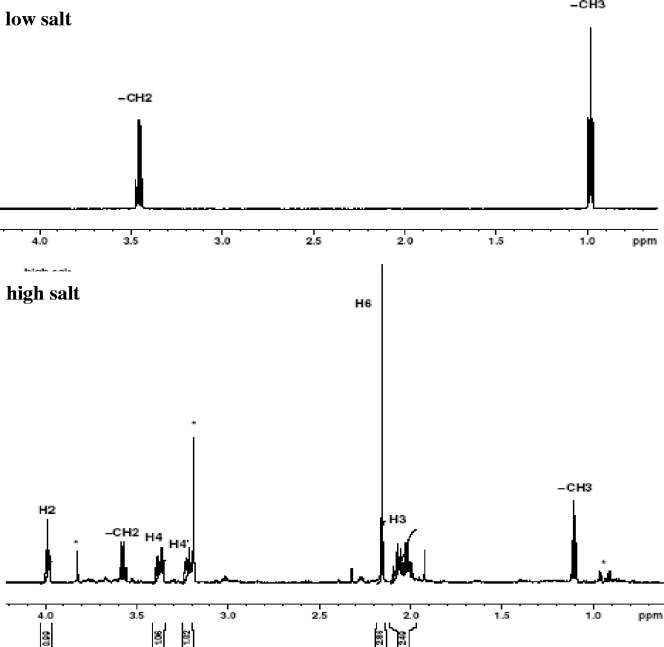
Having observed growth of the wild type in OM supplemented with 1,000 mM NaCl, we suspected that ectoine might be synthesized de novo by the cell in response to increased external osmolarity. To test this, we made ethanol extracts of V. parahaemolyticus cells grown in OM supplemented with 200 mM or 1,000 mM NaCl and determined the one-dimensional H-NMR spectra of these extracts (Fig. 5). Peaks corresponding to the various hydrogen atoms of ectoine are labeled in Fig. 5. Comparative analysis clearly indicates that ectoine-specific peaks are missing from the spectrum of the cell extract derived from V. parahaemolyticus grown in OM supplemented with a low NaCl concentration (200 mM) but present in cells grown at a NaCl high concentration (1,000 mM) (Fig. 5). To demonstrate further that ectABC is required for ectoine synthesis, we determined the one-dimensional H-NMR spectra of extracts from our ΔectB mutant grown at low and high NaCl concentrations. From this analysis, we found no ectoine-specific peaks produced, which was as expected since the organism can no longer synthesize ectoine de novo (see Fig. S1 in the supplemental material).
FIG. 5.
One-dimensional H-NMR spectra of V. parahaemolyticus grown in low (200 mM) and high (1,000 mM) NaCl concentrations. The two-dimensional double-filtered quantum coherence and heteronuclear multiple-quantum coherence spectra of these extracts were also measured and compared with the spectrum of purified ectoine (data not shown). This comparison supported the spectral assignments shown.
Growth analysis of wild-type V. parahaemolyticus and the ΔproU mutant in OM.
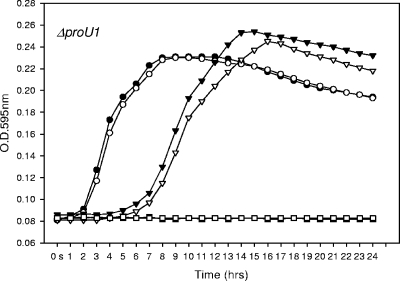
To begin to determine the role of multiple compatible transporters in V. parahaemolyticus, we constructed a knockout mutation in proU1 that clustered with ectABC (Fig. 2). Using SOE PCR and allelic exchange, we created a ΔproU1 mutant (Fig. 1). Our wild-type and ΔproU1 mutant strains were first grown on OM supplemented with 200 mM NaCl and then transferred to OM supplemented with 500 mM, 1,000 mM, or 1,500 mM NaCl. The ΔproU1 mutant did not show a growth defect compared to the wild type (Fig. 6). When grown on OM (1,000 mM NaCl) supplemented with ectoine or aspartic acid, the ΔproU1 mutant had shortened lag times (data not shown). This result suggests that both of these substrates are used as compatible solutes. Neither ectoine nor aspartic acid had an effect on the growth of the mutant in OM containing 500 mM NaCl, similar to the result obtained with the ΔectB mutant under the same conditions. This result indicates that V. parahaemolyticus primarily uses these substrates as osmolytes and not as a carbon source. Overall, our results indicate that under the conditions we examined, the presence of multiple systems within the genome of V. parahaemolyticus results in redundancy. We suggest that under low-salinity conditions, V. parahaemolyticus may use one set of transporters, whereas under high-salinity conditions, it may switch its osmotolerance strategy to a different set of transporters. In addition, V. parahaemolyticus may contain a large number of synthesis and transporter systems that are required for cross-protection against other stresses.
FIG. 6.
Growth analysis of wild-type (solid symbols) and ΔproU1 mutant (open symbols) V. parahaemolyticus following transfer from OM containing 200 mM (1%) NaCl to OM containing 500 mM (3%) NaCl (circles), 1,000 mM (6%) NaCl (triangles), or 1,500 mM (9%) NaCl (squares). O.D., optical density.
Complementation of E. coli MKH13 with bcct2 from V. parahaemolyticus.
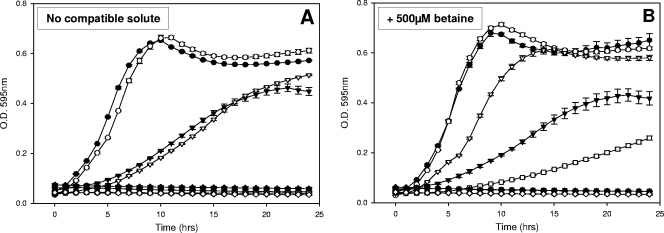
To determine the role of the putative transporter BCCT2, we cloned the bcct2 gene (open reading frame VP1723) into pBBR1MCS and transformed this plasmid into the compatible transport-deficient strain E. coli MKH13, resulting in strain E. coli MC1 (12). We examined the growth of MC1 in 0.5 M NaCl unsupplemented or supplemented with betaine, choline, glutamate, or proline. We found growth in M9 medium supplemented with 6% NaCl only in the presence of betaine, suggesting that BCCT2 has specificity for this substrate, similar to other BCCTs (Fig. 7).
FIG. 7.
Growth of E. coli MKH13 (MKH13/pBBR1MCS, empty symbols) or MC1 (MKH13/pVP1723, filled symbols) in M9 minimal medium supplemented with NaCl to either a 1% (circle), a 3 (triangle), a 6% (square), or a 9% (diamond) final concentration in the absence (A) or presence (B) of 500 μM glycine betaine. O.D., optical density.
Conclusions.
The emergence of the highly virulent V. parahaemolyticus O3:K6 serovar in 1996 with a subsequent increase in the reported cases of V. parahaemolyticus has renewed interest in this organism's ecology and evolution (25, 30). V. parahaemolyticus O3:K6 now has a global distribution and was recovered as far north as Alaska, indicating dissemination to new geographic climes and the organism's ability to survive more extreme fluctuations in environmental conditions (9, 27). Genomic comparisons have identified a large number of regions unique to V. parahaemolyticus, many of these predominantly associated with pathogenic isolates only (2, 14, 16). Through a combination of bioinformatic, molecular genetic, physiological, and biochemical analyses, we have highlighted the role of osmolyte synthesis and transport systems in osmotolerance. The presence of both ectoine and betaine synthesis systems suggests that the ability to synthesize osmolytes de novo is an important survival strategy for this organism and for Vibrio species in general. The present of redundant systems suggests that V. parahaemolyticus may use different osmotolerance strategies under different conditions, a possibility which needs to be examined further. In addition, the presence of so many compatible solute synthesis and transport systems in the cell may be important for cross-protection against other stresses.
Supplementary Material
Acknowledgments
We thank those who kindly provided the V. parahaemolyticus strains used in this study. We thank especially F. Jerry Reen for technical assistance. We thank Steve Bai at the NMR Facility, University of Delaware, for help with the NMR analysis.
This study was supported in part by University of Delaware Research Foundation grant UDRF2007-2008, an NIH COBRE grant, and a Science Foundation Ireland student fellowship to L.M.N.
Footnotes
Published ahead of print on 13 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, E. F., A. L. V. Cohen, L. M. Naughton, D. W. Ussery, T. T. Binnewies, O. C. Stine, and M. A. Parent. 2008. Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 8:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, A. D. 1976. Microbial water stress. Bacteriol. Rev. 40:803-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiou, C. S., S. Y. Hsu, S. I. Chiu, T. K. Wang, and C. S. Chao. 2000. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J. Clin. Microbiol. 38:4621-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Costa, M. S., H. Santos, and E. A. Galinski. 1998. An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv. Biochem. Eng. Biotechnol. 61:117-153. [DOI] [PubMed] [Google Scholar]
- 7.Deepanjali, A., H. S. Kumar, I. Karunasagar, and I. Karunasagar. 2005. Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl. Environ. Microbiol. 71:3575-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePaola, A., C. A. Kaysner, J. Bowers, and D. W. Cook. 2000. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66:4649-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DePaola, A., J. L. Nordstrom, J. C. Bowers, J. G. Wells, and D. W. Cook. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69:1521-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DePaola, A., J. Ulaszek, C. A. Kaysner, B. J. Tenge, J. L. Nordstrom, J. Wells, N. Puhr, and S. M. Gendel. 2003. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microbiol. 69:3999-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galinski, E. A. 1995. Osmoadaptation in bacteria. Adv. Microb. Physiol. 37:272-328. [PubMed] [Google Scholar]
- 12.Haardt, M., B. Kempf, E. Faatz, and E. Bremer. 1995. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol. Gen. Genet. 246:783-786. [DOI] [PubMed] [Google Scholar]
- 13.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurley, C. C., A. Quirke, F. J. Reen, and E. F. Boyd. 2006. Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics 7:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imhoff, J., and F. Rodriguez-Valera. 1984. Betaine is the main compatible solute of halophilic eubacteria. J. Bacteriol. 160:478-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izutsu, K., K. Kurokawa, K. Tashiro, S. Kuhara, T. Hayashi, T. Honda, and T. Iida. 2008. Comparative genomic analysis using microarray demonstrates a strong correlation between the presence of the 80-kilobase pathogenicity island and pathogenicity in Kanagawa phenomenon-positive Vibrio parahaemolyticus strains. Infect. Immun. 76:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph, S. W., R. R. Colwell, and J. B. Kaper. 1982. Vibrio parahaemolyticus and related halophilic vibrios. Crit. Rev. Microbiol. 10:77-124. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko, T., and R. R. Colwell. 1974. Distribution of Vibrio parahaemolyticus and related organisms in the Atlantic Ocean off South Carolina and Georgia. Appl. Microbiol. 28:1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko, T., and R. R. Colwell. 1973. Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J. Bacteriol. 113:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneko, T., and R. R. Colwell. 1975. Incidence of Vibrio parahaemolyticus in Chesapeake Bay. Appl. Microbiol. 30:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapfhammer, D., E. Karatan, K. J. Pflughoeft, and P. I. Watnick. 2005. Role for glycine betaine transport in Vibrio cholerae osmoadaptation and biofilm formation within microbial communities. Appl. Environ. Microbiol. 71:3840-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 23.Louis, P., and E. A. Galinski. 1997. Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in Escherichia coli. Microbiology 143(Pt. 4):1141-1149. [DOI] [PubMed] [Google Scholar]
- 24.Lucht, J. M., and E. Bremer. 1994. Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system proU. FEMS Microbiol. Rev. 14:3-20. [DOI] [PubMed] [Google Scholar]
- 25.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 26.Malli, R., and W. Epstein. 1998. Expression of the Kdp ATPase is consistent with regulation by turgor pressure. J. Bacteriol. 180:5102-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLaughlin, J., A. DePaola, C. Bopp, K. Martinek, N. Napolilli, C. Allison, S. Murray, E. Thompson, M. Bird, and J. Middaugh. 2005. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 353:1463-1470. [DOI] [PubMed] [Google Scholar]
- 28.Murphy, R. A., and E. F. Boyd. 2008. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J. Bacteriol. 190:636-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagata, S. 2001. Growth of Escherichia coli ATCC 9637 through the uptake of compatible solutes at high osmolarity. J. Biosci. Bioeng. 92:324-329. [DOI] [PubMed] [Google Scholar]
- 30.Nair, G., T. Ramamurthy, S. Bhattacharya, B. Dutta, Y. Takeda, and D. Sack. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oren, A. 1999. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63:334-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oren, A. 2002. Halophilic microorganisms and their environments. Kluwer Scientific Publishers, Dordrecht, The Netherlands.
- 33.Pflughoeft, K. J., K. Kierek, and P. I. Watnick. 2003. Role of ectoine in Vibrio cholerae osmoadaptation. Appl. Environ. Microbiol. 69:5919-5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Record, M. T., Jr., E. S. Courtenay, D. S. Cayley, and H. J. Guttman. 1998. Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem. Sci. 23:143-148. [DOI] [PubMed] [Google Scholar]
- 35.Record, M. T., Jr., W. Zhang, and C. F. Anderson. 1998. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv. Protein Chem. 51:281-353. [DOI] [PubMed] [Google Scholar]
- 36.Roberts, M. F. 2005. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Systems 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, M. F. 2004. Osmoadaptation and osmoregulation in archaea: update 2004. Front. Biosci. 9:1999-2019. [DOI] [PubMed] [Google Scholar]
- 38.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Sleator, R. D., and C. Hill. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26:49-71. [DOI] [PubMed] [Google Scholar]
- 41.Ventosa, A., M. Márquez, M. Garabito, and D. Arahal. 1998. Moderately halophilic gram-positive bacterial diversity in hypersaline environments. Extremophiles 2:297-304. [DOI] [PubMed] [Google Scholar]
- 42.Ventosa, A., J. J. Nieto, and A. Oren. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood, J. M. 2007. Bacterial osmosensing transporters. Methods Enzymol. 428:77-107. [DOI] [PubMed] [Google Scholar]
- 44.Wood, J. M., E. Bremer, L. N. Csonka, R. Kraemer, B. Poolman, T. van der Heide, and L. T. Smith. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:437-460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.