Abstract
Acidovorax sp. strain NA3 was isolated from polycyclic aromatic hydrocarbon (PAH)-contaminated soil that had been treated in a bioreactor and enriched with phenanthrene. The 16S rRNA gene of the isolate possessed 99.8 to 99.9% similarity to the dominant sequences recovered during a previous stable-isotope probing experiment with [U-13C]phenanthrene on the same soil (D. R. Singleton, S. N. Powell, R. Sangaiah, A. Gold, L. M. Ball, and M. D. Aitken, Appl. Environ. Microbiol. 71:1202-1209, 2005). The strain grew on phenanthrene as a sole carbon and energy source and could mineralize 14C from a number of partially labeled PAHs, including naphthalene, phenanthrene, chrysene, benz[a]anthracene, and benzo[a]pyrene, but not pyrene or fluoranthene. Southern hybridizations of a genomic fosmid library with a fragment of the large subunit of the ring-hydroxylating dioxygenase gene from a naphthalene-degrading Pseudomonas strain detected the presence of PAH degradation genes subsequently determined to be highly similar in both nucleotide sequence and gene organization to an uncharacterized Alcaligenes faecalis gene cluster. The genes were localized to the chromosome of strain NA3. To test for gene induction by selected compounds, RNA was extracted from amended cultures and reverse transcribed, and cDNA associated with the enzymes involved in the first three steps of phenanthrene degradation was quantified by quantitative real-time PCR. Expression of each of the genes was induced most strongly by phenanthene and to a lesser extent by naphthalene, but other tested PAHs and PAH metabolites had negligible effects on gene transcript levels.
Increasingly, stable-isotope probing (SIP) has been used to identify environmentally relevant bacteria capable of expressing a particular phenotype prior to directed efforts to cultivate those organisms. For instance, RNA-based SIP with [13C6]benzene was used to identify Azoarcus strains in gasoline-contaminated groundwater before targeted cultivation attempts (19). In another example, a Burkholderia strain isolated from soil was matched to the dominant terminal restriction fragment length polymorphism band from heavy DNA resulting from incubation with [13C]benzoic acid (40). The bacterium Polaromonas naphthalenivorans CJ2 was isolated from coal-tar-contaminated sediments after having been identified as a dominant member of clone libraries in SIP experiments with 13C-labeled naphthalene (17). That strain has since been the focus of additional studies, one of which resulted in the discovery of a unique ring-hydroxylating dioxygenase (RHD) (18).
In prior research on polycyclic aromatic hydrocarbon (PAH) degradation, we performed DNA-SIP with uniformly labeled naphthalene, phenanthrene, and pyrene to identify the dominant degraders of those compounds in a bioreactor treating PAH-contaminated soil (45, 46). In those experiments, the most frequently encountered sequences in 13C-enriched DNA fractions from incubations with labeled phenanthrene were associated with the Acidovorax genus (45). Members of that genus have previously been found in high abundance in soils containing PAHs (9, 22) or have been implicated in PAH degradation (12), particularly phenanthrene degradation (7, 35, 42, 45). However, little is known about the genetic determinants for PAH degradation among Acidovorax strains.
There exists wide genetic diversity among organisms capable of PAH degradation. Members of the Pseudomonas genus generally contain the well-studied and commonly encountered nah-type genes and are typically associated with the degradation of naphthalene (14, 56). Other organisms contain homologous but significantly different genes. Examples of these include the nag genes from Polaromonas naphthalenivorans (16) and Ralstonia sp. strain U2 (58), the phn genes of Burkholderia sp. strain RP007 (29), and the nid genes of Mycobacterium vanbaalenii (20). Multiple attempts have been made by researchers to design specific PCR primer sets for the detection of the RHD (or initial dioxygenase) gene, whose product catalyzes the first reaction in the aerobic bacterial metabolism of PAHs (5, 30, 33, 36, 54). More recently, degenerate primers based on the known diversity of dioxygenase sequences from both gram-negative and gram-positive organisms have been designed (8). Yet, despite their frequent association with PAH degradation, Acidovorax sequences do not appear in environmental surveys using these primer sets due to the lack of reference sequences from isolates in the genus.
In this study we describe the isolation and PAH-degradative properties of a phenanthrene-degrading Acidovorax isolate representative of those members of the genus previously detected by SIP in the same soil. We identified and sequenced the putative upper-pathway genes of phenanthrene degradation and determined compounds that induce expression of those genes.
MATERIALS AND METHODS
Chemicals.
PAHs, their purities, and vendors were as follows: naphthalene (99+%; Aldrich, Milwaukee, WI), phenanthrene (>96%; Sigma, St. Louis, MO), anthracene (scintillation grade; Kodak, Rochester, NY), pyrene (product P-2146; Sigma), fluoranthene (98%; Aldrich), chrysene (98%; Aldrich), benz[a]anthracene (1,2-benzanthracene) (99%; Aldrich), and benzo[a]pyrene (97%; Aldrich). Salicylic acid (>99%) was obtained from Aldrich and phthalic acid (99%) from Acros (NJ). 14C-labeled compounds used for mineralization experiments, their purities, specific activities, and vendors were as follows: [5,6-14C]benz[a]anthracene (>98%, 54.6 mCi/mmol; Chemsyn, Lenexa, KA), [7-14C]benzo[a]pyrene (≥98%, 26.6 mCi/mmol; Sigma), [5,6,11,12-14C]chrysene (≥98%, 47.6 mCi/mmol; Chemsyn), [3-14C]fluoranthene (≥98%, 45 mCi/mmol; Sigma), [benzene-UL-14C]naphthalene (>98%, 17.8 mCi/mmol; Sigma), [9-14C]phenanthrene (>98%, 8.3 mCi/mmol; Sigma), [ring-UL-14C]phthalic acid (>98%, 12.7 mCi/mmol; Sigma), [4,5,9,10-14C]pyrene (∼95%, 61 mCi/mmol; Sigma), and [ring-UL-14C]salicylic acid (>98%, 10 mCi/mmol; Sigma).
Isolation and growth media.
Contaminated soil from the site of a former manufactured gas plant, which had been treated in a lab-scale, aerobic bioreactor (45), was enriched for phenanthrene degraders by adding crystalline phenanthrene in excess of its aqueous solubility to a flask containing slurry and incubating the vessel for 2 weeks. Strain NA3 was isolated by plating serial dilutions of the enriched slurry on Difco nutrient agar plates (pH 7.0) (Becton, Dickinson, and Co., Sparks, MA). The identity of the isolated organism was confirmed by 16S rRNA gene sequencing (see below), and it was maintained on nutrient agar or in Difco nutrient broth (NB) (pH 7.0) (Difco Laboratories, Detroit, MI). To test for indigo production, indole (99+%; Aldrich) was added to nutrient agar at a final concentration of 1 mM. For testing of growth substrates, strain NA3 was grown in liquid MM2 medium (24) containing 2 g·liter−1 of the carbon source added in solid form postautoclaving. Substrates for these tests included naphthalene, phenanthrene, anthracene, fluoranthene, pyrene, phthalate, and salicylate.
Mineralization.
Strain NA3 was grown to high turbidity in 300 ml of NB (pH 7.0) at 30°C. The volume was divided in half and each half centrifuged at 3,000 × g for 5 min to pellet cells. The liquid was discarded and the pellets resuspended in a total of 75 ml of fresh M9 medium (43). One of those volumes was then treated with 85% phosphoric acid to a pH of <2 as a killed control. The experimental and acidified suspensions were allow to sit at room temperature for 1 h prior to dispensing 2.5 ml (approximately 2.4 × 109 cells) into sterile 40-ml amber-glass EPA vials. The number of cells in the live sample was determined by plating serial dilutions of cells on nutrient agar plates. An additional 2.5 ml of M9 medium was added to each vial to bring the total volume to 5 ml. Resuspended and acid-killed cells were created in triplicate for each of the tested compounds. Approximately 20,000 dpm of each test compound was then added directly to the cell suspension. A sterile 12- by 75-mm glass test tube containing a piece of filter paper saturated with 60 μl of 2 M KOH to act as a CO2 trap was placed in each vial. The vial was sealed with an aluminum foil-covered, Teflon-seal cap and incubated at 30°C with gentle agitation for 12 h. At the end of the incubation, the filter paper was removed and the captured 14C counted on a Packard (Meriden, CT) Tri-Carb liquid scintillation analyzer, model 1900 TR. Differences in captured 14C between live and acidified controls were tested for significance using Student's t test (ProStat for Windows, v.4.02; Poly Software International, Inc., Pearl River, NY). To determine the percentage of added 14C mineralized, the mean of the values for the acidified controls for each compound was subtracted from each of the triplicate experimental values and divided by the total dpm of 14C added.
Genomic fosmid library construction and analysis.
A genomic library was constructed from strain NA3 DNA using the CopyControl fosmid library production kit with the pCC1FOS vector and the phage T1-resistant EPI300 Escherichia coli plating strain (Epicentre, Madison, WI). Briefly, high-molecular-weight DNA was isolated from cells grown in NB using a Wizard genomic DNA purification kit (Promega, Madison, WI) and sheared to ∼40 kb by pipetting. The fragments were end repaired, ligated into the fosmid vector, and packaged, and the host cells were grown overnight following the kit instructions. Colonies were selected and transferred to LB plates containing chloramphenicol.
Colonies on plates were then tested for the presence of genes homologous to the naphthalene RHD gene of Pseudomonas putida G7. A digoxigenin (DIG)-labeled nucleotide probe was generated by PCR using as the template DNA extracted from Pseudomonas putida G7 and with primers specific for a portion of the nahAc gene (primers NAH-F and NAH-R) (5).
Colonies were transferred to nylon membranes (Roche, Indianapolis, IN) per the manufacturer's instructions. Hybridizations with the DIG-labeled Pseudomonas nahAc amplicon were carried out per the manufacturer's instructions using DIG Easy Hyb (Roche) and DIG Wash and Block buffer sets (Roche) with final stringency washes at 45°C. Pseudomonas putida G7 DNA was added to each membrane as a positive control. The presence of DNA that bound the probe was detected by enzyme immunoassay and enzyme-catalyzed color reaction with nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate as part of the DIG nucleic acids detection kit (Roche). Positive colonies were transferred to LB broth containing chloramphenicol and induced to high copy number with the CopyControl induction solution (Epicentre) before fosmid isolation with the FosmidMAX DNA purification kit (Epicentre).
Molecular analyses.
The 16S rRNA gene of strain NA3 was amplified by PCR with primers 8f (10) and 1492r (27). The resulting product was then cloned into the plasmid PCR4-TOPO using the TOPO-TA cloning kit for sequencing (Invitrogen, Carlsbad, CA). The insert was sequenced with primers M13R, M13F, and 341F (37); 338R (2); 907F (28); and 939R (4) at the University of North Carolina Genome Analysis Facility. Sequences were assembled using the program Sequencher 4.8 (Gene Codes Corp., Ann Arbor, MI).
An isolated fosmid which hybridized the nahAc gene of Pseudomonas (designated phnNA3.1) was digested with BamHI (New England Biolabs, Ipswich, MA), Sau3A1, and a combination of the two enzymes. The resulting fragments were separated by agarose gel electrophoresis and the DNA transferred to a membrane and analyzed by Southern hybridization as described above. A new fosmid digest was performed solely with BamHI, the resulting fragments were ligated into plasmid pCR2.1 (Invitrogen) using standard methods (43), inserts were screened by PCR with M13F and M13R, and one insert of approximately 800 bp was sequenced with primer M13R. Subsequent cloning and sequencing efforts employed a combination of primer walking and PCR/sequencing primer design based on GenBank entry AB024945 (Alcaligenes faecalis phenanthrene degradative gene cluster) to recover the Acidovorax gene sequences of interest. To avoid the introduction of erroneous base calls from the Alcaligenes sequence, all primer sequences were trimmed from the recovered sequences prior to assembly of the fragments in Sequencher.
For comparisons of gene organization, open reading frames (ORFs) from NA3 and other organisms whose sequences were recovered from GenBank were visualized with the frames program of the GCG suite of programs (Accelrys Inc., San Diego, CA). ORFs were identified and labeled according to the results of BLAST searches (1) and GenBank entry header information. The resulting images were then scaled based on the size of the RHD large-subunit gene and aligned according to the same.
To generate phylogenetic trees, sequences were first aligned with the Pileup program of the GCG suite of programs prior to constructing neighbor-joining trees with the ClustalW program (48). Trees were bootstrapped 1,000 times, and gaps in the alignment were ignored.
Pulsed-field gel electrophoresis (PFGE) was combined with Southern blotting to determine whether the identified genes were located on a chromosome or plasmid. Agarose-embedded NA3 DNA was prepared according to the instructions provided in the CHEF-DR III pulsed-field electrophoresis systems (Bio-Rad, Hercules, CA) manual. S1 nuclease treatment was performed according to the method of Basta et al. (6). Gels were run under the following conditions on the CHEF-DR III pulsed-field electrophoresis system: (i) separation of high-molecular-weight DNA, with Saccharomyces cerevisiae chromosomal standards (Bio-Rad), 1% agarose gel, 60- to 120-s switch, 20 h, 6 V/cm, 105° angle, and 0.5× Tris-borate-EDTA running buffer, or (ii) separation of mid-range DNA, with a λ concatemer ladder and same conditions as above except for a 120° angle. Transfer of DNA for Southern hybridizations was performed as described in the CHEF-DR III instruction manual, and hybridizations were performed as described above using a DNA fragment spanning the phnAcd genes of strain NA3 as a probe.
Primer sets suitable for quantitative real-time PCR (qPCR) were designed to amplify fragments of the phnAc, phnB, and phnC genes of Acidovorax sp. strain NA3 (Table 1). Standard curves for the quantification of phn genes were created by digesting fosmid phnNA3.1 with SmaI (New England Biolabs), quantifying the amount of DNA using a NanoDrop ND-3300 fluorospectrometer (Thermo Fisher Scientific, Wilmington, DE), and performing qPCR with serial dilutions of the digested fosmid. Standard curves for the quantification of Acidovorax 16S rRNA genes were generated with BamHI-digested 16S rRNA gene clone PHE7d8 (45) as described previously (44). Amplification efficiencies (39) of the primer sets for phnAc, phnB, phnC, and Acidovorax 16S rRNA genes were 1.92, 1.92, 1.94, and 1.90, respectively. The 16S rRNA and phnAc reactions were run with an annealing temperature of 55°C, and the phnB and phnC reactions were run at 60°C.
TABLE 1.
Quantitative real-time PCR primers
| Primer | Target | Sequence (5′→3′) | Amplicon size (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| phnAc-F | Ring-hydroxylating dioxygenase gene, phnAc | GAC AGC TTG ATT CCG | 171 | 55 | This study |
| phnAc-R | TGC AAC TGA ACG CAC | ||||
| phnB-F | Dihydrodiol dehydrogenase gene, phnB | TGT CCC CCT TGT CGA CC | 187 | 60 | This study |
| phnB-R | TAT AGA GCA CGC CGC CG | ||||
| phnC-F | Ring cleavage dioxygenase gene, phnC | CAT TCT GCG ATC CGT AGA CC | 148 | 60 | This study |
| phnC-R | CCA CGG AAT GCT CAC GG | ||||
| AcidF | Acidovorax 16S rRNA gene | TAA CGG AGC GAA AGC TT | 75 | 55 | 44 |
| AcidR | GTC CGC GCA AGG CCT T |
Fosmid phnNA3.1 was used as a template to test whether degenerate PCR primers for the amplification of the initial RHD would be successful for NA3 using the methods of Cébron et al. (8).
phn gene induction.
Strain NA3 was grown overnight in NB (pH 7.0) at 30°C with shaking at 225 rpm. To 5 ml of fresh NB was added 100 μl of the overnight culture in triplicate for each compound tested. Cultures were allowed to grow several hours at 30°C with shaking to an optical density at 600 nm of approximately 0.12 before 100 μl of a methanol solution containing a given compound was added. Each PAH was added at or slightly above its aqueous solubility (32). The compounds and their nominal concentrations in the medium were as follows: naphthalene,21 mg/liter; phenanthrene, 1 mg/liter; pyrene, 0.13 mg/liter; fluoranthene, 0.2 mg/liter; chrysene, 0.002 mg/liter; benz[a]anthracene, 0.01 mg/liter; benzo[a]pyrene, 0.003 mg/liter; salicylate, 100 mg/liter; and phthalate, 100 mg/liter. Additionally, methanol-only (100 μl) and no-carbon-added controls were also performed in triplicate. After addition of each compound, tubes were returned to the shaker for 10 min before 500 μl of each culture was removed and added to 1 ml of RNAprotect Bacteria Reagent (Qiagen, Valencia, CA). Each sample was vortexed, centrifuged, and stored at −20°C as suggested by the manufacturer (41).
Total RNA was extracted and purified from each cell pellet individually according to protocols 4 and 7 of reference 41 using an RNeasy Mini Kit (Qiagen). RNA was eluted in a total of 30 μl of RNase-free water (Fisher Scientific, Pittsburgh, PA). For removal of genomic DNA and creation of cDNA, 12 μl of extracted total RNA from each cell pellet was first treated with genomic DNA wipeout buffer before addition of reverse transcriptase, buffer, and the RT primer mix included with the QuantiTect reverse transcription kit (Qiagen), per the manufacturer's instructions.
The transcript number was quantified with a SmartCycler (Cepheid, Sunnydale, CA) and QuantiTect SYBR green PCR kit (Qiagen). For the template, 1 μl of the cDNA solution was used for each reaction containing primers for the phn transcripts, and 1 μl of cDNA diluted 1:1,000 was used in the qPCR for 16S rRNA transcripts. All qPCR reactions were run at least in duplicate. The quantity of phn transcripts for each sample was normalized to 108 16S rRNA transcripts per μl of cDNA to allow direct comparison.
Nucleotide sequence accession numbers.
Sequences generated in this study were deposited in GenBank with the accession numbers EU910093 and EU910094.
RESULTS
Growth.
Strain NA3 was isolated on nutrient agar from phenanthrene-enriched PAH-contaminated soil (44). The addition of indole to nutrient agar plates resulted in purple colonies from the production of indigo and was indicative of an active dioxygenase (11). In MM2 medium, strain NA3 grew on phenanthrene as a sole source of carbon and energy after 6 days but did not display visibly turbid growth with naphthalene, anthracene, fluoranthene, pyrene, phthalic acid, or salicylic acid.
Phylogeny.
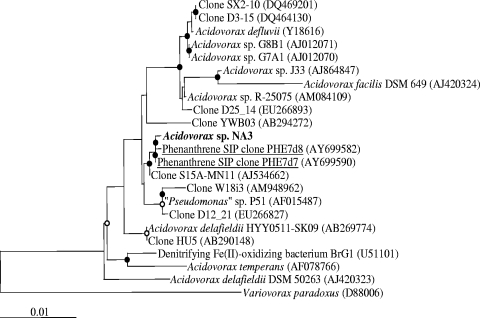
The nearly complete 16S rRNA gene sequence of strain NA3 indicated that the bacterium was a member of the Acidovorax genus (Fig. 1). It was most similar to two sequences from the same bioreactor-treated, PAH-contaminated soil recovered during an SIP experiment with [U-13C]phenanthrene (45), with 99.93% sequence similarity to clone PHE7d7 and 99.80% similarity to clone PHE7d8. It also shared similarity to clone sequences from Siberian deep-well groundwater (clone S15A-MN11) (unpublished), BTEX contaminated groundwater (clone W18i3) (unpublished), and a tar-oil-impacted aquifer BTEX plume (clone D12_21) (55).
FIG. 1.
Neighbor-joining phylogenetic tree of 16S rRNA gene sequences showing the relationship of strain NA3 to highly similar clone and isolate sequences. Strain NA3 is shown in bold, while clonal sequences from the same environmental sample are underlined. The tree was constructed from 1,285 aligned bases. Open and closed circles at nodes represent >50 and >95% bootstrap support, respectively. GenBank accession numbers are indicated in parentheses. The scale bar indicates the number of substitutions per position. The chlorinated aromatic compound degrader Pseudomonas sp. strain P51 was originally classified as a pseudomonad based on physiological data (51). However, based on the 16S rRNA gene sequence of the organism, strain P51 appears to be a member of the Acidovorax genus (47).
Mineralization.
Cells of NA3 in M9 medium were incubated in the presence of 14C-labeled PAHs and PAH metabolites. After 12 h of incubation, significant levels of CO2 production from labeled substrates compared to acidified controls (Student's t test, P < 0.05) were observed for naphthalene (5.9% ± 0.2% of added 14C mineralized), phenanthrene (23.7% ± 10.7%), chrysene (3.9% ± 2.1%), benz[a]anthracene (6.2% ± 0.5%), benzo[a]pyrene (0.2% ± 0.01%), and phthalic acid (0.05% ± 0.01%). Strain NA3 did not mineralize pyrene, fluoranthene, or salicylate to a greater extent than acidified controls. In separate experiments, longer incubations of labeled benzo[a]pyrene and phthalic acid resulted in higher levels of 14CO2 production than observed during this test (data not shown).
Genetics.
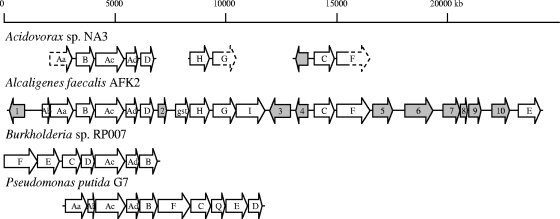
Southern hybridizations using the nahAc gene of Pseudomonas putida G7 as a probe successfully identified a fosmid clone from the genomic library (designated phnNA3.1) containing a homologous gene. Digestion of the fosmid and subcloning and sequencing of an approximately 800-bp piece of DNA identified a gene fragment with nearly 100% nucleotide identity to the phnC gene of Alcaligenes faecalis AFK2 (GenBank accession number AB024945). The sequences of the putative large and small subunits of the RHD (phnAcd), dihydrodiol dehydrogenase (phnB), and 3,4-dihydroxyphenanthrene dioxygenase (phnC) genes were specifically targeted and determined through a combination of primer walking from the phnC gene fragment and use of the AFK2 operon as a template to design specific sequencing primers for recovering the other genes of interest. A total of 8,286 nucleotides in three discontinuous regions of 4343, 1344, and 2599 bases were sequenced, and all of the recovered sequences were present on fosmid phnNA3.1. The arrangement of recovered genes and gene fragments suggests that the phn operon of Acidovorax strain NA3 is organized similarly to that of Alcaligenes faecalis AFK2 (Fig. 2). Partial sequences of other genes from NA3 (phnH, phnG, and phnF) supported this finding as well, and although the sequences between the three fragments were not determined, PCR analyses confirmed their arrangement as presented in Fig. 2 (data not shown). This organization of genes in strains NA3 and AFK2 differs from that of other well-known PAH degradation operons. Notably, the putative dihydrodiol dehydrogenase gene appears upstream of the large subunit of the initial RHD, and the ring cleavage dioxygenase (phnC) is separated from the other genes of the upper pathway.
FIG. 2.
Organization of sequenced genes from Acidovorax strain NA3 aligned to homologous genes from Alcaligenes faecalis AFK2 (GenBank accession number AB024945), Pseudomonas putida G7 (AB237655), and Burkholderia sp. strain RP007 (AF061751). Letters indicate the gene designations for each PAH degradation operon (gst, glutathione S-transferase). Numbered or shaded ORFs are presumably unaffiliated with PAH degradation. Sequenced ORFs for NA3 are shown with solid arrows, while segments of genes for which the complete sequence was not obtained are indicated with dashed lines. Unsequenced areas of the NA3 operon lack a horizontal line. Operons were aligned by the RHD large subunit (Ac).
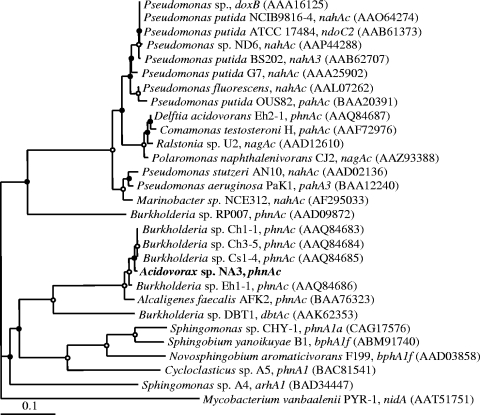
The large subunit of the RHD is the most well studied of the PAH degradation genes. While the predicted protein sequence of the large subunit of the RHD in NA3 was similar to that of Alcaligenes faecalis AFK2, the protein sequences with the highest similarity were found in several recently cultivated Burkholderia strains (Fig. 3).
FIG. 3.
Neighbor-joining phylogenetic tree based on predicted amino acid sequences of the large subunit of the RHD for Acidovorax strain NA3 and other gram-negative bacteria. The tree was based on 189 aligned amino acids. The nidA gene of Mycobacterium vanbaalenii was used as an outgroup. Other notation is as described in Fig. 1.
After the identification of the phn genes in strain NA3, fosmid phnNA3.1 was used to test whether degenerate PCR primers for the RHD gene designed by Cébron et al. could amplify the Acidovorax phnAc gene (8). Despite significant predicted mismatches (4 of 24 bases and 8 of 28 bases for the forward and reverse gram-negative organism RHD primers, respectively), a weak PCR amplicon of the expected size was produced (data not shown).
Localization of phn genes.
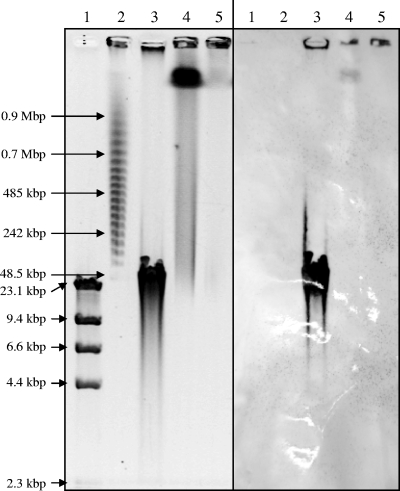
PFGE was used to determine whether the detected phn genes were located on the chromosome or on a plasmid. No low-molecular-weight DNA that might be indicative of a plasmid was detected in any PFGE runs, and Southern hybridization with a DIG-labeled phnAcd fragment showed hybridization only with very high-molecular-weight DNA in or near the wells (Fig. 4 and data not shown), suggesting that the genes were located on the chromosome.
FIG. 4.
(Left) Pulsed field electrophoresis gel of λHindIII digest markers (lane 1), λ concatemer marker (lane 2), S1 nuclease-digested fosmid phnNA3.1 (lane 3), S1 nuclease-digested NA3 genomic DNA (lane 4), and undigested NA3 genomic DNA (5). (Right) Southern blot of the same gel using the phnAcd genes of NA3 as a probe. Sizes of the markers are indicated on the left.
Induction.
To test the effect of compound addition on the expression of three genes putatively involved in PAH degradation, selected PAHs were added at or near their aqueous solubility limits to NA3 cells grown in NB. Two PAH metabolites, salicylate and phthalate, were also added at a 100-mg/liter final concentration. Of the compounds tested only two, naphthalene and phenanthrene, strongly induced the expression of the phnAc, phnB, and phnC genes compared to unamended controls (Table 2). All other compounds, including methanol (the carrier solvent for the addition of test compounds), pyrene, chrysene, fluoranthene, benz[a]anthracene, benzo[a]pyrene, salicylate, and phthalate, produced much smaller or no responses. For the two compounds that did elicit a pronounced response, the levels of induction were similar for the phnAc and phnB genes, and although the levels of phnC induction were lower, they were still significantly higher than basal levels of expression as determined by the quantification of transcripts in unamended cultures.
TABLE 2.
Increases in expression of strain NA3 phn genes in response to addition of PAH or PAH metabolites compared to unamended controlsa
| Compound | Fold increase in expression
|
||
|---|---|---|---|
| phnAc | phnB | phnC | |
| Methanol | 0 | 0 | 1 |
| Naphthalene | 18 | 17 | 8 |
| Phenanthrene | 34 | 34 | 12 |
| Pyrene | 1 | 1 | 0 |
| Chrysene | 0 | 0 | 0 |
| Fluoranthene | 1 | 1 | 0 |
| Benz[a]anthracene | 0 | 0 | 0 |
| Benzo[a]pyrene | −1 | 0 | 0 |
| Salicylate | 0 | 0 | 0 |
| Phthalate | 0 | 0 | 0 |
Calculated levels in the unamended controls for phnAc, phnB, and phnC transcripts per 108 16S rRNA transcripts were 1.2 × 104 ± 4.0 × 103, 2.8 × 104 ± 5.8 × 103, and 2.4 × 104 ± 3.1 × 103, respectively.
DISCUSSION
Members of the Acidovorax genus are frequently encountered in association with PAH degradation, notably that of phenanthrene, but prior to this work little was known of the underlying genetic determinants behind the phenotype. The particular Acidovorax strain used in this study has genes very similar in sequence and arrangement to those in Alcaligenes faecalis AFK2, and the initial dioxygenase sequence is also highly similar to those in several Burkholderia strains, although not to that of Burkholderia sp. strain RP007. These observations suggest that this particular genotype may be widespread among some PAH-degrading members of the order Burkholderiales.
Unfortunately for comparative purposes, while the genes determined in this study bear significant resemblance to the Alcaligenes faecalis AFK2 sequences deposited in GenBank, there does not appear to be a publication associated with the entry (GenBank accession number AB024945). However, other previously published research on AFK2 revealed the presence of two plasmids, the larger of which (named pHK2, 42.5 kb) conferred the phenanthrene degradation phenotype to a Pseudomonas strain (25). Presumably the deposited gene sequences originated from that plasmid. In contrast to the case for Alcaligenes faecalis AFK2, the phenanthrene degradation genes of Acidovorax strain NA3 appear to be located on the chromosome, as no evidence for plasmids was found during this study. However, this appears to be one of few significant differences between the clusters, as even intergenic regions and genes unaffiliated with phenanthrene degradation in NA3 are highly similar to equivalent regions of the AFK2 sequence deposited in GenBank.
Due to the high similarity of the phn operon between the NA3 and AFK2 strains, it might be presumed that Alcaligenes faecalis AFK2 may actually be a member of the Acidovorax genus. The Acidovorax genus was created in 1990 (53), well after AFK2 was first described in 1982 (23). Not only did the newly created Acidovorax genus include organisms previously characterized as Alcaligenes strains, but many traits used to define AFK2 as an Alcaligenes species (e.g., flagellated, oxidase positive, and catalase positive) were also shared by the Acidovorax genus (15, 53). In support of its current classification, however, AFK2 was reported as being unable to grow on d-glucose (23), a ubiquitous trait in Acidovorax strains (53). Additionally, numerous other examples of PAH-degrading Alcaligenes species, including some associated with phenanthrene degradation, have been reported recently (see, e.g., references 3, 49, and 52). Unfortunately, the ribosomal sequences of AFK2 are not available in public databases, and the organism does not appear to be available in either the ATCC or DSMZ culture collections in order to verify its genus affiliation.
Organisms containing the type of phn genes present in NA3 and other related strains appear to be associated primarily with the degradation of three-ring PAHs (notably phenanthrene). Of the compounds tested, only phenanthrene supported the growth of NA3, and the highest level of mineralization was observed for phenanthrene. Similarly, the highest levels of gene induction were observed when phenanthrene was added to the culture medium. Alcaligenes faecalis AFK2 was described as a phenanthrene- and anthracene-degrading strain (23), and the Burkholderia isolates with phnAc genes highly similar to that in NA3 were isolated on humic acid-absorbed phenanthrene (50). Additional support for this apparent association comes from a recent survey of RHDs from PAH-contaminated sediments that found only AFK2-type or closely related phnAc genes in samples containing solely phenanthrene (31).
It was expected that transcription of the phnAc, phnB, and phnC genes in Acidovorax strain NA3 would be induced by the addition of phenanthrene to the culture media. There is evidence for increased protein expression of RHDs in Mycobacterium strains when exposed to various PAHs (21, 26), and phenanthrene has been shown to increase the expression of the nahAc gene in Pseudomonas putida G7 (34). In environmental samples, increases in both nidA and nahAc transcripts could be detected in sediments when PAHs were added (57). The nearly equivalent levels of increased expression of both the phnAc and phnB genes in NA3 when exposed to either phenanthrene or naphthalene suggest that the genes are cotranscribed. The phnAabcd genes encode the four subunits of the initial RHD, and the phnB gene is located in the middle of that cluster, so it is likely that the parent compound induces the expression of both phnAc and phnB. However, in both Acidovorax strain NA3 and Alcaligenes faecalis AFK2, the gene encoding the third protein in the upper pathway of PAH degradation (phnC) is distinct from the phnABD cluster and is likely controlled by separate transcriptional regulators. For both phenanthrene and naphthalene, the level of induction for phnC was lower than that for phnAc and phnB, and it is uncertain whether the expression of phnC is influenced by the parent compound or a metabolite such as 3,4-dihydroxyphenanthrene. It was also interesting that the presence of naphthalene could induce the expression of phnAc, phnB, and phnC in NA3, because the organism could not utilize naphthalene as a sole source of carbon and energy in the defined media tested and Acidovorax sequences did not appear in 13C-enriched DNA fractions during previous SIP experiments with naphthalene (45).
Phenanthrene is typically degraded through a pathway that utilizes either salicylate or phthalate as an intermediate (13). While the pathway utilized by NA3 was not explicitly tested during this experiment, we found it incapable of growth on either compound under the conditions tested, and neither salicylate nor phthalate induced expression of the upper pathway phn genes. Additionally, it did not mineralize salicylate but was capable of weakly mineralizing phthalate. Based solely on these results, it would be difficult to speculate which pathway NA3 might utilize in the metabolism of phenanthrene. However, there are indications that Alcaligenes faecalis AFK2 utilizes the phthalate pathway (23, 38), and given the similarities in the phn genes of NA3 and AFK2 and the weak mineralization of phthalate by NA3, metabolism of phenanthrene through phthalate is probably the case for this Acidovorax strain as well.
Acknowledgments
Liza Guzmán Ramirez was supported by a grant from the National Institute of General Medical Sciences’ Bridges to the Doctorate Program. This work was supported by the National Institute of Environmental Health Sciences (grant 5 P42 ES05948).
We thank Beatriz Zayas, Alberto Rivera, and Donald Fox for their contributions to the summer internship program.
Footnotes
Published ahead of print on 6 March 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreoni, V., L. Cavalca, M. A. Rao, G. Nocerino, S. Bernasconi, E. Dell'Amico, M. Colombo, and L. Gianfreda. 2004. Bacterial communities and enzyme activities of PAHs polluted soils. Chemosphere 57:401-412. [DOI] [PubMed] [Google Scholar]
- 4.Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin, B. R., C. H. Nakatsu, and L. Nies. 2003. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl. Environ. Microbiol. 69:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basta, T., A. Keck, J. Klein, and A. Stolz. 2004. Detection and characterization of conjugative degradative plasmids in xenobiotic-degrading Sphingomonas strains. J. Bacteriol. 186:3862-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogan, B. W., L. M. Lahner, W. R. Sullivan, and J. R. Paterek. 2003. Degradation of straight-chain aliphatic and high-molecular-weight polycyclic aromatic hydrocarbons by a strain of Mycobacterium austroafricanum. J. Appl. Microbiol. 94:230-239. [DOI] [PubMed] [Google Scholar]
- 8.Cébron, A., M.-P. Norini, T. Beguiristain, and C. Leyval. 2008. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 73:148-159. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y.-T., J.-F. Lee, and H.-P. Chao. 2007. Variability of communities and physiological characteristics between free-living bacteria and attached bacteria during the PAH biodegradation in a soil/water system: In Situ Bioremediation, Third European Bioremediation Conference. Eur. J. Soil Biol. 43:283-296. [Google Scholar]
- 10.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ensley, B. D., B. J. Ratzkin, T. D. Osslund, M. J. Simon, L. P. Wackett, and D. T. Gibson. 1983. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science 222:167-169. [DOI] [PubMed] [Google Scholar]
- 12.Eriksson, M., E. Sodersten, Z. Yu, G. Dalhammar, and W. W. Mohn. 2003. Degradation of polycyclic aromatic hydrocarbons at low temperature under aerobic and nitrate-reducing conditions in enrichment cultures from northern soils. Appl. Environ. Microbiol. 69:275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson, D. T., and S. Venkiteswaran. 1984. Microbial degradation of aromatic hydrocarbons, p. 181-252. In D. T. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, Inc., New York, NY.
- 14.Habe, H., and T. Omori. 2003. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 67:225-243. [DOI] [PubMed] [Google Scholar]
- 15.Holt, J. G., N. R. Krieg, H. A. Sneath, J. T. Staley, and S. T. Williams. 1994. Bergey's manual of determinative bacteriology. Williams and Wilkins, Baltimore, MD.
- 16.Jeon, C. O., M. Park, H. S. Ro, W. Park, and E. L. Madsen. 2006. The naphthalene catabolic (nag) genes of Polaromonas naphthalenivorans CJ2: evolutionary implications for two gene clusters and novel regulatory control. Appl. Environ. Microbiol. 72:1086-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon, C. O., W. Park, W. C. Ghiorse, and E. L. Madsen. 2004. Polaromonas naphthalenivorans sp. nov., a naphthalene-degrading bacterium from naphthalene-contaminated sediment. Int. J. Syst. Evol. Microbiol. 54:93-97. [DOI] [PubMed] [Google Scholar]
- 18.Jeon, C. O., W. Park, P. Padmanabhan, C. DeRito, J. R. Snape, and E. L. Madsen. 2003. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. USA 100:13591-13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasai, Y., Y. Takahata, M. Manefield, and K. Watanabe. 2006. RNA-based stable isotope probing and isolation of anaerobic benzene-degrading bacteria from gasoline-contaminated groundwater. Appl. Environ. Microbiol. 72:3586-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan, A. A., R. F. Wang, W. W. Cao, D. R. Doerge, D. Wennerstrom, and C. E. Cerniglia. 2001. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:3577-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, S. J., R. C. Jones, C. J. Cha, O. Kweon, R. D. Edmondson, and C. E. Cerniglia. 2004. Identification of proteins induced by polycyclic aromatic hydrocarbon in Mycobacterium vanbaalenii PYR-1 using two-dimensional polyacrylamide gel electrophoresis and de novo sequencing methods. Proteomics 4:3899-3908. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S.-I., J. J. Kukor, K.-H. Oh, and H.-Y. Kahng. 2006. Evaluating the genetic diversity of dioxygenases for initial catabolism of aromatic hydrocarbons in Pseudomonas rhodesiae KK1. Enzyme Microb. Technol. 40:71-78. [Google Scholar]
- 23.Kiyohara, H., K. Nagao, K. Kouno, and K. Yano. 1982. Phenanthrene-degrading phenotype of Alcaligenes faecalis AFK2. Appl. Environ. Microbiol. 43:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiyohara, H., K. Nagao, and K. Yana. 1982. Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates. Appl. Environ. Microbiol. 43:454-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiyohara, H., N. Takizawa, H. Date, S. Torigoe, and K. Yano. 1990. Characterization of a phenanthrene degradation plasmid from Alcaligenes faecalis AFK2. J. Ferment. Bioeng. 69:54-56. [Google Scholar]
- 26.Krivobok, S., S. Kuony, C. Meyer, M. Louwagie, J. C. Willison, and Y. Jouanneau. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J. Bacteriol. 185:3828-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid sequencing techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 28.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laurie, A. D., and G. Lloyd-Jones. 2000. Quantification of phnAc and nahAc in contaminated New Zealand soils by competitive PCR. Appl. Environ. Microbiol. 66:1814-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozada, M., J. P. Riva Mercadal, L. D. Guerrero, W. D. Di Marzio, M. A. Ferrero, and H. M. Dionisi. 2008. Novel aromatic ring-hydroxylating dioxygenase genes from coastal marine sediments of Patagonia. BMC Microbiol. 8:50. doi: 10.1186/1471-2180-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackay, D., W. Y. Shiu, and J. C. Ma. 1992. Illustrated handbook of physical-chemical properties and environmental fate for organic chemicals. Lewis Publishers, Chelsea, MI.
- 33.Margesin, R., D. Labbe, F. Schinner, C. W. Greer, and L. G. Whyte. 2003. Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine alpine soils. Appl. Environ. Microbiol. 69:3085-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marlowe, E. M., J. M. Wang, I. L. Pepper, and R. M. Maier. 2002. Application of a reverse transcription-PCR assay to monitor regulation of the catabolic nahAc gene during phenanthrene degradation. Biodegradation 13:251-260. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, S., R. Moser, A. Neef, U. Stahl, and P. Kämpfer. 1999. Differential detection of key enzymes of polyaromatic-hydrocarbon-degrading bacteria using PCR and gene probes. Microbiology 145:1731-1741. [DOI] [PubMed] [Google Scholar]
- 36.Moser, R., and U. Stahl. 2001. Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria. Appl. Microbiol. Biotechnol. 55:609-618. [DOI] [PubMed] [Google Scholar]
- 37.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagao, K., N. Takizawa, and H. Kiyohara. 1988. Purification and properties of cis-phenanthrene dihydrodiol dehydrogenase in Alcaligenes faecalis AFK2. Agric. Biol. Chem. 52:2621-2623. [Google Scholar]
- 39.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pumphrey, G. M., and E. L. Madsen. 2008. Field-based stable isotope probing reveals the identities of benzoic acid-metabolizing microorganisms and their in situ growth in agricultural soil. Appl. Environ. Microbiol. 74:4111-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiagen. 2005. RNAprotect bacteria reagent handbook, 2nd ed. Qiagen, Valencia, CA.
- 42.Samanta, S. K., A. K. Chakraborti, and R. K. Jain. 1999. Degradation of phenanthrene by different bacteria: evidence for novel transformation sequences involving the formation of 1-naphthol. Appl. Microbiol. Biotechnol. 53:98-107. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Singleton, D. R., M. Hunt, S. N. Powell, R. Frontera-Suau, and M. D. Aitken. 2007. Stable-isotope probing with multiple growth substrates to determine substrate specificity of uncultivated bacteria. J. Microbiol. Methods 69:180-187. [DOI] [PubMed] [Google Scholar]
- 45.Singleton, D. R., S. N. Powell, R. Sangaiah, A. Gold, L. M. Ball, and M. D. Aitken. 2005. Stable-isotope probing of bacteria capable of degrading salicylate, naphthalene, or phenanthrene in a bioreactor treating contaminated soil. Appl. Environ. Microbiol. 71:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singleton, D. R., R. Sangaiah, A. Gold, L. M. Ball, and M. D. Aitken. 2006. Identification and quantification of uncultivated proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ. Microbiol. 10:1736-1745. [DOI] [PubMed] [Google Scholar]
- 47.Tchelet, R., R. Meckenstock, P. Steinle, and J. R. van der Meer. 1999. Population dynamics of an introduced bacterium degrading chlorinated benzenes in a soil column and in sewage sludge. Biodegradation 10:113-125. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toledo, F. L., C. Calvo, B. Rodelas, and J. González-López. 2006. Selection and identification of bacteria isolated from waste crude oil with polycyclic aromatic hydrocarbons removal capacities. Syst. Appl. Microbiol. 29:244-252. [DOI] [PubMed] [Google Scholar]
- 50.Vacca, D. J., W. F. Bleam, and W. J. Hickey. 2005. Isolation of soil bacteria adapted to degrade humic acid-sorbed phenanthrene. Appl. Environ. Microbiol. 71:3797-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Meer, J. R., W. Roelofsen, G. Schraa, and A. J. B. Zehnder. 1987. Degradation of low concentrations of dichlorobenzenes and 1,2,4-trichlorobenzene by Pseudomonas sp. strain P51 in nonsterile columns. FEMS Microbiol. Ecol. 45:333-341. [Google Scholar]
- 52.Vinas, M., J. Sabate, M. J. Espuny, and A. M. Solanas. 2005. Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl. Environ. Microbiol. 71:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willems, A., E. Falsen, B. Pot, E. Jantzen, B. Hoste, P. Vandamme, M. Gillis, K. Kersters, and J. De Ley. 1990. Acidovorax, a new genus for Pseudomonas facilis, Pseudomonas delafieldii, E. Falsen (EF) group 13, EF group 16, and several clinical isolates, with the species Acidovorax facilis comb. nov., Acidovorax delafieldii comb. nov., and Acidovorax temperans sp. nov. Int. J. Syst. Evol. Microbiol. 40:384-398. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, M. S., C. Bakermans, and E. L. Madsen. 1999. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl. Environ. Microbiol. 65:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winderl, C., B. Anneser, C. Griebler, R. U. Meckenstock, and T. Lueders. 2008. Depth-resolved quantification of anaerobic toluene degraders and aquifer microbial community patterns in distinct redox zones of a tar-oil contaminant plume. Appl. Environ. Microbiol. 74:792-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yen, K. M., and C. M. Serdar. 1988. Genetics of naphthalene catabolism in pseudomonads. Crit. Rev. Microbiol. 15:247-268. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, H. W., T. G. Luan, F. Zou, and N. F. Y. Tam. 2008. Different bacterial groups for biodegradation of three- and four-ring PAHs isolated from a Hong Kong mangrove sediment. J. Hazard. Mater. 152:1179-1185. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, N. Y., S. L. Fuenmayor, and P. A. Williams. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]