Abstract
Small-insert and large-insert metagenomic libraries were constructed from glacial ice of the Northern Schneeferner, which is located on the Zugspitzplatt in Germany. Subsequently, these libraries were screened for the presence of DNA polymerase-encoding genes by complementation of an Escherichia coli polA mutant. Nine novel genes encoding complete DNA polymerase I proteins or domains typical of these proteins were recovered.
DNA polymerases are essential for DNA replication and DNA repair. Based on sequence similarities and phylogenetic relationships, DNA polymerases are grouped into six different families (A, B, C, D, X, and Y) (17). In this study, we used a DNA polymerase I (polA) mutant of Escherichia coli as a host for the screening of metagenomic libraries. PolA belongs to family A and contains three different domains: a 5′-3′ exonuclease domain at the N terminus, a central proofreading 3′-5′ exonuclease domain, and a polymerase domain at the C terminus of the enzyme (11). These polymerases are employed as tools in molecular biology, including probe labeling, DNA sequencing, and mutagenic PCR (13). To improve their suitability for such applications, various family A DNA polymerases have been modified; e.g., the Klenow fragment of E. coli DNA polymerase I has been redesigned by the removal of the 5′-3′ exonuclease domain (12). Nevertheless, expanding the known DNA polymerase sequence space and discovery of polymerases with novel properties are required for the development of novel or improved molecular methods and tools (13, 20).
Metagenomics based on direct isolation of DNA from environmental samples, generation of metagenomic libraries from the isolated DNA, and function-based screening of the constructed libraries has led to identification and characterization of a variety of novel biocatalysts, such as lipases, amylases, amidases, nitrilases, and oxidoreductases (for reviews, see references 6, 7, and 10). In particular, the use of host strains or mutants of host strains that require heterologous complementation for growth under selective conditions has proven to be an efficient strategy to screen complex metagenomic libraries. This approach has been applied to, e.g., the isolation of genes encoding Na+/H+ antiporters (14), antibiotic resistance (18), or enzymes involved in poly-3-hydroxybutyrate metabolism (21).
In this study, we employed the last-named strategy to recover functional genes encoding DNA polymerases. To our knowledge, this is the first report of identification of polymerases or other DNA-modifying enzymes by function-driven screening of metagenomes. For this purpose, we constructed small-insert and large-insert metagenomic libraries from DNA isolated from glacial ice. The employment of glacial ice samples for metagenomic library construction has not been reported by other researchers. The screening for the targeted genes was based on complementation of a cold-sensitive lethal mutation in the polA gene of E. coli (16).
Sample collection and construction of metagenomic glacier ice libraries.
Glacier ice samples were collected in June 2005 at the Northern Schneeferner (47°25′N, 10°59′E), which is located on the Zugspitzplatt in Germany. In order to avoid contamination by surface melt water, the first 30 cm of glacier ice was removed and discarded. Ice up to a depth of approximately 0.5 m was collected and transported frozen to the laboratory. To extract DNA from such a low-biomass environment, the ice was melted at 4°C, and portions of 1 to 2 liters were filtered using a sterile filter unit with a removable cellulose acetate membrane (Whatman, Dassel, Germany) (pore size, 0.2 μm). The cell-containing membrane filters were used as starting material for DNA isolation. Several DNA isolation methods and kits were tested. The performance of a NucleoSpin tissue kit (Macherey-Nagel, Düren, Germany) was best with respect to yield and purity of the isolated DNA (data not shown). Approximately 5 μg of DNA per kg of melted glacier ice was recovered. Since the DNA yield of glacial ice is much lower than that of high-biomass environments such as soils (6), starting material for the construction of small-insert metagenomic libraries was generated by multiple displacement amplification (MDA) of glacial DNA. For this purpose, a GenomiPhi V2 DNA amplification kit (GE Healthcare, Munich, Germany) was used as recommended by the manufacturer. To improve cloning efficiency and to avoid abnormal insert size distribution of the amplified DNA, hyperbranched structures introduced by MDA were resolved and the DNA was inserted into pCR-XL-TOPO (Invitrogen, Karlsruhe, Germany) as suggested by Zhang et al. (22). In this way, a small-insert library, which comprised 230,000 E. coli DH5α clones with an average insert size of 4 kb, was constructed. The proportion of plasmids containing inserts was approximately 97%. A large-insert fosmid library was constructed by using the fosmid pCC1FOS as vector and a CopyControl fosmid library production kit (Epicenter Biotechnologies, Madison, WI) as recommended by the manufacturer. For this purpose, the purified DNA (2 μg) was directly inserted into the fosmid vector without prior amplification. The fosmid library consisted of approximately 4,000 fosmids with an average insert size of 36 kb. In summary, the two constructed metagenomic libraries harbored approximately 1.07 Gb of cloned glacial DNA.
Screening for genes encoding DNA polymerase I.
The function-based detection of plasmids and fosmids harboring polymerase-encoding genes was based on complementation of the cold-sensitive E. coli mutant CSH26 fcsA29 [F− ara (lac-pro) thi fcsA29 met::Tn5] (16). This strain carries a temperature-sensitive lethal mutation in the 5′-3′ exonuclease domain of DNA polymerase I; this mutation causes filamentation of the cells with dispersed nuclei. The mutation is lethal at temperatures below 20°C (16). The screening was initiated by transfer of the glacial DNA-containing recombinant plasmids and fosmids into the mutant. Subsequently, the resulting E. coli CSH26 fcsA29 clones were plated onto Luria-Bertani agar (3) containing 50 μg of kanamycin/ml (plasmids) or 12.5 μg of chloramphenicol/ml (fosmids) and incubated at 18°C. Thus, only recombinant E. coli strains harboring a gene conferring polymerase activity could grow under the employed conditions. Positive clones with a colony diameter of >3 mm were visible after 48 to 72 h of incubation. The E. coli CSH26 fcsA29 negative control harboring the cloning vector without an insert showed no growth under the employed conditions. To identify one positive clone during the initial screening, approximately 1,000 clones (plasmid library) and 200 clones (fosmid library) needed to be tested. Seventeen plasmid-containing positive clones and one fosmid-containing clone were randomly chosen for further analysis. In order to confirm that complementation of the cold-sensitive mutation was encoded by the inserts of the vectors, the fosmid and plasmids were isolated from the positive clones, retransformed into E. coli CSH26 fcsA29, and screened again under selective conditions. The fosmid (fCS1) and 15 of the plasmids (pCS1 to pCS15) conferred stable phenotypes at 18°C and were subjected to further analyses.
Identification and characterization of polymerase-encoding genes.
The inserts of all recombinant plasmids and the fosmid were sequenced by the Göttingen Genomics Laboratory (Göttingen, Germany). All other manipulations of DNA, PCR, and transformation of vectors into E. coli were done according to routine procedures (3) unless otherwise specified. The sequences were analyzed with the gap4 program of the Staden software package (4) and were compared to entries in the database of the National Center for Biotechnology Information (1). Analysis of the fosmid fCS1 insert (32 kb) revealed the presence of 26 open reading frames (ORFs), including one putative polA gene (see GenBank accession number in nucleotide sequence accession number paragraph below). Most of the predicted ORFs (15) were similar to genes from Rhodoferax ferrireducens or Polaromonas sp., both of which belong to the Comamonadaceae (Betaproteobacteria). Members of this family are known to be abundant in cold environments (8). Partial sequencing of the flanking regions of the inserts from plasmids pCS9 to pCS15 revealed that they were identical to the inserts of pCS1, pCS4, pCS7, and pCS8. Therefore, plasmids pCS9 to pCS15 were not studied further. The high number of duplicates was probably a result of the amplification of the DNA by MDA. The insert sizes of plasmids pCS1 to pCS8 ranged from 3.5 to 15 kb (Table 1). Sequencing of the complete plasmid inserts by primer walking was possible for pCS1, pCS5, and pCS6 but not for the five remaining plasmids. This result was caused by the presence of repeat structures. The formation of these chimeric artifacts is a known drawback of MDA (22). To circumvent this problem, shotgun libraries of the plasmids with insert sizes of approximately 1 kb were constructed using pCR2.1-TOPO (Invitrogen, Karlsruhe, Germany) as the vector and were sequenced.
TABLE 1.
Sequence similarities of the presumptive DNA polymerase activity-conferring gene products encoded by pCS1 to pCS8 and by fCS1 to sequences of gene products from other organismsa
| Plasmid or fosmid | Insert size (kb) | No. of. amino acids of polymerase gene product | Detected domain(s)b | Closest similar protein, no. of amino acids (accession no. of similar protein) | Organism | Region of similar amino acids (% identity) | E value |
|---|---|---|---|---|---|---|---|
| pCS1 | 3.5 | 557 | 53 exo, 35 exo | Putative DNA polymerase I, 941 (ZP_01718371) | Algoriphagus sp. strain PR1 | 8-521 (52) | 1e-155 |
| pCS2 | 15.0 | 944 | 53 exo, 35 exo, DNA_polA | DNA polymerase I, 955 (ZP_01689558) | Microscilla marina ATCC 23134 | 1-955 (58) | 0 |
| pCS3 | 6.3 | 803 | 53 exo, DNA_polA | DNA polymerase I, 833 (YP_144320) | Thermus thermophilus HB8 | 7-827 (35) | 5e-124 |
| pCS4 | 4.8 | 282 | 53 exo | Exodeoxyribonuclease, 276 (AAX12058) | Bacteriophage T5 | 2-257 (43) | 8e-52 |
| pCS5 | 6.4 | 927 | 53 exo, 35 exo, DNA_polA | DNA polymerase I, 923 (YP_001845275) | Acinetobacter baumannii ACICU | 1-923 (65) | 0 |
| pCS6 | 4.6 | 942 | 53 exo, 35 exo, DNA_polA | DNA polymerase I, 937 (ZP_01884419) | Pedobacter sp. strain BAL39 | 2-937 (57) | 0 |
| pCS7 | 6.2 | 646 | 53 exo, 35 exo, DNA_polA | DNA polymerase I, 920 (AAG43148) | Rhodococcus erythropolis | 1-647 (82) | 0 |
| pCS8 | 9.0 | 927 | 53 exo, 35 exo, DNA_polA | DNA polymerase I, 923 (YP_001845275) | Acinetobacter baumannii ACICU | 1-923 (66) | 0 |
| fCS1 | 32.0 | 962 | 53 exo, 35 exo, DNA_polA | Putative DNA polymerase I, 941 (ZP_01718371) | Algoriphagus sp. strain PR1 | 8-941 (57) | 0 |
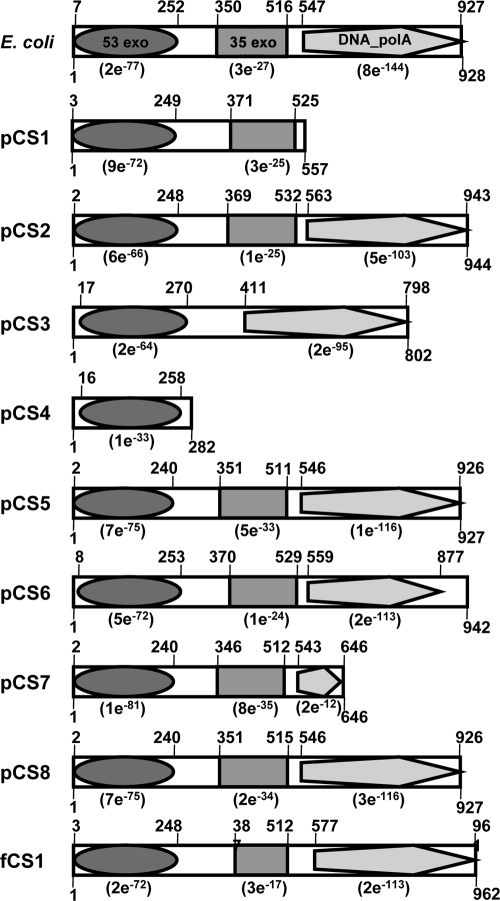
Sequence analyses of plasmids pCS1 to pCS8 and fosmid fCS1 revealed that all inserts contained ORFs that exhibited similarities to known PolA-encoding genes (Table 1; see also GenBank nucleotide sequence accession number paragraph below). Four of the plasmids (pCS2, pCS5, pCS6, and pCS8) and the fosmid (fCS1) contained a putative polA gene that encodes all three domains typical of DNA polymerase I (Fig. 1 and Table 1). The numbers of amino acids deduced from analysis of the corresponding proteins (927 to 962 amino acids) are similar to that of DNA polymerase I from E. coli (928 amino acids), which is the prototype for this type of enzyme (19). In addition, the pCS7 plasmid contained an almost complete version of the polA gene, lacking part of the C-terminal polymerase domain (Fig. 1). The amino acid sequence of the putative polA gene product encoded by pCS3 is slightly shorter (803 amino acids) than that of E. coli. The central region of the deduced enzyme showed no significant similarities to central 3′-5′ exonuclease domains of other DNA polymerases.
FIG. 1.
Domain structures of the DNA polymerase activity-conferring gene products of pCS1 to pCS8 and fCS1. The localizations and similarities of the domains were determined by searches of the Conserved Domain Database (15). The domain structure of DNA polymerase I of E. coli was taken from Riley et al. (19). The amino acid length of the gene products is given below each box. The localizations of the detected domains are indicated above the boxes. The recorded E values for the 5′-3′ exonuclease domain (53 exo [cd00008]), the central 3′-5′ exonuclease domain (35 exo [cd00007]), and the polymerase domain (DNA_polA [pfam00476]) are given in parentheses.
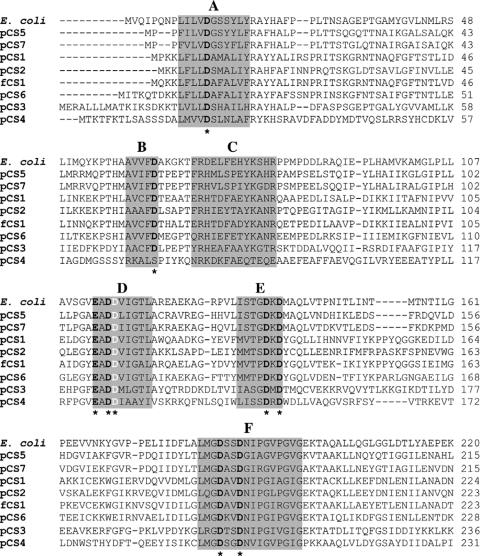
The remaining two plasmids (pCS1 and pCS4) harbored complete ORFs which encode shorter versions of PolA (Fig. 1). The gene product encoded by pCS1 (557 amino acids) contained a putative 5′-3′ exonuclease domain and a 3′-5′ exonuclease domain. The protein encoded by pCS4 (282 amino acids) was the smallest of all and contained solely a 5′-3′ exonuclease domain. The mutation of the complemented E. coli host strain is located in the 5′-3′ exonuclease domain of DNA polymerase I (16). Correspondingly, the identified genes located on the inserts of pCS1 to pCS8 and of fCS1 encoded at least this domain. Growth experiments revealed that the growth rates of all recombinant strains containing plasmids pCS1 to pCS8 were in the same range (0.18 h−1 to 0.2 h−1). These results indicated that the complementation of the mutant is independent of the presence of a 3′-5′ exonuclease domain or of a polymerase domain. Furthermore, all amino acid sequences of these domains harbored the six regions characteristic of the 5′-3′ exonuclease domains of DNA polymerase I proteins (Fig. 2). In addition, these regions contain nine conserved aspartate or glutamate residues (9). It has been suggested that some of these residues are involved in binding of metal ligands that are indispensable for nuclease activity (2, 11). Amino acid residues corresponding to these residues were also conserved in the 5′-3′ exonuclease domains derived from pCS1 to pCS3, pCS5 to pCS8, and fCS1. The protein encoded by pCS4 showed a replacement of aspartate by serine at one position (residue 72) (Fig. 2). All predicted polA gene products exhibited amino acid sequences that were 35% (pCS3) to 82% (pCS7) identical to those of DNA polymerases from other organisms (Table 1). These proteins were derived from species of a variety of different genera such as Algoriphagus, Pedobacter, Microscilla, Thermus, Acinetobacter, and Rhodococcus (Table 1). In general, polA genes are conserved and can be used as a phylogenetic marker gene (5). Taking this into account, the recorded degree of similarity to known DNA polymerases was low in most cases and indicated that the polA genes were recovered from uncharacterized and novel microorganisms.
FIG. 2.
Alignment of the deduced amino acid sequences of putative 5′-3′ exonuclease domains encoded by pCS1 to pCS8 and by fCS1. The amino acid sequence of the 5′-3′ exonuclease domain of E. coli DNA polymerase I (19) was used as a reference. The six conserved regions of 5′-3′ exonuclease domains described by Gutman and Minton (9) are marked with the letters A to F and are shaded. Positions of conserved aspartate and glutamate resides that are putatively involved in binding of metals (9) are marked by asterisks and are shown in bold letters. The aspartate residue (D116 [E. coli]) which was replaced by asparagine in the cold-sensitive E. coli mutants (16) is indicated by white letters.
In order to verify that the identified polA genes were responsible for complementation of the cold-sensitive E. coli mutant, the genes were amplified by PCR and cloned into the expression vector pBAD Myc/His A (Invitrogen, Karlsruhe, Germany), thereby placing the genes under the control of the arabinose-inducible araBAD promoter. Since arabinose is toxic for E. coli CSH26 fcsA29, this strain was not a suitable host for these experiments. Alternatively, the E. coli strain cs2-29 was used as a host. This strain carries the same cold sensitivity mutation of polA as E. coli CSH26 fcsA29 but is able to grow in the presence of arabinose (16). Recombinant E. coli cs2-29 clones containing the original recombinant plasmids (pCS1 to pCS8) or the fosmid (fCS1) were indistinguishable from the corresponding E. coli CSH26 fcsA29 clones with respect to growth at 18°C.
The pBAD Myc/His A constructs harboring the different identified polA genes were used to transform E. coli cs2-29. Subsequently, the resulting recombinant strains were plated on L agar plates (16) supplemented with 20 mg of thymine/liter and 0.1% arabinose. Growth of all strains was detected after 5 to 6 days of incubation at 18°C. The negative control containing the expression vector without an insert was not able to grow under the employed conditions. Thus, these results confirmed that the identified genes were responsible for complementation of the cold-sensitive E. coli mutants.
In conclusion, the chosen function-driven strategy was found to be an efficient way to identify the targeted DNA polymerase-encoding genes. The complementation of the cold-sensitive E. coli mutant allowed simple and rapid screening of both metagenomic libraries derived from glacial ice. Since almost no false positives were encountered, the high selectivity of this approach was evident. In this way, large gene banks consisting of genes conferring polymerase activity can be prepared rapidly. These gene banks or the corresponding clones can serve as starting material for the development of novel products. Sequence analysis of the first metagenome-derived DNA polymerase-encoding genes revealed that all encoded domains are typical of DNA polymerases belonging to family A. Most of the protein sequences exhibited low similarities to sequences of DNA polymerases from a variety of different microorganisms. This indicated that libraries derived from a permanently frozen habitat are a rich resource for the discovery of genes that originate from uncharacterized organisms.
Nucleotide sequence accession numbers.
The nucleotide sequences of the inserts of pCS1 to pCS8 and of fCS1 have been deposited in the GenBank database under accession numbers FJ384787 to FJ384794 and accession number FJ384795, respectively.
Acknowledgments
We thank Masaaki Wachi (Department of Bioengineering, Tokyo Institute of Technology, Yokohama, Japan) for providing the E. coli mutants CSH26 fcsA29 and cs2-29.
This work was supported by a grant from the Bundesministerium für Bildung und Forschung.
Footnotes
Published ahead of print on 6 March 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amblar, M., M. G. de Lacoba, M. A. Corrales, and P. Lopez. 2001. Biochemical analysis of point mutations in the 5′-3′ exonuclease of DNA polymerase I of Streptococcus pneumoniae: functional and structural implications. J. Biol. Chem. 276:19172-19181. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 4.Bonfield, J. K., K. Smith, and R. Staden. 1995. A new DNA sequence assembly program. Nucleic Acids Res. 23:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croan, D. G., D. A. Morrison, and J. T. Ellis. 1997. Evolution of the genus Leishmania revealed by comparison of DNA and RNA polymerase gene sequences. Mol. Biochem. Parasitol. 89:149-159. [DOI] [PubMed] [Google Scholar]
- 6.Daniel, R. 2005. The metagenomics of soil. Nat. Rev. Microbiol. 3:470-478. [DOI] [PubMed] [Google Scholar]
- 7.Ferrer, M., A. Beloqui, K. N. Timmis, and P. N. Golyshin. 2009. Metagenomics for mining new genetic resources of microbial communities. J. Mol. Microbiol. Biotechnol. 16:109-123. [DOI] [PubMed] [Google Scholar]
- 8.Foght, J., J. Aislabie, S. Turner, C. E. Brown, J. Ryburn, D. J. Saul, and W. Lawson. 2004. Culturable bacteria in subglacial sediments and ice from two southern hemisphere glaciers. Microb. Ecol. 47:329-340. [DOI] [PubMed] [Google Scholar]
- 9.Gutman, P. D., and K. W. Minton. 1993. Conserved sites in the 5′-3′ exonuclease domain of Escherichia coli DNA polymerase. Nucleic Acids Res. 21:4406-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyce, C. M., and T. A. Steitz. 1994. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 63:777-822. [DOI] [PubMed] [Google Scholar]
- 12.Klenow, H., and I. Henningsen. 1970. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc. Natl. Acad. Sci. USA 65:168-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loh, E., and L. A. Loeb. 2005. Mutability of DNA polymerase I: implications for the creation of mutant DNA polymerases. DNA Repair (Amsterdam) 4:1390-1398. [DOI] [PubMed] [Google Scholar]
- 14.Majerník, A., G. Gottschalk, and R. Daniel. 2001. Screening of environmental DNA libraries for the presence of genes conferring Na+ (Li+)/H+ antiporter activity on Escherichia coli: characterization of the recovered genes and the corresponding gene products. J. Bacteriol. 183:6645-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchler-Bauer, A., J. B. Anderson, M. K. Derbyshire, C. DeWeese-Scott, N. R. Gonzales, M. Gwadz, L. Hao, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, D. Krylov, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, N. Thanki, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2007. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35:D237-D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagano, K., M. Wachi, A. Takada, F. Takaku, T. Hirasawa, and K. Nagai. 1999. fcsA29 mutation is an allele of polA gene of Escherichia coli. Biosci. Biotechnol. Biochem. 63:427-429. [DOI] [PubMed] [Google Scholar]
- 17.Ohmori, H., E. C. Friedberg, R. P. Fuchs, M. F. Goodman, F. Hanaoka, D. Hinkle, T. A. Kunkel, C. W. Lawrence, Z. Livneh, T. Nohmi, L. Prakash, S. Prakash, T. Todo, G. C. Walker, Z. Wang, and R. Woodgate. 2001. The Y-family of DNA polymerases. Mol. Cell 8:7-8. [DOI] [PubMed] [Google Scholar]
- 18.Riesenfeld, C. S., R. M. Goodman, and J. Handelsman. 2004. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 6:981-989. [DOI] [PubMed] [Google Scholar]
- 19.Riley, M., T. Abe, M. B. Arnaud, M. K. Berlyn, F. R. Blattner, R. R. Chaudhuri, J. D. Glasner, T. Horiuchi, I. M. Keseler, T. Kosuge, H. Mori, N. T. Perna, G. Plunkett III, K. E. Rudd, M. H. Serres, G. H. Thomas, N. R. Thomson, D. Wishart, and B. L. Wanner. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 34:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tvermyr, M., B. E. Kristiansen, and T. Kristensen. 1998. Cloning, sequence analysis and expression in E. coli of the DNA polymerase I gene from Chloroflexus aurantiacus, a green nonsulfur eubacterium. Genet. Anal. 14:75-83. [DOI] [PubMed] [Google Scholar]
- 21.Wang, C., D. J. Meek, P. Panchal, N. Boruvka, F. S. Archibald, B. T. Driscoll, and T. C. Charles. 2006. Isolation of poly-3-hydroxybutyrate metabolism genes from complex microbial communities by phenotypic complementation of bacterial mutants. Appl. Environ. Microbiol. 72:384-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, K., A. C. Martiny, N. B. Reppas, K. W. Barry, J. Malek, S. W. Chisholm, and G. M. Church. 2006. Sequencing genomes from single cells by polymerase cloning. Nat. Biotechnol. 24:680-686. [DOI] [PubMed] [Google Scholar]