Abstract
We compared dideoxy sequencing of cloned chaperonin-60 universal target (cpn60 UT) amplicons to pyrosequencing of amplicons derived from vaginal microbial communities. In samples pooled from a number of individuals, the pyrosequencing method produced a data set that included virtually all of the sequences that were found within the clone library and revealed an additional level of taxonomic richness. However, the relative abundances of the sequences were different in the two datasets. These observations were expanded and confirmed by the analysis of paired clone library and pyrosequencing datasets from vaginal swabs taken from four individuals. Both for individuals with a normal vaginal microbiota and for those with bacterial vaginosis, the pyrosequencing method revealed a large number of low-abundance taxa that were missed by the clone library approach. In addition, we showed that the pyrosequencing method generates a reproducible profile of microbial community structure in replicate amplifications from the same community. We also compared the taxonomic composition of a vaginal microbial community determined by pyrosequencing of 16S rRNA amplicons to that obtained using cpn60 universal primers. We found that the profiles generated by the two molecular targets were highly similar, with slight differences in the proportional representation of the taxa detected. However, the number of operational taxonomic units was significantly higher in the cpn60 data set, suggesting that the protein-encoding gene provides improved species resolution over the 16S rRNA target. These observations demonstrate that pyrosequencing of cpn60 UT amplicons provides a robust, reliable method for deep sequencing of microbial communities.
Scientific interest in human microbial communities is growing, and basic concepts about the “human microbiome” are evolving rapidly (3, 34). Molecular phylogenetic analysis of 16S rRNA-encoding DNA sequences has revealed a vast diversity of uncultured microbial symbionts that influence animal physiology in ways only beginning to be understood. In particular, microbial species inhabiting the human vagina are thought to play an important role in host health (10). A shift in the composition of the vaginal microbiota from “normal” (Lactobacillus dominated) to a state defined as bacterial vaginosis (BV; increased abundance of gram-negative organisms) is associated with a range of negative outcomes, including pelvic inflammatory disease, preterm births, and the acquisition of sexually transmitted diseases (21, 22, 37). This observation has led to an increased interest in determining the composition of the vaginal microbiota by culture-independent methods (8, 11, 17, 25, 30, 35, 36). However, established cloning and sequencing techniques remain time- and labor-intensive, severely limiting the reach of phylogenetic or functional surveys of microbial communities across body sites, individuals, geographic areas, and scales of time.
The advent of next-generation ultra-high-throughput sequencing technologies, in particular, the GS FLX (454 Life Sciences, Branford, CT), has removed an important quantitative barrier in molecular analysis by increasing the number of reads from a gene or genome by orders of magnitude in a single run (20). Unfortunately, the short average length of pyrosequencing reads (∼200 bp compared to ∼700 bp using dideoxy sequencing) presents a new set of problems. The results of recent application of this technology to analysis of 16S rRNA gene sequences from microbes in vaginal samples have demonstrated that short reads are more likely to generate matches to multiple sequences in the rRNA sequence database and that taxonomic and phylogenetic resolution was limited due to strong similarities between 16S rRNA sequences from closely related species (32).
An alternative molecular target for microbial identification and phylogenetic analysis is cpn60, a gene that encodes the 60-kDa chaperonin or heat shock protein (HSP60/GroEL) (13). The cpn60 gene is universal in eubacteria and eukaryotes and an extensive, curated reference database is available (13) (http://cpndb.cbr.nrc.ca). The cpn60 universal target (UT) offers key advantages, including short target length (549 to 567 bp), sufficient resolving power to distinguish closely related species and subspecies, and a relatively uniform distribution of variability across the entire length of the target (9, 12). The use of the cpn60 UT has been well established for phylogenetic analysis of complex samples (4, 14) and has recently been applied to vaginal microbial communities (11). In the present study, we examined the feasibility of pyrosequencing for determining the composition of the vaginal microbiota using the cpn60 UT. We compared the microbial community structure generated by pyrosequencing of cpn60 amplicons using the GS FLX with dideoxy sequencing based on clone libraries generated from the same samples. In addition, we evaluated the microbial community profiles generated by pyrosequencing of cpn60 UT amplicons and 16S rRNA amplicons from the same vaginal samples.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Vaginal samples were collected as part of a larger study in a cohort of commercial sex workers in Nairobi, Kenya (18). Midvaginal swabs were collected by a single physician during 6-month follow-up examinations. A separate swab was taken and rolled onto a glass slide for evaluation of BV using the criteria of Nugent (24). Swab heads were snipped into freezing buffer and stored at −80°C until DNA extraction as described previously (27). Sample DNA was extracted by using Instagene Matrix (Bio-Rad, Mississauga, Ontario, Canada). Initially, equal volumes of DNA extracted from 20 individuals with or without BV was pooled prior to being amplified as described below for cpn60 clone library construction and pyrosequencing (“20-pool”; see Table 1). For subsequent samples, extracts were analyzed on an individual basis (Table 1).
TABLE 1.
Samples chosen for clone library construction and pyrosequencing
| Sample | Individual(s) | BV status | Use/comment |
|---|---|---|---|
| 20-pool | Pooled template DNA from 20 individuals | BV/normal | Paired cpn60 clone library/pyrosequencing |
| ind001 | 001 | Normal | Paired cpn60 clone library/pyrosequencing |
| ind006 | 006 | Normal | Paired cpn60 clone library/pyrosequencing |
| ind054 | 054 | BV | Paired cpn60 clone library/pyrosequencing |
| ind027 | 027 | BV | Paired cpn60 clone library/pyrosequencing |
| 4-pool | 001, 006, 054, 027 | BV/normal | 16S rRNA pyrosequencing |
| ind166 | 166 | BV | Technical replicate of cpn60 pyrosequencing |
Clone libraries.
The cpn60 UT was amplified and clone libraries were prepared from each pooled or individual sample as previously described (11), using two primer sets at a 1:3 ratio to improve representation of complex microbiota (14).
Pyrosequencing of cpn60 UT amplicons.
For high throughput sequencing using the GS FLX platform, cpn60 UT amplicons were prepared using the same primers and templates as for the clone libraries and all amplicons were purified on a 1.5% agarose gel. The sequencing libraries were prepared using the GS DNA library preparation kit and emulsion PCR (emPCR) was performed with a GS emPCR kit I or kit II as suggested by the manufacturer (Roche Diagnostics, Laval, Canada), except that the amplicons were treated as sheared DNA so the nebulization step normally performed in the library procedure was omitted. For the analysis of the 20-pool (Table 1), the GS FLX run was set up in one region of a 16-region gasket. Amplicons from the 20-pool were ligated to the linkers used for emPCR and sequencing. For the analysis of individual samples, a set of cpn60 UT primers containing unique sequence tags was used (see Table S1 in the supplemental material). The set of upstream primers (hybridizing to the 5′ end of the cpn60 UT) each contained at their 5′ ends the primer used in the subsequent emPCR and sequencing (primer A), followed by a unique four-base sequence tag and the cpn60 UT primer sequence. Each of the upstream primers was paired with a downstream primer that contained the other emPCR primer sequence immediately upstream of the cpn60 UT primer sequence. All primers were HPLC purified. The sequence tags enabled a multiplexed run in which individual samples were each amplified with a primer set containing a unique sequence tag. The resulting sequence data was then sorted according to the tag prior to subsequent analysis.
Pyrosequencing of 16S rRNA amplicons.
Amplicons were generated from each of four individuals (“4-pool”; Table 1) using the untagged broad-range PCR primers L27F and 355R, which target variable regions V1 and V2 (30). Amplicons from the four individuals were pooled by volume, purified on a 1.5% agarose gel, and ligated to PCR linkers containing a Roche multiplexing ID sequence prior to pyrosequencing. The 16S rRNA amplicons were sequenced on the same picotiter plate used for the individual cpn60 samples and identified after pyrosequencing by the unique multiplexing ID.
Data management and taxonomic assignment.
Clone library and pyrosequencing data were analyzed using a bioinformatic pipeline to evaluate each sequencing read and determine the optimal possible taxonomic label(s). Reads derived from Sanger sequencing were base-called using Phred (6, 7). Cloned insert sequences were identified and vector sequences removed by using Lucy (2). Pyrosequencing data was processed by using the default on-rig procedures from 454/Roche. Filter-passing reads were used in the subsequent analyses for each of the pyrosequencing libraries. All sequencing data was imported and warehoused using the APED software package (http://aped.sourceforge.net).
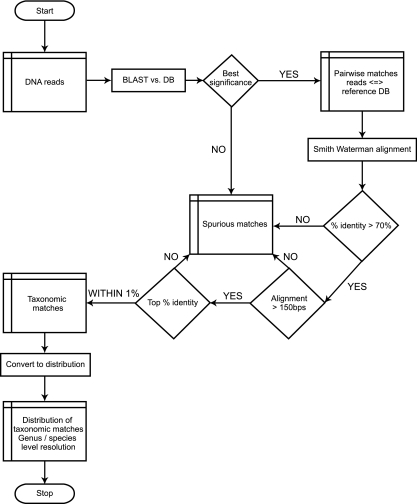
A combination of BLAST and Smith-Waterman alignments (watered-BLAST) was used to identify the most significant matches between each individual sequence read and an appropriate reference sequence database. As outlined in Fig. 1, each read was initially compared to the reference database using BLAST (nonstandard parameters: −F F) (1). The BLAST hits at the best significance level were examined further by performing a Smith-Waterman alignment of the sequence read and each hit in the reference database that was at the best significance level (29). Smith-Waterman alignments were generated by using the water program from EMBOSS (26). The optimal local alignments produced from water were examined, and those which had a percent identity of <70% or a length of <150 bp were deemed spurious matches and were rejected from further analysis. In order to identify the best putative taxonomic assignment possible for each read, the water alignments were limited to those within 1% identity of the top percent identity match to the reference database. In cases where there were multiple hits to the reference database that were within 1% of the best match to the reference database, a read was annotated as matching each of the reference database sequences. The resulting taxonomic assignments were used to calculate distributions of organism abundance at the genus and species levels. To account for variation between the sizes of pyrosequencing libraries, organism abundance was normalized to the respective library size as follows: normalized abundance = (no. of taxonomic matches/no. of total matches) × 100.
FIG. 1.
Data analysis flowchart of the watered-BLAST pipeline used to assign a taxonomic label for each sequence from the Sanger and GS FLX datasets.
The watered-BLAST analysis was implemented in Perl by using BioPerl (31). The code is maintained as part of the APED software package (http://sourceforge.net/projects/aped).
Reference databases for watered-BLAST matching.
For cpn60 data, a nonredundant, customized database of cpn60 UT sequences was created and was comprised of a single reference strain for each species (cpnDB_nr). Unique sequences (did not match anything in cpnDB_nr) obtained from 25 cultured isolates derived from vaginal swabs of women from the cohort were added to complete the customized database cpnDB_NR_vag (1,373 sequences). For 16S rRNA data, each read was compared by the same watered-BLAST method to a database of 16S rRNA sequences from RDP (19). The 16S rRNA database (RDP_isolates) consisted of 66,304 full-length or nearly full-length (>1,200 bp) sequences from RDP that were annotated as “good quality” data from isolates (metagenomic and uncultured data excluded). To facilitate direct pairwise comparison of 16S rRNA and cpn60 pyrosequencing libraries derived from the same samples, customized reference databases were generated for each gene target. These databases contained 505 nonredundant type strain sequences representing species that are in common between RDP and cpnDB and for which the entire V1 to V8 region of the 16S rRNA gene was available. The V1 to V8 region was defined as the region corresponding to nucleotides 8 to 1406 of E. coli 16S rRNA.
Rarefaction.
Rarefaction curves and richness estimators (Chao1 and ACE) were calculated from the pyrosequencing and Sanger data using EstimateS (version 8.0.0; R. Colwell, University of Connecticut [http://purl.oclc.org/estimates]) as described previously (15, 16). EstimateS analyses were performed on 100 random samplings, without replacement, and where appropriate the classical method for Chao1 calculations was used.
Pairwise comparison of libraries.
In order to evaluate whether there were differences observed in the taxonomic composition of libraries, the relative abundance for each genus/species in the comparison was calculated as follows: relative abundance = log2 (normalized abundance library A/normalized abundance library B). When species were represented by a single read in one library and by >1 read in the paired library, that species was considered unrepresented in the single-read library.
Phylogenetic trees.
The phylogenetic tree from the 20-pool (Table 1) was drawn based on a CLUSTALW (33) alignment of the cpn60 UT using PHYLIP (Phylogeny Inference Package) version 3.5c (J. Felsenstein, distributed by the author, Department of Genetics, University of Washington, Seattle). The alignment was sampled using bootstrap, and distances were calculated using the F84 distance method.
Sequence clustering and assembly.
Assemblies of the pyrosequencing data were generated by using a gsAssembler/newbler (454/Roche) with the default parameters. The resulting number of contigs from the assembly of the pyrosequencing data was used to give an approximation of the number of distinct sequences sampled for a given library.
OTU diversity calculation for Prevotella spp.
For each of the pyrosequencing libraries generated for cpn60 and 16S rRNA, the reads identified by watered-BLAST as Prevotella spp. were processed by using t_coffee (23) (nonstandard parameters: −mode quickaln) to generate a PHYLIP format output file of their multiple sequence alignment. Distance matrix files suitable for input to DOTUR (28) were created with dnadist from the PHYLIP package. The number of operational taxonomic units (OTUs) was calculated at various sampling depths and percent identity cutoffs using the farthest-neighbor algorithm of DOTUR (28).
RESULTS
Clone library and pyrosequencing analysis of the 20-pool.
We initially compared cpn60 UT sequences generated from cloned cpn60 UT fragments using the dideoxy sequencing method (“cpn60 Sanger” data set) to sequences generated by using the library-independent pyrosequencing approach (“cpn60 GS FLX” data set) using cpn60 UT amplicons generated from pooled DNA from 20 individuals (Table 1). A total of 324 library clones were analyzed from this pool, which represented 72 distinct cpn60 UT sequences. The taxonomic distribution of the cpn60 Sanger data set included Lactobacillales, Clostridiales, Bacteroidetes, and Actinobacteria (see Table S2 in the supplemental material), consistent with the expected composition of the vaginal microbiota in normal and BV individuals (11, 17).
The cpn60 GS FLX data set generated from the 20-pool contained 5,938 unassembled individual filter pass reads in a single run with a mean length of 197 bp. Most of these sequences were categorized as cpn60 (4,410 of 5,938 reads or 74%), with 1,129 sequences discarded due to insufficient length (<150 bp) and the remainder identified as human non-cpn60, bacterial non-cpn60, or unknown. BLAST comparison of the sequences from the cpn60 GS FLX data set to the Sanger data set revealed that 3,509 of the sequences matched a sequence found in the latter data set (where a match is defined as having ≥97% identity over ≥150 nucleotides). The remaining 901 sequences were unique to the cpn60 GS FLX data set and could be reduced to 72 different partial cpn60 sequences (see Table S2 in the supplemental material). Therefore, the total cpn60 GS FLX data set for the 20-pool included 144 partial cpn60 sequences, comprised of 72 that were also found in the smaller Sanger data set and an additional 72 sequences that were found only in the larger GS FLX data set.
Overlap of Sanger and GS FLX data for the 20-pool.
A comparison of the paired Sanger and GS FLX datasets from the 20-pool showed that the Sanger data was essentially entirely included with the pyrosequencing data (Fig. 2). Although there were 18 sequences in the Sanger data set that had no identical match in the GS FLX data set, 17 of these sequences were found to reside in the phylogeny with closely neighboring with sequences from the GS FLX data set. Only one Sanger sequence (FJ594055) was completely distinct from anything found in the GS FLX data set. In contrast, the 901 sequences found only in the GS FLX data set ranged from 96 to only 80% identical to their nearest match in the Sanger data (range, 80 to 96%; mean, 92%), indicating that the deeper pyrosequencing had resulted in the identification of novel sequences. These pyrosequencing-only reads included sequences representing previously described vaginal organisms such as Lactobacillus delbrueckii, Lactobacillus crispatus, and Mobiluncus curtsii, as well as sequences with weak similarity to anything in the Sanger data set or in the cpnDB reference database (see Table S2 in the supplemental material).
FIG. 2.
Phylogenetic tree of sequences found in the Sanger data set generated from the 20-pool sample. The numbers in brackets after each sequence indicate the frequency with which each sequence is represented in the GS FLX data set compared to the Sanger data set, as described in the text (positive numbers indicate relatively greater frequency in the GS FLX data set). Sequences found only in the Sanger data set are indicated. The tree is a consensus of 100 neighbor-joined trees. Numbers at the nodes are bootstrap values out of 100. Sequences are labeled with their GenBank accession numbers.
Proportional representation of taxa in Sanger and GS FLX datasets.
Although the taxonomic profiles generated for the 20-pool by the two methods were nearly identical (Fig. 2), the relative proportions of the sequences represented were different between the two datasets (Fig. 2 and 3). To quantify the relative proportions of each sequence in each data set while normalizing for library size, we determined the log2 of the ratio of the percentage of each sequence in its corresponding data set (see Materials and Methods). According to this calculation, a value of “0” indicates the same proportion in the two datasets, while a positive value indicates that a sequence is more abundant in the GS FLX data set, and a negative value indicates that it is less abundant in the GS FLX data set. We calculated frequency ratios for each bacterial family in the Sanger data set (which were also found in 3,509 of the sequences from the GS FLX data set), as well as in the total data set (including the additional 901 sequences that were found only in the GS FLX data set).
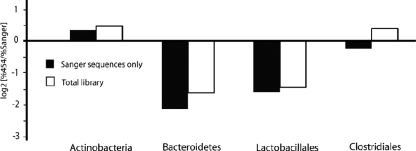
FIG. 3.
Relative representation of bacterial families in the Sanger/pyrosequencing and total datasets for the 20-pool. The proportions of sequences representing each family were compared among the sequences found in both datasets (▪) and among the total data set (□).
In the Sanger data set, the Actinobacteria were the only family to be overrepresented in the GS FLX data, while the Bacteroidetes and Lactobacillales were found less frequently in the GS FLX data; Clostridiales were represented with approximately equal frequency in the two datasets (Fig. 3). When all of the sequences were considered (the sequences found in both datasets plus the sequences found only in the GS FLX data set), a similar pattern emerged, although the Bacteroidetes and Lactobacillales were slightly less underrepresented in the GS FLX data. However, Clostridiales were overrepresented in the total data set (Fig. 3), which is consistent with the fact that many of the sequences that were unique to the GS FLX data set were Clostridiales (see Table S2 in the supplemental material). This observation suggests that many of the lower-abundance organisms in this sample are Clostridiales.
Sampling depth.
Rarefaction analysis of the number of OTUs observed in each of the Sanger and GS FLX datasets for the 20-pool showed the increased sampling depth obtained with the pyrosequencing method (see Fig. S1 in the supplemental material). The species accumulation curve for the Sanger data exactly followed the curve generated for the GS FLX data but stopped far short of the depth obtained in the GS FLX data set. However, the sampling was evidently not complete even in the larger GS FLX data set; the Chao1 and ACE richness estimators yielded values of 178.5 and 77.6 OTUs, respectively, for this pooled sample (data not shown). For each of the individual samples, pyrosequencing of the cpn60 UT appeared to result in nearly complete sampling of the taxonomic richness of the samples (see Fig. S1 in the supplemental material).
Paired clone libraries and pyrosequencing of individuals.
To further evaluate the efficacy of metagenomic profiling by pyrosequencing of the cpn60 UT, we prepared larger clone libraries and matching larger GS FLX datasets for four individuals with normal or BV vaginal microbiota (Table 1). The numbers of reads generated for each individual, along with the proportion of reads that were retained after the watered-BLAST analysis (showed > 70% identity to a sequence in the reference database), are shown in supplemental Table S3 in the supplemental material. The taxonomic distributions of the sequences identified by both methods were consistent with the clinical diagnosis of the individuals. For example, individuals 001 and 006 both had normal Nugent scores (Table 1), and both sequencing methods showed a predominance of Lactobacillales (Fig. 4). Similarly, individuals 027 and 054 were diagnosed with BV and showed a more diverse vaginal microbiota with Actinobacteria and Bacteroidetes predominant and Lactobacillales being less abundant (Fig. 4). Consistent with observations in the 20-pool (Fig. 3), each individual sample showed essentially the same taxonomic distribution in the paired datasets; however, differences were apparent in the proportions of the different taxa identified. For all four individuals, the majority (81 to 97%) of the sequences identified in the GS FLX datasets were also identified in the corresponding Sanger datasets (Fig. 5). In addition, each individual showed a substantial number of sequences that were unique to the GS FLX data set; although the percentages were relatively small, the very large numbers of reads generated by the pyrosequencing method resulted in a large number of taxa identified uniquely in the GS FLX data set (Fig. 5). For example, individual 006 had only a single taxon (Lactobacillus iners) represented in 862 sequences identified by dideoxy sequencing of clones, while the GS FLX data set revealed an additional 39 taxa in this sample (Fig. 5). Similarly, the vaginal microbiota of individual 001 consisted mostly of L. crispatus in the Sanger data set, and the GS FLX data set revealed an additional 18 taxa in this sample. The two individuals with BV each contained more taxa than the normal individuals in the Sanger datasets and showed a gain of 31 and 33 taxa in the corresponding GS FLX datasets (Fig. 5).
FIG. 4.
Proportional representation of taxonomic categories in Sanger, Sanger overlap, (sequences in common to both datasets) and all sequences for each of four individuals. Individuals 001 and 006 were normal by microscopy, while individuals 027 and 054 were diagnosed with BV (Table 1).
FIG. 5.
Taxonomic composition of individual vaginal microbiota as determined by clone libraries and pyrosequencing. The taxonomic assignments of sequences found in the Sanger (A, C, E, and G) and GS FLX (B, D, F, and H) datasets are shown. For B, D, F, and H, additional taxa found in the GS FLX datasets are shown as stacked bar graphs, while the Sanger-overlap data set is shown as a pie chart. Colors are used to indicate bacterial families: yellow, Firmicutes; blue, Actinobacteria; red, Bacteroidetes; green, Proteobacteria. Species abbreviations: Lin, L. iners; Lcr, L. crispatus; L6, Lactobacillus sp. strain L6; Lje, L. jensenii; Pbu, P. buccalis; Gva, G. vaginalis; N156, Nairobi isolate 156 (Actinobacteria spp.); Afa, Acidovorax facilis; Pme, P. melaninogenica; Ava, A. vaginae; Mel, Megasphaera eldensii; Pin, P. intermedia; N137, Nairobi isolate 137 (Actinobacteria spp.); N160, Nairobi isolate 160 (Actionobacteria spp.); Lsa, Lactobacillus salivarius; Fma, Finegoldia magna; Tca, Thermosinus carboxydivorans.
Reproducibility of pyrosequencing using cpn60.
To determine whether the same taxonomic profile is generated from the same sample in multiple pyrosequencing runs, we analyzed technical replicates of a single sample (individual 166; Table 1), with each sample amplified independently using different sequence-tagged primers. As shown in Fig. 6A, the relative abundances of the 21 taxa identified in each of the two runs were virtually identical. In addition, of the 18 species that were uniquely identified in one or the other datasets, only one (L. iners) represented more than 0.03% of the data for the library (0.29%). Similar results were obtained with a technical replicate of ind027 (data not shown).
FIG. 6.
Relative abundances of genera and species found in the technical replicates of individual 166 (A) and in the cpn60 and 16S rRNA GS FLX datasets for the four individuals pooled (B). For panel B, the shaded area represents the maximum observed variability expected from technical replicates of the same sample (A). Abbreviations: N137, Actinobacteria sp. strain N137; N156. Actinobacteria sp. strain N156; N160, Actinobacteria sp. strain N160; Gva, G. vaginalis; Ava, A. vaginae; Bov, Bacteroides ovatus; Mhy, Megamonas hypermegale; Pgi, Porphyromonas gingivalis; Pbi, Prevotella bivia; Pbu, P. buccalis; Pco, P. corporis; Pdi, P. disiens; Pin, P. intermedia; Pme, P. melaninogenica; Por, P. oralis; Pru, P. ruminocola; Prevotella sp., all Prevotella species; Fma, Finegoldia magna; Mel, Megasphaera elsdenii; Tca, Thermosinus carboxydivorans; Afa, Acidovorax facilis; Lcr, Lactobacillus crispatus; Lga, L. gasseri; Lin, L. iners; Lre, L. reuteri; Lsa, L. salivarius; Lactobacillus sp., all Lactobacillus species; Ssa, Streptococcus salivarius.
Pyrosequencing of 16S rRNA amplicons.
We generated amplicons from the 16S rRNA target for individuals 001, 006, 027, and 054 (4-pool; Table 1) and pooled the amplicons prior to generating sequence data on a GS FLX instrument. These data were analyzed by watered-BLAST using a reference database of sequences from RDP that represented full-length or nearly full-length 16S rRNA sequences from cultured isolates. The 16S rRNA-based taxonomic profile of the vaginal microbiota of this sample was generally consistent with the profile generated by cpn60 UT sequencing (data not shown).
Comparison of 16S rRNA and cpn60 pyrosequencing data.
To compare directly the data obtained by cpn60 pyrosequencing to that obtained for the 16S rRNA target, we aligned by watered-BLAST the 16S rRNA and cpn60 pyrosequencing reads from the same four individuals to the paired databases representing 505 nonredundant type strain reference sequences found in both the RDP and cpnDB. This approach resulted in a taxonomic profile of the matched samples using exactly analogous reference databases. This analysis revealed that 15 taxa were identified in these samples by both targets (Fig. 6B) and that the relative abundances of most taxa were similar in the two datasets. However, the 16S rRNA data set contained a relatively higher abundance of Atopobium vaginae, Prevotella spp., and Lactobacillus gasseri and a lower abundance of Gardnerella spp. relative to the cpn60 data set. Only six species were found uniquely in each of the cpn60 or 16S rRNA pyrosequencing datasets at an abundance of >0.1% and combined were less than ∼2% of the total data (Table 2).
TABLE 2.
Taxa that were uniquely found in 16S rRNA or cpn60 pyrosequencing data and represented at >0.1% of the matches
| Taxon | No. of matches | % of library |
|---|---|---|
| Unique to cpn60 | ||
| Megamonas hypermegale | 547 | 1.27 |
| Acidovorax facilis | 380 | 0.88 |
| Prevotella intermedia | 236 | 0.55 |
| Campylobacter lari | 134 | 0.31 |
| Sphingomonas wittichii | 64 | 0.15 |
| Comamonas terrigena | 59 | 0.14 |
| Unique to 16S rRNA | ||
| Fusobacterium nucleatum | 265 | 2.13 |
| Anaerococcus vaginalis | 194 | 1.56 |
| Lactobacillus ruminis | 22 | 0.18 |
| Peptoniphilus asaccharolyticus | 17 | 0.14 |
| Clostridium aldrichii | 14 | 0.11 |
| Mycoplasma genitalium | 13 | 0.10 |
To determine whether the numbers of OTUs in the paired datasets were similar, we assembled the sequences within each data set using the default parameters of the assembly program Newbler (see Materials and Methods). This rough estimate of sequence similarity identified 47 contigs within the 16S rRNA data and 253 within the cpn60 data, suggesting that the taxonomic richness of the cpn60 data is greater. To address this question directly, we used DOTUR (28) to calculate the number of OTUs present within a subset of each of the two datasets. We chose sequences that were identified as Prevotella spp. because they were identified in equal proportions in the 16S rRNA and cpn60 datasets (14.3 and 16.6%, respectively; Fig. 6B) and because the sizes of these subsets were computationally tractable to this analysis. At the two identity cutoffs chosen (95 and 97%), DOTUR calculated significantly more OTUs in the cpn60 data than in the 16S rRNA data (P < 0.05) at sampling depths greater than 100 sequences (Fig. 7).
FIG. 7.
Calculation of the number of OTUs for each of the 16S rRNA and cpn60 GS FLX subsets identified as Prevotella spp. The number of OTUs calculated by the farthest-neighbor algorithm of DOTUR are reported at various sampling depths for percent identity cutoffs of 3% (cpn60, ▪; 16S rRNA, □) and 5% (cpn60, ░⃞; 16S rRNA, ▧). Error bars represent 95% confidence intervals.
DISCUSSION
We compared clone library and pyrosequencing data generated from the vaginal microbiota of individuals and pools using the cpn60 UT. In addition, we directly compared the results generated by cpn60 UT pyrosequencing to those obtained using 16S rRNA. We also describe a novel bioinformatic analysis pipeline for assignment of putative taxonomic identities using “watered-BLAST”. To our knowledge, this is the first description of the application of high throughput pyrosequencing for analysis of amplicons derived from microbial communities using a target other than 16S rRNA.
As a pilot experiment, we compared the taxonomic profile of pooled vaginal microbiota samples (20-pool) generated from a small clone library to that obtained by a small GS FLX data set generated from a single region of a 16-region run. In general, the taxa that were identified were consistent with what would be expected from a human vaginal microbial community (11, 17). In addition, we found that the microbial profiles generated by the two sequencing methods agreed very well with the smaller Sanger data set virtually entirely contained within the larger GS FLX data set. A substantial amount of taxonomic depth was gained with the pyrosequencing method, essentially doubling the number of taxa identified in the same samples. These results are consistent with those obtained by Edwards et al. (5), who found similar taxonomic distributions of 16S rRNA sequences in clone libraries and GS FLX datasets in samples taken from deep mines. We also found that the proportions of sequences represented in the two datasets were somewhat different, with certain taxa, especially Clostridiales, present in a higher proportion of the reads in the GS FLX data set. The fact that the proportions of the sequences represented were different between the Sanger data and the GS FLX data is not entirely surprising, given the different biases that apply to each of these methods. Although both methods are equally subject to representational biases that can arise in the PCR step since the same primers, templates, and amplification conditions were used for both methods, the library method has the additional bias of cloning the PCR products that are generated. The cloning step could introduce biases into the Sanger data set; for example, colonies with different inserts may not grow equally well on the selection plates. We have observed in previous work that the frequency with which clones are represented in cpn60 UT libraries does not always reflect the abundance of the organism as measured by methods such as quantitative PCR (4). Therefore, we expect that the pyrosequencing data reflect more accurately than the clone libraries the composition of the PCR product pool.
Since the number of reads generated in this pilot experiment was low for a 1/16 region on the GS FLX (which typically generates in excess of 12,000 sequences in this format), we generated expanded datasets containing paired Sanger and GS FLX data for four individuals. The results of this larger analysis also showed that the pyrosequencing method consistently revealed a far richer taxonomic composition of the vaginal microbiota in each individual than was shown with the clone library approach. This trend was particularly notable in the samples that were scored as normal by microscopy (individuals 001 and 006), which increased from 1 to 3 taxa in the clone libraries to 21 to 40 taxa in the corresponding GS FLX datasets. Samples from individuals with BV (027 and 054), which were more diverse in composition in their Sanger datasets, showed the same trends (4 to 12 taxa in the Sanger datasets versus 37 to 43 taxa in the GS FLX datasets). In four individual samples, we found that the Sanger data were nearly completely contained within the GS FLX data and that additional taxonomic richness was revealed with the GS FLX sequencing method.
We investigated the reproducibility of the pyrosequencing approach using cpn60 amplicons generated independently from the same sample and analyzed in two separate pyrosequencing reactions. We found that the taxonomic profile generated with this method was highly reproducible, including the proportions of reads represented at the species level. Since the maximal variation between runs for a given species was ∼2-fold, we suggest that this is the normal range of variation within technical replicates using this method. Although 18 species were found specifically in one of the two repeats, none of these represented more than 0.2% of the total data for a library. We conclude that the taxonomic profile generated using the sequence-tagged GS FLX approach is sufficiently robust that a single sample can be used for community analysis.
We also compared pyrosequencing data obtained from the same samples using the cpn60 UT and the more widely used 16S rRNA. In order to provide a valid, easily interpreted comparison of the taxonomic assignments given by the two targets, we prepared reference databases containing data for 505 isolates for which paired cpn60 UT and (near) full-length, good quality 16S rRNA sequence data are available. Using these databases for taxonomic assignments by watered-BLAST, we found that the profiles generated by the two targets on the same samples were virtually identical. A total of 16 species were found in both datasets, and while a few were represented with different proportional abundances within their respective datasets, the majority were represented with nearly equal abundances. Species with a proportional difference of more than two- to threefold likely represented real differences in their representation in the two datasets, since this is the maximal variation that was seen in the technical replicates. The different representation of some of the targets might be explained by differences in the efficiency with which various species are amplified by the universal primers. We found 12 species that were specifically represented in one or the other of the datasets generated by the 16S rRNA or cpn60 primers. However, none of these were more than ca. 2% of the sequences of the respective datasets, and most (10 of 12) were less than 1%. We can therefore conclude that the taxonomic profiles within the two datasets were essentially in agreement with one another.
It has been noted that protein-encoding genes may provide an increased level of resolution compared to the structural 16S rRNA-encoding gene (13, 28). However, we did not specifically address this question with the approach used above. Therefore, to compare the taxonomic richness of the data generated from the two targets, we used the farthest-neighbor algorithm of DOTUR to calculate the number of OTUs at various sampling depths for a genus whose relative abundance was similar across target libraries. The fact that cpn60 sequences consistently yielded a higher number of OTUs at each cutoff suggests that the sequences identified as Prevotella are more different from one another within the cpn60 data set than are the sequences within the 16S rRNA data set. We could not expand this observation to other genera since the sizes of the datasets made the generation of the distance matrices computationally intractable.
In summary, we found that pyrosequencing of cpn60 UT amplicons compared very favorably to the clone library approach as a method of characterizing a complex microbial system. Moreover, pyrosequencing of cpn60 amplicons yielded a taxonomic profile of a microbial community that was very similar to that generated by the 16S rRNA molecular target but with a higher level of taxonomic resolution. The very high number of reads that are generated by pyrosequencing resulted in a total data set that included essentially all of the sequences that were represented using the library method, along with additional sequences that greatly increased the number of distinct taxa that were identified. Since the pyrosequencing method does not require the cloning of amplicons, it is much less labor-intensive than sequencing of clone libraries, and it avoids the representational biases that can result from the cloning step. Pyrosequencing of cpn60 UT PCR products offers the ability to probe much deeper into the compositions of microbial ecosystems than is feasible using the library approach, making the detection of lower-abundance organisms possible. We conclude that generating microbial community profiles by pyrosequencing of cpn60 UT amplicons results in a reliable, reproducible taxonomic profile of a microbial community that can be used to identify low-abundance organisms that are typically missed by the clone library approach.
Supplementary Material
Acknowledgments
We thank Josh Neufeld for help with the rarefaction analysis and Erin Cadieu for help with the illustrations.
Footnotes
Published ahead of print on 6 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chou, H. H., and M. H. Holmes. 2001. DNA sequence quality trimming and vector removal. Bioinformatics 17:1093-1104. [DOI] [PubMed] [Google Scholar]
- 3.Dethlefsen, L., M. McFall-Ngai, and D. A. Relman. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449:811-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumonceaux, T. J., J. E. Hill, S. M. Hemmingsen, and A. G. Van Kessel. 2006. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 72:2815-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards, R. A., B. Rodriguez-Brito, L. Wegley, M. Haynes, M. Breitbart, D. M. Peterson, M. O. Saar, S. Alexander, E. C. Alexander, Jr., and F. Rohwer. 2006. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 7.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 8.Fredricks, D. N., T. L. Fiedler, K. K. Thomas, B. B. Oakley, and J. M. Marrazzo. 2007. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J. Clin. Microbiol. 45:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goh, S. H., S. Potter, J. O. Wood, S. M. Hemmingsen, R. P. Reynolds, and A. W. Chow. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hay, P. 2005. Life in the littoral zone: lactobacilli losing the plot. Sex. Transm. Infect. 81:100-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill, J. E., S. H. Goh, D. M. Money, M. Doyle, A. Li, W. L. Crosby, M. Links, A. Leung, D. Chan, and S. M. Hemmingsen. 2005. Characterization of vaginal microflora of healthy, nonpregnant women by chaperonin-60 sequence-based methods. Am. J. Obstet. Gynecol. 193:682-692. [DOI] [PubMed] [Google Scholar]
- 12.Hill, J. E., A. Paccagnella, K. Law, P. L. Melito, D. L. Woodward, L. Price, A. H. Leung, L. K. Ng, S. M. Hemmingsen, and S. H. Goh. 2006. Identification of Campylobacter spp. and discrimination from Helicobacter and Arcobacter spp. by direct sequencing of PCR-amplified cpn60 sequences and comparison to cpnDB, a chaperonin reference sequence database. J. Med. Microbiol. 55:393-399. [DOI] [PubMed] [Google Scholar]
- 13.Hill, J. E., S. L. Penny, K. G. Crowell, S. H. Goh, and S. M. Hemmingsen. 2004. cpnDB: a chaperonin sequence database. Genome Res. 14:1669-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, J. E., J. R. Town, and S. M. Hemmingsen. 2005. Improved template representation in cpn60 polymerase chain reaction (PCR) product libraries generated from complex templates by application of a specific mixture of PCR primers. Environ. Microbiol. 8:741-746. [DOI] [PubMed] [Google Scholar]
- 15.Hughes, J. B., and B. J. Bohannan. 2004. Application of ecological diversity statistics in microbial ecology, p. 1321-1344. In G. A. Kowalchuck, F. J. de Bruijn, I. M. Head, A. D. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishing, London, England.
- 16.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyman, R. W., M. Fukushima, L. Diamond, J. Kumm, L. C. Giudice, and R. W. Davis. 2005. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. USA 102:7952-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimani, J., R. Kaul, N. J. Nagelkerke, M. Luo, K. S. MacDonald, E. Ngugi, K. R. Fowke, B. T. Ball, A. Kariri, J. Ndinya-Achola, and F. A. Plummer. 2008. Reduced rates of HIV acquisition during unprotected sex by Kenyan female sex workers predating population declines in HIV prevalence. AIDS 22:131-137. [DOI] [PubMed] [Google Scholar]
- 19.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Money, D. 2005. The laboratory diagnosis of bacterial vaginosis. Can. J. Infect. Dis. Med. Microbiol. 16:77-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris, M., A. Nicoll, I. Simms, J. Wilson, and M. Catchpole. 2001. Bacterial vaginosis: a public health review. Br. J. Obstet. Gynecol. 108:439-450. [DOI] [PubMed] [Google Scholar]
- 23.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 24.Nugent, R. P., M. A. Krohn, and S. L. Hillier. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J. Clin. Microbiol. 29:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oakley, B. B., T. L. Fiedler, J. M. Marrazzo, and D. N. Fredricks. 2008. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 74:4898-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 27.Schellenberg, J., T. Blake Ball, M. Lane, M. Cheang, and F. Plummer. 2008. Flow cytometric quantification of bacteria in vaginal swab samples self-collected by adolescents attending a gynecology clinic. J. Microbiol. Methods 73:216-226. [DOI] [PubMed] [Google Scholar]
- 28.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195-197. [DOI] [PubMed] [Google Scholar]
- 30.Spear, G. T., M. Sikaroodi, M. R. Zariffard, A. L. Landay, A. L. French, and P. M. Gillevet. 2008. Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J. Infect. Dis. 198:1131-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stajich, J. E., D. Block, K. Boulez, S. E. Brenner, S. A. Chervitz, C. Dagdigian, G. Fuellen, J. G. Gilbert, I. Korf, H. Lapp, H. Lehvaslaiho, C. Matsalla, C. J. Mungall, B. I. Osborne, M. R. Pocock, P. Schattner, M. Senger, L. D. Stein, E. Stupka, M. D. Wilkinson, and E. Birney. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12:1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundquist, A., S. Bigdeli, R. Jalili, M. L. Druzin, S. Waller, K. M. Pullen, Y. Y. El-Sayed, M. M. Taslimi, S. Batzoglou, and M. Ronaghi. 2007. Bacterial flora-typing with targeted, chip-based pyrosequencing. BMC Microbiol. 7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turnbaugh, P. J., R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, and J. I. Gordon. 2007. The human microbiome project. Nature 449:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verhelst, R., H. Verstraelen, G. Claeys, G. Verschraegen, J. Delanghe, L. Van Simaey, C. De Ganck, M. Temmerman, and M. Vaneechoutte. 2004. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis, and bacterial vaginosis. BMC Microbiol. 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verstraelen, H., R. Verhelst, G. Claeys, M. Temmerman, and M. Vaneechoutte. 2004. Culture-independent analysis of vaginal microflora: the unrecognized association of Atopobium vaginae with bacterial vaginosis. Am. J. Obstet. Gynecol. 191:1130-1132. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, J. 2004. Managing recurrent bacterial vaginosis. Sex. Transm. Infect. 80:8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.