Abstract
The soil saprophyte Bacillus cereus forms biofilms at solid-liquid interfaces. The composition of the extracellular polymeric matrix is not known, but biofilms of other bacteria are encased in polysaccharides, protein, and also extracellular DNA (eDNA). A Tn917 screen for strains impaired in biofilm formation at a solid-liquid interface yielded several mutants. Three mutants deficient in the purine biosynthesis genes purA, purC, and purL were biofilm impaired, but they grew planktonically like the wild type in Luria-Bertani broth. Biofilm populations had higher purA, purC, and purL transcript ratios than planktonic cultures, as measured by real-time PCR. Laser scanning confocal microscopy (LSCM) of BacLight-stained samples indicated that there were nucleic acids in the cell-associated matrix. This eDNA could be mobilized off the biofilm into an agarose gel matrix through electrophoresis, and it was a substrate for DNase. Glass surfaces exposed to exponentially growing populations acquired a DNA-containing conditioning film, as indicated by LSCM. Planktonic exponential-phase cells released DNA into an agarose gel matrix through electrophoresis, while stationary-phase populations did not do this. DNase treatment of planktonic exponential-phase populations rendered cells more susceptible than control populations to the DNA-interacting antibiotic actinomycin D. Exponential-phase purA cells did not contain detectable eDNA, nor did they convey a DNA-containing conditioning film to the glass surface. These results indicate that exponential-phase cells of B. cereus ATCC 14579 are decorated with eDNA and that biofilm formation requires DNA as part of the extracellular polymeric matrix.
Bacteria in natural surroundings are able to grow as matrix-enclosed multicellular communities called biofilms (17). This multicellular, polymer-encased mode of growth is now accepted to be a preferred lifestyle option for prokaryotes. Early biofilm studies focused on aquatic ecosystems (12), and Pseudomonas aeruginosa was adopted as the model bacterium of choice to study biofilm development (13, 14). Subsequently, biofilms of a range of pathogenic bacteria allochthonous to aquatic environments have been studied. Like their gram-negative counterparts, many gram-positive bacteria are also able to form biofilms (1, 26). The gram-positive soil bacteria Bacillus subtilis and Bacillus cereus form biofilms at both solid-liquid interfaces (28) and liquid-air interfaces (7, 57), and they are often termed pellicles. Biofilm populations display a distinct phenotype, as revealed by proteomic studies (43, 52, 53), and have also proved to be more impervious to antibiotic treatment (27, 31). Planktonic cells of B. cereus growing in the presence of a biofilm display a unique phenotype that is distinct from the phenotype of true planktonic populations (35). We have termed planktonic cells that grow in proximity to a biofilm biofilm and surface-exposed planktonic cells (52).
Extracellular polymeric substances (EPS) produced by a biofilm community form the microenvironment for cells in the biofilm (18). The EPS matrix is highly hydrated and has various roles, including adhesion of the biofilm to surfaces, sequestering of substances from the environment, and protection from predators (49). EPS is also thought to contribute to the increased antibiotic resistance often reported for bacteria in biofilms (32). Early studies focused on polysaccharide components of EPS. It is now clear that the specific polysaccharide composition varies between strains and is also determined in part by growth conditions and the age of the biofilm (18, 50). Exopolysaccharide biosynthesis in B. subtilis biofilms is encoded by a 15-gene operon (epsA to epsO), but the chemical nature of the polymer is not known (9, 25). The only genes predicted to encode polysaccharide biosynthesis in B. cereus ATCC 14579 occur in a 17-gene operon (BC5263 to BC5279) and produce a putative galactose-containing polymer (22). In addition to polysaccharides, biofilm EPS may contain proteins and nucleic acids (18). B. subtilis excretes the protein TasA, which occurs in the EPS and is required for biofilm formation (6).
DNA was first shown to occur in the EPS of P. aeruginosa biofilms, and young biofilms could be dislodged by treatment with DNase (56). The nucleic acid present in the EPS matrix of biofilms has been termed extracellular DNA (eDNA) (47, 56). eDNA is required for the structural integrity of biofilms of P. aeruginosa, Variovorax paradoxus, and Rhodococcus erythropolis (47). Biofilms of Haemophilus influenzae and the gammaproteobacterium F8 are held together by a distinct filamentous meshwork of double-stranded DNA (dsDNA) (4, 24). Although the emphasis in eDNA research has been on gram-negative bacteria, the gram-positive pathogens Staphylococcus aureus, Staphylococcus epidermidis, and Streptococcus pneumoniae have also been reported to require eDNA to maintain biofilm integrity (23, 33, 39). However, eDNA has not been reported to be present in biofilms of Bacillus and other gram-positive rods previously.
By screening a Tn917 library for biofilm-impaired mutants of B. cereus ATCC 14579, we found several transposons located in purA, purC, and purL, which are genes involved in purine biosynthesis. Despite being biofilm-impaired, the mutants obtained were not growth impaired when they were growing planktonically in Luria-Bertani (LB) broth. Here we report that the EPS of B. cereus biofilms contains eDNA and that biofilm formation is dependent on the presence of purine biosynthesis genes.
MATERIALS AND METHODS
Culture conditions.
B. cereus ATCC 14579 was cultured in a 250-ml Erlenmeyer flask containing 100 ml of LB broth (pH 7.0) (Fisher Biotech). The broth was inoculated to a density of 105 CFU/ml with a washed, calibrated inoculum from an overnight culture and incubated at either 37 or 28°C with shaking at 200 rpm. Biofilms of B. cereus were allowed to develop either on the walls of acid-washed glass tubes and glass beakers or on glass wool fibers by adding 2 g of glass wool per 100 ml broth in a 250-ml Erlenmeyer flask as described previously (52). The growth of suspended populations was characterized by periodically determining the optical density at 546 nm. All determinations were performed with three separate samples.
Transposon mutagenesis.
B. cereus ATCC 14579 was transformed with pLTV1 (8) by electroporation as described previously (5), and transposon mutagenesis was also performed as described previously (11), with the following modifications. A stationary-phase culture of B. cereus(pLTV1), obtained by culturing the bacterium overnight at 28°C in LB broth containing tetracycline (50 μg/ml), erythromycin (1 μg/ml), and chloramphenicol (10 μg/ml), was diluted 1:800 in prewarmed (43°C) LB broth that contained erythromycin (1 μg/ml) and chloramphenicol (5 μg/ml). Following incubation at 43°C for 24 h with shaking at 200 rpm, the culture was again diluted and incubated as described above. Transposon mutants were subsequently selected on LB agar containing erythromycin (1 μg/ml) and chloramphenicol (5 μg/ml) and incubated at 37°C.
Identification of biofilm-impaired mutants.
A total of 3,500 mutants from five independent libraries were screened by inoculating bacteria into 200 μl of LB broth in Durham tubes (25 by 6.5 mm) and incubating the tubes statically at 25°C for 32 h. The tubes were visually inspected for the presence of biofilm-impaired mutants, as shown by diminished biofilm growth at the solid-liquid-air interface compared to the growth of wild-type B. cereus. Selected mutants were cultured in 25-ml glass beakers containing 15 ml of LB broth for 72 h at 25°C without agitation. The planktonic phase was removed by aspiration, and the biofilms were removed from the walls of the beakers by repeated rinsing with 15 ml of phosphate-buffered saline and were quantified relative to wild-type B. cereus biofilms by measuring the optical density at 600 nm. Chromosomal DNA flanking transposon insertion points was recovered by plasmid rescue (8). The DNA flanking Tn917-LTV1 insertions were sequenced with primer 917S (5′-CTCACAATAGAGAGATGTCACC-3′).
Quantitation of transcripts.
Biofilms of B. cereus wild-type and mutant strains were cultured in 25-ml glass beakers as described above. Total RNA was isolated from both biofilm and planktonic populations using the EZ RNA reagent (Bio Basic, Ontario, Canada), treated with RNase-free DNase I (Fermentas, St. Leon-Rot, Germany), and then reverse transcribed with a Quantitect reverse transcription kit (Qiagen, Hilden, Germany). Real-time PCR was carried out with quadruplicate 15-μl reaction mixtures containing 400 nM of each gene-specific sense and antisense oligonucleotide and a 10-fold dilution of cDNA (Table 1) using a Lightcycler 1 (Roche Diagnostics, Mannheim, Germany) and a QuantiTect SYBR green PCR kit (Qiagen). Template denaturation at 95°C for 15 min was followed by 45 cycles of denaturation at 94°C for 15 s, annealing at 60°C (57°C for purA) for 30 s, and extension at 72°C for 20 s. The PCR efficiency for each reaction was calculated with LinRegPCR (40), and the expression of each gene relative to that of 16S rRNA was quantified with REST 2005 (38).
TABLE 1.
Primers used for real-time PCR
| Gene | Amplicon size (bp) | Forward primer | Reverse primer |
|---|---|---|---|
| purA | 169 | CCCAATTGCTGGTGGTGTAAC | TCCATACTCGCGACCAACTTC |
| purC | 165 | TTTCGCAAGCTGTCGTGTAAG | CCAAGATCACGACGGAATACG |
| purL | 162 | AAGCACAGTTGTTACGCCAG | CATACGATATTACGCGCTGCC |
| 16S rRNA | 162 | TAGGTGGCAAGCGTTATCCG | GCATTTCACCGCTACACATGG |
Fluorescence and confocal microscopy.
Biofilms on glass wool were prepared for laser scanning confocal microscopy (LSCM) by staining with Syto 9 and propidium iodide (Live/Dead BacLight stain; catalog no. L7012; Molecular Probes) or 4′,6′-diamidino-2-phenylindole·2HCl (DAPI) (10 ng/ml). Bacteria on glass wool fibers were directly stained on a glass slide by adding a 1/1,000 dilution in 50 μl of stain in phosphate buffer (10 mM, pH 7.0). The Syto 9 stain conveys green fluorescence to intact cells, while propidium iodide is taken up by cells with impaired membranes, making them fluoresce in the red spectrum. LSCM was performed using an Olympus Fluoview FV300 LSCM system interfaced with an inverted microscope (Olympus IX70). Three detection channels were available: blue argon (488 nm), green helium neon (543 nm), and differential interference contrast.
Assay for extracellular nucleic acid.
Planktonic and biofilm populations were cultured in 100 ml of LB broth in 250-ml Erlenmeyer flasks with shaking at 28°C. Inoculation of flasks to an optical density at 546 nm of 0.005 with washed, exponentially growing planktonic populations was staggered to obtain populations that were different ages at the same time point. Planktonic populations were harvested from flasks with or without glass wool by centrifugation (12,000 × g, 15 min). Biofilm populations were dislodged from glass wool using glass beads as described previously (52) and were harvested by centrifugation. The pellets were washed twice in cold phosphate buffer (100 mM, pH 7.0) and resuspended in sterile water to an optical density at 546 nm of 5 in order to compensate for different initial cell densities. Culture supernatants obtained by centrifugation were filtered through 0.22-μm-pore-size filters (Millipore) to remove all cells, and each 2-ml sample was concentrated to 20 μl by vacuum centrifugation. Prior to electrophoresis, resuspended pellets and crude concentrates were treated with 2 U RNase-free DNase and its buffer, 10 U RNase ONE and its buffer (both obtained from Promega), or 2 μg proteinase K (Sigma) for 1 h at 37°C. Concentrated cells and supernatants were loaded into wells of an agarose gel (0.8% [wt/vol] agarose in Tris-borate-EDTA) and electrophoresed at 60 V for 60 min. Gels were stained using ethidium bromide and were imaged using a ChemiDoc XRS (Bio-Rad).
Southern blot analysis.
Southern blot analysis was performed using standard protocols. eDNA of biofilm and exponentially growing planktonic populations was obtained by electrophoresing washed cell suspensions through a low-melting-point agarose gel and extracting the DNA using GELase (Epicentre Biotechnologies). Biofilm and planktonic eDNA and chromosomal DNA were digested with EcoRI, resolved by electrophoresis, and transferred by vacuum blotting to Hybond-N+ (Amersham). Hybridization and detection were performed using standard protocols.
The presence of cell-associated DNA was investigated by exposing exponentially growing (2-h-old) planktonic populations to 50 μg/ml of the antibiotic actinomycin D (Fisher) for 30 min. The culturable counts for treated and untreated populations were determined by the droplet plate technique (29), and the log decline was calculated. Parallel populations were pretreated with 5 U DNase in 1× buffer for 30 min prior to exposure to actinomycin D.
RESULTS
Screening for biofilm-impaired mutants.
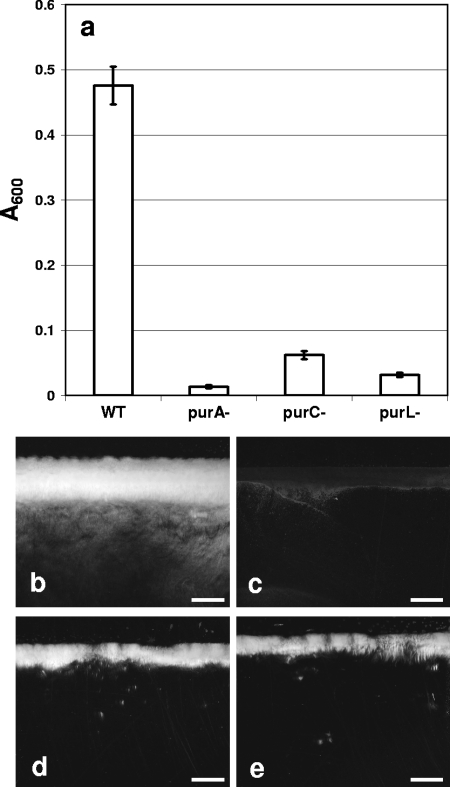
We screened a Tn917-LTV1 library for biofilm-impaired mutants using a semiquantitative, glass tube-based assay that differs from the standard approach (36). Preliminary trials in which biofilms of wild-type B. cereus were cultured by the standard approach in polystyrene and polyvinyl chloride 96-well microtiter plates yielded highly variable quantities of biofilm (data not shown). The biofilms of the wild type did not appear to adhere well to the plastic walls, leading to variable loss of biofilm during removal of the planktonic phase and the subsequent staining with crystal violet. A recent report of development of biofilms by strains of B. cereus supports our observations (57). We therefore decided to use a modified approach in which the biofilm formed at the solid-liquid-air interface is scored visually without any sample handling. As this initial screen is semiquantitative, mutants of interest were assayed using a 25-ml glass beaker and quantifying the biofilm biomass formed at the glass surface by removal of the liquid phase by aspiration, suspension of the biofilm, and spectrophotometry. A number of Tn917-LTV1 mutants of B. cereus ATCC 14579 were impaired in biofilm development on glass. The mutations of three biofilm-impaired mutants mapped to genes involved in purine biosynthesis, purA, purC, and purL encoding adenylosuccinate synthetase, phosphoribosylaminoimidazole succinocarboxamide synthetase, and phosphoribosylformylglycinamidine synthetase II, respectively (Fig. 1a). All three mutants formed much thinner biofilms than the wild type formed, as determined by stereomicroscopy (Fig. 1b to e).
FIG. 1.
Biofilm formation by B. cereus ATCC 14579 and selected transposon mutants. (a) Biofilm formation was quantified by dislodging the biomass from the walls of glass beakers following static incubation in LB broth at 25°C for 72 h. The error bars indicate the standard deviations of the means (P < 0.01, n = 5). WT, wild type. (b to e) Stereophotomicrographs of biofilms of the wild type (b) and purA (c), purC (d), and purL (e) mutants on glass slides at the air-liquid interface. Scale bars = 2 mm.
Expression of purine biosynthesis genes in biofilms.
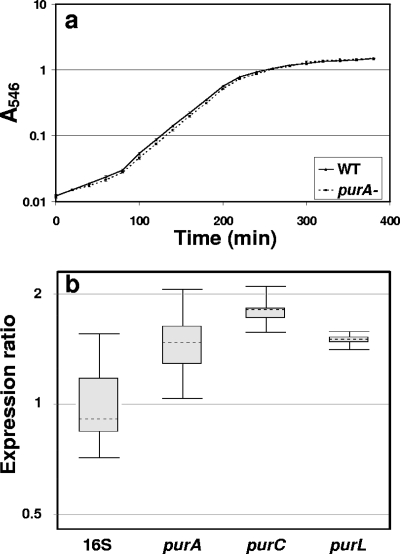
Purines are anabolites that are required for DNA and RNA synthesis (51), which means that an exogenous supply is needed when auxotrophs such as the pur mutants are cultured. The purA mutant grew in LB broth without supplemental nucleotides at a rate similar to the rate of the wild type and to the same density as the wild type (Fig. 2a). Supplementation of minimal medium with the AMP precursor adenosine restored the growth of the purA mutant to wild-type rates (data not shown). No information on the nucleotide contents of the LB broth components (yeast extract or tryptone) is available from the various suppliers. Yeast extract is an autolysate of Saccharomyces cerevisiae and contains ribonucleotides (3, 45), the most likely source of purines that supply the needs of these auxotrophic mutants. Agarose gel electrophoresis of 100-fold-concentrated uninoculated LB broth also showed the presence of nucleic acid molecules smaller than 500 bp (data not shown). Collectively, these results indicated that the biofilm formation deficiency of the pur mutants was not due to a planktonic growth deficiency caused by a purine requirement. The exogenous nucleotide source in LB broth was apparently sufficient for planktonic growth but was not sufficient to support biofilm development. The three pur genes were all upregulated approximately 1.5-fold in the biofilm compared with planktonic populations, indicating that there was an elevated requirement for purines in biofilm-associated cells (Fig. 2b). The purine biosynthesis genes were therefore expressed in the presence of the exogenous nucleotide source present in LB broth, prompting us to investigate the distribution and possible sources of nucleic acids in biofilms of B. cereus.
FIG. 2.
(a) Growth of the wild-type and purA mutant strains in LB broth at 28°C. (b) Box plot indicating the ratio of the pur gene transcripts in a biofilm to the pur gene transcripts in planktonic populations quantified by real-time PCR using the 16S rRNA gene as an endogenous control. WT, wild type. 16S, 16S rRNA gene.
eDNA surrounding surface-associated cells.
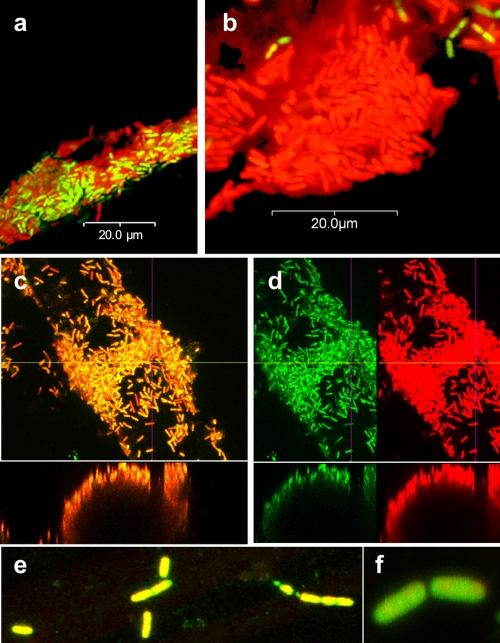
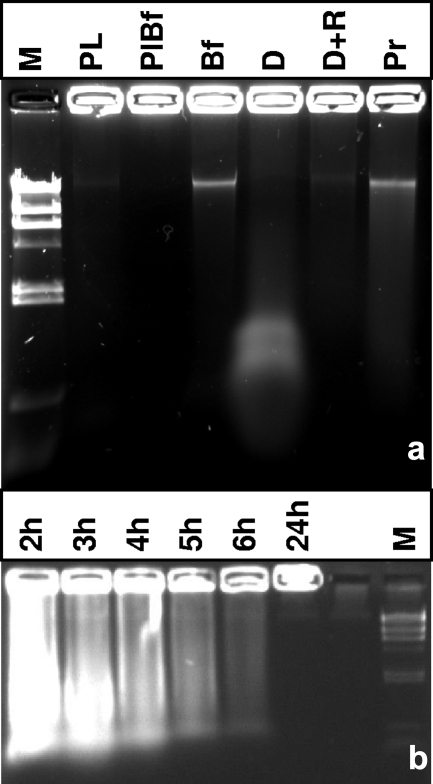
Biofilms of B. cereus on glass wool were stained with the nucleic acid stains Syto 9 and propidium iodide (BacLight). LSCM indicated that there was a mixture of green and red cells on the glass wool (Fig. 3a). Red cells are widely believed to be dead due to perfusion of propidium iodide across nonpolarized cell membranes of nonrespiring cells. When viewed in single-channel mode, the same cells that appeared to be red also appeared to be green in the green channel, indicating that there were green centers surrounded by intense red fluorescence (Fig. 3c and d). This indicated that nucleic acid was associated with the cell exterior, where propidium iodide had free access. Suspensions of biofilm cells placed in the wells of an agarose gel released ethidium bromide-fluorescing material with low electrophoretic mobility (Fig. 4a, lane Bf). Treatment of biofilm populations with DNase prior to electrophoresis eliminated appearance of this band and resulted in a substance with much greater electrophoretic mobility. The complete removal of this substance following DNase and RNase treatment indicated that DNA complexed with RNA was present (Fig. 4a, lanes D and D+R). Protease treatment indicated that some amount of protein contributed to the observed complex. Stationary-phase planktonic populations released a small amount of DNA, both from cells as determined by electrophoresis (Fig. 4a and b) and into the culture supernatant (data not shown). The apparent release of nucleic acid from biofilm cells was therefore not likely release from the cytosol through gating of the cell membrane in the electrical field; instead, the origin of the nucleic acid was extracellular.
FIG. 3.
LSCM of a 24-h BacLight-stained B. cereus biofilm cultured on glass wool at 28°C for 24 h. (a and b) Some sections of the biofilm contained cells that appeared to be both red and green (a), while the majority of the cells appeared to be red and to be surrounded by a less dense red area (propidium iodide fluorescence) (b). (c and d) Cells that appeared to be red (c) did contain green-fluorescing centers (Syto 9) when they were viewed in separate channels (d). Images for green and red channels set at identical detection values are shown separately, and images of x-z cross sections are shown below the large images. (e and f) The purA mutant cells formed very sparse biofilms and did not have an apparent red exterior.
FIG. 4.
Biofilms and exponential-phase planktonic cells of B. cereus contain eDNA. (a) A biofilm population (lane Bf), a planktonic population (lane Pl), and a planktonic stationary-phase population from a biofilm culture flask (lane PlBf) were placed in wells of a 0.8% agarose gel, along with biofilm treated with DNase (lane D), with RNase plus DNase (lane D+R), and with proteinase K (lane P). Lane M contained HindIII-digested λ DNA. (b) Ethidium bromide-stained agarose gel containing planktonic populations from cultures that were different ages. Populations were harvested by centrifugation after they were cultured in LB broth for 2, 3, 4, 5, 6, and 24 h and normalized for cell density, and cell suspensions were loaded directly into the wells for electrophoresis. Lane M contained HindIII-digested λ DNA as a size marker.
eDNA as a conditioning film.
Biofilms on glass wool were mostly surrounded by thin layers fluorescing red with propidium iodide (Fig. 3b) and blue when they were stained with DAPI (not shown). Some attached cells appeared to be green without an associated underlying nucleic acid film. Staining with the protein-specific stain Sypro Red indicated that protein was present in the film (not shown). This implied that DNA and some RNA and protein were components of the EPS.
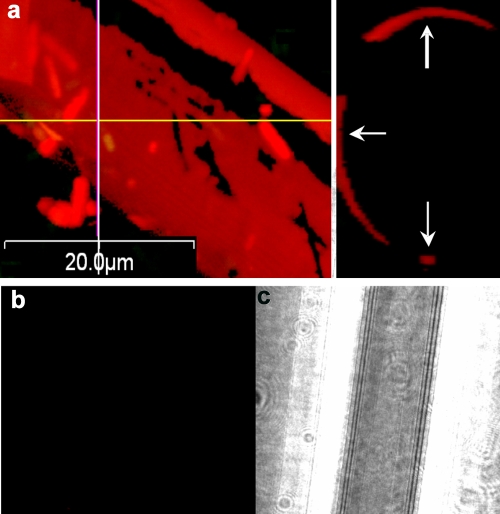
Glass wool exposed to an exponential planktonic population for 30 min and subsequently stained with propidium iodide fluoresced red, indicating that it was coated with nucleic acid (Fig. 5a and Table 2). Exposure of glass wool to a genomic DNA extract and separately to cell lysate prepared by ultrasonication also led to coating with nucleic acid (Table 2). Staining of culture-exposed glass wool with the dsDNA stain Pico Green yielded green fluorescence, supporting the hypothesis that dsDNA was present on the glass surface (not shown). Pico Green staining of biofilms on glass wool did not reveal the presence of much biofilm, but free-floating cells and debris could be observed after staining, possibly due to dislodgement by dimethyl sulfoxide, the Pico Green solvent. Glass wool exposed to sterile LB broth was not coated with DNA (Fig. 5b and c), indicating that the nucleic acid in LB broth did not adhere to the glass surface and therefore was not the source of the conditioning film. Glass wool exposed to exponential- or stationary-phase cultures of the purA mutant did not fluoresce (not shown), indicating that the mutant did not serve as a source of the DNA attached to the glass. Addition of genomic DNA extract and cell lysates of the purA mutant did lead to coating of the glass surface with nucleic acid. The exponentially growing populations used throughout this study were initiated by inoculation of washed cells from an exponentially growing culture, eliminating the possibility that DNA that originated from previously lysed cells was transferred with the inoculum. The DNA observed on the glass surface, therefore, originated from the exponentially growing population and not from the LB broth or from carryover from a previous culture. Microscopic investigation did not reveal any obviously broken cells.
FIG. 5.
(a) Glass wool exposed for 30 min to an exponentially growing B. cereus culture before removal and staining with propidium iodide, viewed by LSCM. The left panel is a composite x-y image, and the right panel is a y-z image of the in silico preparation at the position of the line. (b and c) Glass wool exposed to sterile LB broth for 2 h and then stained with propidium iodide and viewed by LSCM in the red channel (b) and by differential interference contrast microscopy (c).
TABLE 2.
Detection of nucleic acid on glass wool fibers by fluorescence microscopy using propidium iodide and DAPI
| Treatment | Presence of nucleic acid
|
||
|---|---|---|---|
| LB only | B. cereus in LB broth | purA mutant in LB broth | |
| 2-h culture | − | + | − |
| 24-h culture | − | + | − |
| Lysed exponential-phase (2 h) cells | NDa | + | + |
| Genomic DNA | ND | + | + |
ND, not determined.
eDNA is cell associated.
The glass-associated DNA appeared to be derived from growing cells. Exposure of washed planktonic cell suspensions from the early through mid-exponential, transition, and stationary phases to an electric field in an agarose gel released nucleic acids into the matrix (Fig. 4b). Exponentially growing wild-type cells harvested after 2 and 3 h of incubation yielded the largest amounts of nucleic acid, while cells entering stationary phase yielded very little nucleic acid. The wells were loaded with similar amounts of biomass, standardized by spectrophotometric determination, from cultures inoculated at 1-h intervals to obtain samples for electrophoresis at the same time point. Late-stationary-phase populations released only a very small amount of nucleic acid with high electrophoretic mobility into the gel, suggesting that a proportion of these cells had lysed by this point (24 h) (Fig. 4b).
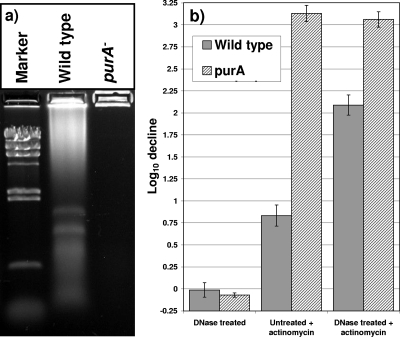
In contrast to the wild type, exponentially growing purA mutant populations did not release any detectable nucleic acid into the gel matrix (Fig. 6a). Furthermore, LSCM of attached purA mutant cells did not reveal detectable levels of red fluorescence around the cells (Fig. 3e and f). Very few attached cells were found, supporting the quantitative data. This apparent lack of nucleic acid release by the mutant supported the notion that exponential-phase cells exposed to a 60-V field in Tris-borate-EDTA did not release DNA from their cytosol. Rather, the origin of the wild-type-derived nucleic acid was extracellular. Exponentially growing populations lost culturability after exposure to the DNA-interacting antibiotic actinomycin D (46). DNase treatment did not affect the culturability of the untreated cells (Fig. 6b). Yet DNase-treated populations were 10 times more susceptible to actinomycin than the untreated cells (Fig. 6b). Untreated purA mutant populations were highly susceptible to actinomycin D, and DNase treatment did not further increase the susceptibility (Fig. 6b). This result indicated that the cell-associated DNA in wild-type cells acted as a layer that protected against the action of actinomycin D.
FIG. 6.
(a) The purA mutant does not appear to be decorated with eDNA like the wild type. The presence of eDNA was determined electrophoretically by placing 20-μl portions of washed exponential-phase suspensions (optical density at 546 nm, 5.0) of wild-type and purA mutant populations in a 0.8% agarose gel, which was resolved by electrophoresis at 60 V for 60 min. The marker was HindIII-digested λ DNA. (b) Susceptibility of exponential-phase wild-type and purA mutant populations to actinomycin D determined with and without prior treatment with DNase. Mid-exponential-phase planktonic cultures were washed and treated either with 5 U/ml RNase-free DNase or with water for 30 min prior to exposure to 50 μg·ml−1 actinomycin D.
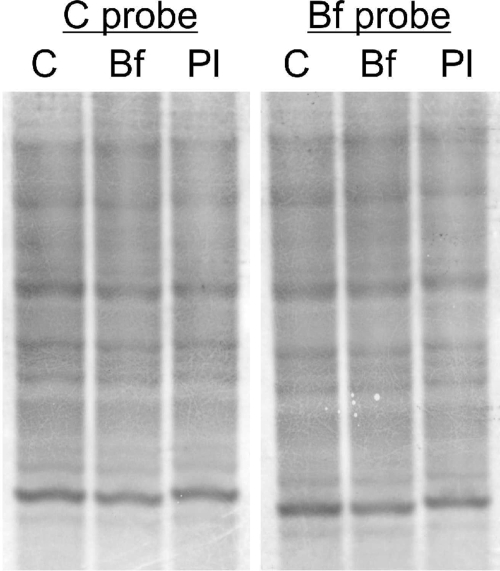
The nature of the eDNA of biofilm and exponential-phase planktonic populations was investigated by hybridizing this DNA to chromosomal DNA using Southern blotting. EcoRI-digested eDNA and chromosomal DNA were electrophoresed, blotted, and probed with digoxigenin-labeled chromosomal DNA and biofilm eDNA. Both types of eDNA produced the same pattern as chromosomal DNA (Fig. 7), indicating that both biofilm eDNA and planktonic eDNA are similar to chromosomal DNA.
FIG. 7.
Southern hybridization of biofilm eDNA (lane Bf), exponential-phase planktonic eDNA (lane Pl), and chromosomal DNA (lane C). Biofilm and planktonic eDNA were obtained by electrophoresis from 24-h biofilm and 2-h planktonic populations, followed by extraction from the gel. DNA was digested with EcoRI, and all three extracts were hybridized to digoxigenin-labeled chromosomal DNA (C probe) or biofilm eDNA (Bf probe).
DISCUSSION
Biofilms of several bacterial species, including some gram-positive cocci, such as S. aureus, S. epidermidis, and S. pneumoniae, have previously been shown to contain eDNA (23, 33, 39). Here we report eDNA in biofilms of B. cereus, the first example of a gram-positive rod whose biofilms contain eDNA. Growing cultures of B. cereus convey a DNA-containing conditioning film to the glass surface, and the majority of cells in the biofilm appear to be encased in a DNA-containing matrix. The microscopic and electrophoretic analyses described here show that DNA is deposited as a conditioning film on glass in the presence of exponential-phase populations of B. cereus. Similarly, the majority of cells in a growing biofilm are encased in eDNA-containing EPS. The eDNA is therefore more homogeneously distributed, as it is in biofilms of P. aeruginosa (55) and S. pneumoniae (33), in contrast to biofilms of H. influenzae and the gammaproteobacterium F8, where eDNA occurs as filamentous strands (4, 24).
The origin of the eDNA occurring in the EPS matrix of biofilms is not clear. Three hypotheses for this origin that are supported by experimental evidence from P. aeruginosa are lysis of a subpopulation of the biofilm (2), release through membrane vesicles from live cells (41, 44), and active secretion (20). Neisseria gonorrhoeae excretes DNA using a type IV secretion system (16). B. subtilis is reported to excrete DNA during late log phase (30). Yet DNA appears to be released through cell lysis in some species, including Acinetobacter calcoaceticus (37) and S. pneumoniae (48).
Biofilm eDNA was found to be similar to chromosomal DNA by Southern blotting. Yet B. cereus biofilm eDNA did not appear to originate primarily through cell lysis. While Bacillus cells are widely known to lyse during stationary phase in rich medium, most of the eDNA of planktonic cells was detected during the exponential phase in axenic cultures washed between transfers. In B. subtilis, transition-phase excretion of DNA coincides with natural competence (30). It seems unlikely that the multigene-encoded process of DNA uptake would have evolved to be dependent on DNA released at random from other cells. A correlation between DNA release and competence has been suggested for various bacterial species, including S. pneumoniae (48), B. subtilis (30), and N. gonorrhoeae (42). Planktonic populations of B. subtilis and P. aeruginosa contain the most eDNA per cell, primarily during late exponential phase and not during stationary phase (2, 30), suggesting that lysed cells are not the primary source of eDNA. Colonies of various strains of P. aeruginosa have been shown to produce extracellular slime containing DNA (34). Admittedly, B. cereus is not known to take up DNA through natural competence, nor are there any reported indications in the genome sequence of a DNA export system. No obviously lysed cells could be found, and all biofilm cells showed only green fluorescence in the cytosol upon staining with BacLight. Protein gel electrophoresis of concentrated culture supernatant did not indicate that there were detectable levels of cytosolic proteins, arguing against lysis. The bulk of eDNA detected was not free-floating in the culture broth but was cell associated. The purA mutant strain, while growing like the wild type in LB broth, did not show any cell-associated or free DNA in planktonic cultures.
B. cereus is a soil saprophyte that forms multicellular structures that appear to be bundles of chains when it is growing in its natural soil environment (54). Cell surface-associated eDNA would provide a selective advantage to B. cereus in its natural environment in the soil. The maintenance of stable populations of soil bacteria is challenged, inter alia, by a range of antibacterial agents produced by competing microflora, such as the actinomycetes (10). The majority of well-known antibiotics target prokaryote-specific aspects of transcription, translation, or cell wall synthesis. Yet certain antibiotics produced by soil streptomycetes, such as Streptomyces antibioticus (19), appear to target nucleic acids directly (15). Actinomycin D binds with the DNA double helix (46), and DNA repair systems in Escherichia coli reportedly protect cells from the effect of actinomycin (21), indicating that the genome is a target. Our data indicate that cell surface-associated eDNA acted as a protective layer, protecting B. cereus, while DNase-pretreated cells and purA mutant cells were more susceptible to actinomycin D.
In conclusion, the data presented here indicate eDNA as an integral component of the EPS of the biofilms of B. cereus ATCC 14579. While they grew like the wild type in planktonic phase, mutants deficient in the purine biosynthesis genes purA, purC, and purL were biofilm impaired. DNA in the conditioning film was derived from exponentially growing populations of the wild type, but not the purA mutant, indicating that adenylosuccinate synthetase activity is required for biofilm formation.
Acknowledgments
We thank Anne Moir for providing pLTV1 and W. F. Burkholder for helpful suggestions.
This research was supported by the South Dakota Agricultural Experiment Station, by National Science Foundation/EPSCoR grant EPS-0091948, and by the State of South Dakota (V.S.B.), as well as by National Research Foundation of South Africa grant 2046811 (V.S.B. and J.T.). We acknowledge use of the SDSU-FGCF, which is supported in part by NSF/EPSCoR grant 0091948 and by the State of South Dakota. J.M.P. was supported by a National Research Foundation of South Africa scholarship.
Footnotes
Published ahead of print on 27 February 2009.
Journal series publication 3626 from the South Dakota Agricultural Experiment Station.
REFERENCES
- 1.Aguilar, C., H. Vlamakis, R. Losick, and R. Kolter. 2007. Thinking about Bacillus subtilis as a multicellular organism. Curr. Opin. Microbiol. 10:638-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114-1128. [DOI] [PubMed] [Google Scholar]
- 3.Belem, M. A. F., B. F. Gibbs, and B. H. Lee. 1997. Enzymatic production of ribonucleotides from autolysates of Kluyveromyces marxianus grown on whey. J. Food Sci. 62:851-854+857. [Google Scholar]
- 4.Böckelmann, U., A. Janke, R. Kuhn, T. R. Neu, J. Wecke, J. R. Lawrence, and U. Szewzyk. 2006. Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiol. Lett. 262:31-38. [DOI] [PubMed] [Google Scholar]
- 5.Bone, E. J., and D. J. Ellar. 1989. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol. Lett. 49:171-177. [DOI] [PubMed] [Google Scholar]
- 6.Branda, S. S., F. Chu, D. B. Kearns, R. Losick, and R. Kolter. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59:1229-1238. [DOI] [PubMed] [Google Scholar]
- 7.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camilli, A., D. A. Portnoy, and P. Youngman. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J. Bacteriol. 172:3738-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chai, Y., F. Chu, R. Kolter, and R. Losick. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 67:254-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clardy, J., M. A. Fischbach, and C. T. Walsh. 2006. New antibiotics from bacterial natural products. Nat. Biotechnol. 24:1541-1550. [DOI] [PubMed] [Google Scholar]
- 11.Clements, M. O., and A. Moir. 1998. Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J. Bacteriol. 180:6729-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costerton, J. W., G. G. Geesey, and K. J. Cheng. 1978. How bacteria stick. Sci. Am. 238:86-95. [DOI] [PubMed] [Google Scholar]
- 13.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, D. G., and G. G. Geesey. 1995. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 61:860-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeMarini, D. M., K. H. Brock, C. L. Doerr, and M. M. Moore. 1987. Mutagenicity of actinomycin D in mammalian cells due to clastogenic effects. Mutat. Res. 192:151-155. [DOI] [PubMed] [Google Scholar]
- 16.Dillard, J. P., and H. S. Seifert. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 41:263-277. [DOI] [PubMed] [Google Scholar]
- 17.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flemming, H. C., T. R. Neu, and D. J. Wozniak. 2007. The EPS matrix: the “house of biofilm cells.” J. Bacteriol. 189:7945-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goss, W. A., E. Katz, and S. A. Waksman. 1956. Changes in the composition of an actinomycin complex during growth of a Streptomyces culture. Proc. Natl. Acad. Sci. USA 42:10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hara, T., and S. Ueda. 1981. A study on the mechanism of DNA excretion from P. aeruginosa KYU-1. Effect of mitomycin C on extracellular DNA production. Agric. Biol. Chem. 45:2457-2461. [Google Scholar]
- 21.Holmalahti, J., H. Santa, H. Laatsch, and A. von Wright. 1997. Comparison of the lethal effects of different actinomycins on a repair-deficient strain of Escherichia coli. Mutat. Res. 391:33-38. [DOI] [PubMed] [Google Scholar]
- 22.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 23.Izano, E. A., M. A. Amarante, W. B. Kher, and J. B. Kaplan. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurcisek, J. A., and L. O. Bakaletz. 2007. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol. 189:3868-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearns, D. B., F. Chu, S. S. Branda, R. Kolter, and R. Losick. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55:739-749. [DOI] [PubMed] [Google Scholar]
- 26.Lazazzera, B. A. 2005. Lessons from DNA microarray analysis: the gene expression profile of biofilms. Curr. Opin. Microbiol. 8:222-227. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, K. 2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322:107-131. [DOI] [PubMed] [Google Scholar]
- 28.Lindsay, D., V. S. Brözel, and A. Von Holy. 2005. Spore formation in Bacillus subtilis biofilms. J. Food Prot. 68:860-865. [DOI] [PubMed] [Google Scholar]
- 29.Lindsay, D., and A. Von Holy. 1999. Different responses of planktonic and attached Bacillus subtilis and Pseudomonas fluorescens to sanitizer treatment. J. Food Prot. 62:368-379. [DOI] [PubMed] [Google Scholar]
- 30.Lorenz, M. G., D. Gerjets, and W. Wackernagel. 1991. Release of transforming plasmid and chromosomal DNA from two cultured soil bacteria. Arch. Microbiol. 156:319-326. [DOI] [PubMed] [Google Scholar]
- 31.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 32.Mah, T. F. C., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 33.Moscoso, M., E. Garcia, and R. Lopez. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 188:7785-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murakawa, T. 1973. Slime production by Pseudomonas aeruginosa. IV. Chemical analysis of two varieties of slime produced by Pseudomonas aeruginosa. Jpn. J. Microbiol. 17:513-520. [DOI] [PubMed] [Google Scholar]
- 35.Oosthuizen, M. C., B. Steyn, J. Theron, P. Cosette, D. Lindsay, A. Von Holy, and V. S. Brözel. 2002. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl. Environ. Microbiol. 68:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 37.Palmen, R., and K. J. Hellingwerf. 1995. Acinetobacter calcoaceticus liberates chromosomal DNA during induction of competence by cell lysis. Curr. Microbiol. 30:7-10. [DOI] [PubMed] [Google Scholar]
- 38.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin, Z., Y. Ou, L. Yang, Y. Zhu, T. Tolker-Nielsen, S. Molin, and D. Qu. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083-2092. [DOI] [PubMed] [Google Scholar]
- 40.Ramakers, C., J. M. Ruijter, R. H. Deprez, and A. F. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62-66. [DOI] [PubMed] [Google Scholar]
- 41.Renelli, M., V. Matias, R. Y. Lo, and T. J. Beveridge. 2004. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150:2161-2169. [DOI] [PubMed] [Google Scholar]
- 42.Salgado-Pabon, W., S. Jain, N. Turner, C. van der Does, and J. P. Dillard. 2007. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol. Microbiol. 66:930-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schooling, S. R., and T. J. Beveridge. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188:5945-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sezonov, G., D. Joseleau-Petit, and R. D'Ari. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sobell, H. M. 1973. The stereochemistry of actinomycin binding to DNA and its implications in molecular biology. Prog. Nucleic Acid Res. Mol. Biol. 13:153-190. [DOI] [PubMed] [Google Scholar]
- 47.Steinberger, R. E., and P. A. Holden. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 71:5404-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinmoen, H., E. Knutsen, and L. S. Havarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 99:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutherland, I. W. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3-9. [DOI] [PubMed] [Google Scholar]
- 50.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 51.Switzer, R. L., H. Zalkin, and H. H. Saxild. 2002. Purine, pyrimidine, and pyridine nucleotide metabolism, p. 255-269. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its relatives: from genes to cells. ASM Press, Washington, DC.
- 52.Vilain, S., and V. S. Brozel. 2006. Multivariate approach to comparing whole-cell proteomes of Bacillus cereus indicates a biofilm-specific proteome. J. Proteome Res. 5:1924-1930. [DOI] [PubMed] [Google Scholar]
- 53.Vilain, S., P. Cosette, I. Zimmerlin, J. P. Dupont, G. A. Junter, and T. Jouenne. 2004. Biofilm proteome: homogeneity or versatility? J. Proteome Res. 3:132-136. [DOI] [PubMed] [Google Scholar]
- 54.Vilain, S., Y. Luo, M. B. Hildreth, and V. S. Brözel. 2006. Analysis of the life cycle of the soil saprophyte Bacillus cereus in liquid soil extract and in soil. Appl. Environ. Microbiol. 72:4970-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 57.Wijman, J. G., P. P. de Leeuw, R. Moezelaar, M. H. Zwietering, and T. Abee. 2007. Air-liquid interface biofilms of Bacillus cereus: formation, sporulation, and dispersion. Appl. Environ. Microbiol. 73:1481-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]