Abstract
Studies of phytoplasma-insect vector interactions and epidemiological surveys of plant yellows associated with the stolbur phytoplasma (StolP) require the identification of relevant candidate genes and typing markers. A recent StolP genome survey identified a partial coding sequence, SR01H10, having no homologue in the “Candidatus Phytoplasma asteris” genome but sharing low similarity with a variable surface protein of animal mycoplasmas. The complete coding sequence and its genetic environment have been fully characterized by chromosome walking. The vmp1 gene encodes a protein of 557 amino acids predicted to possess a putative signal peptide and a potential C-terminal transmembrane domain. The mature 57.8-kDa VMP1 protein is likely to be anchored in the phytoplasma membrane with a large N-terminal hydrophilic part exposed to the phytoplasma cell surface. Southern blotting experiments detected multiple sequences homologous to vmp1 in the genomes of nine StolP isolates. vmp1 is variable in size, and eight different vmp1 RsaI restriction fragment length polymorphism types could be distinguished among 12 StolP isolates. Comparison of vmp1 sequences revealed that insertions in largest forms of the gene encode an additional copy of a repeated domain of 81 amino acids, while variations in 11-bp repeats led to gene disruption in two StolP isolates. vmp1 appeared to be much more variable than three housekeeping genes involved in protein translation, maturation, and secretion and may therefore be involved in phytoplasma-host interactions.
The stolbur phytoplasma (StolP) is a phloem-restricted, noncultivable plant pathogen which infects a wide range of cultivated plants in Europe and in the Mediterranean Basin, such as solanaceous crops, grapevine, celery, sugar beet, strawberry, and lavender (17). Symptoms of stolbur disease, observed in annual crops since 1933 (26), are leaf discoloration, stunting, and abnormal floral development leading to sterility. In European vineyards, StolP causes grapevine yellows, the bois noir disease. StolP belongs to the 16SrXII-A group of the “Candidatus Phytoplasma” genus taxonomy, which is based mainly on 16S rRNA gene phylogeny, and its designation as “Candidatus Phytoplasma solani” has been proposed but not yet formally established (13, 28). The main reservoirs of StolP in France, Germany, and Italy are weeds such as bindweeds (Convolvulus arvensis and Calystegia sepium) or nettles (Urtica dioica), from which it is transmitted by planthoppers to other weeds or cultivated plants (5, 16, 27). According to tuf gene typing, StolP genotype VKI is associated with nettles and genotype VKII is associated with bindweed (27), while no clear association between StolP genotypes or plant hosts and genetically distinct insect vector populations has yet been shown (22). StolP is naturally transmitted by polyphagous Fulgoromorpha planthoppers of the Cixiidae family such as Hyalesthes obsoletus Signoret (16, 31, 39), Pentastiridius leporinus Linnaeus (4, 18), and Reptalus panzeri Löw (23). Interestingly the other phytoplasma species, members of the 16SrXII phylogenetic group, are also transmitted by Fulguromorpha planthoppers (1, 30). Therefore, some specific phytoplasma genetic determinant may be associated with the ability to interact with this particular clade of insects. As phytoplasmas have a complex life cycle in their insect vectors that implies adhesion and invasion to the cells of the insect midgut epithelium and salivary glands, as well as trophic interactions during intracellular multiplication, surface proteins have more chance to play major roles during the invasion process. As a consequence, a search for species-specific genes encoding StolP surface proteins has been undertaken. A recent StolP genome survey pointed out a partial coding sequence with some similarity to variable surface proteins of animal mycoplasmas (7). In this work, we present the characterization of this gene, formerly described as stol1H10 and now named vmp1, and show its remarkable variability by comparison to housekeeping genes.
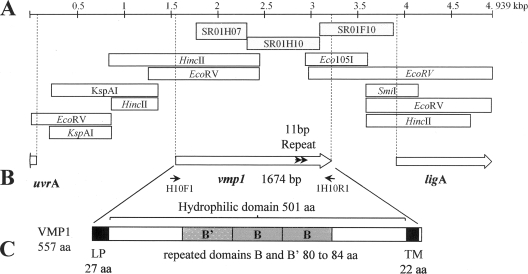
The StolP partial gene sequence SR01H10, issued from a suppression subtractive hybridization (SSH) survey of the StolP isolate PO genome, shared low homology with the gene encoding the variable surface lipoprotein VPMA of Mycoplasma agalactiae (identity, 22%; E value, 4 × 10−8) but had no homologous gene in the genome of “Candidatus Phytoplasma asteris” (OY-M). To complete the sequence of the gene, four primers directed toward the region neighboring the DNA fragment (1H10D, 1H10DN, 1H10G, and 1H10GN) (Table 1) were designed from the extremities of SR01H10 in order to perform genome walking amplification using the GenomeWalker universal kit (BD Biosciences Clontech) (Fig. 1A). PCR was performed on nine GenomeWalker DNA libraries consisting of digested fragments obtained from DNA of StolP-infected periwinkle and linked to adaptors. Four PCR products were obtained and sequenced. Their sequences were assembled with SR01H10 together with available SSH sequences to produce a larger sequence from which two new rounds of genome walking were realized and new PCR products obtained and sequenced. The final sequence and assembly, performed using the Phred, Phrap, and Consed software programs (11, 12, 20), shown in Fig. 1A produced a consensus sequence of 4,939 bp which was subjected to coding sequence (CDS) prediction. Three CDSs were predicted by frameD (37) and analyzed for sequence similarity by using BLASTX (http://www.ncbi.nlm.nih.gov/BLAST/). The larger CDS, which was 1,674 bp long (557 amino acids) and encoded the protein homologous to VPMA from Mycoplasma agalactiae, was named vmp1 for variable membrane protein 1 (Fig. 1B). Downstream of vmp1, a CDS encoded a peptide of 333 amino acids showing 72% identity with the N-terminal part of the NAD-dependent DNA ligase (ligA) of “Ca. Phytoplasma asteris” (OY-M) (PAM438). The beginning of a third CDS was identified on the minus strand corresponding to the 20 first amino acids and was 84% identical to the N-terminal part of excinuclease ATPase subunit UVRA (PAM450) of “Ca. Phytoplasma asteris” (OY-M).
TABLE 1.
Sequences of the primers used in this study
| Use and primer name | Nucleotide sequence (5′ → 3′) |
|---|---|
| Genome walking, vmp1 | |
| 1H10D | GCATCAGTATTTACGGTTAATGAACCAGC |
| 1H10DN | GCTTGTGTAACAGTAATTGTTTCAGTTGTAGG |
| 1H10D2 | TCTTAGACTTAATAATATACAACTTTAATGCTTGA |
| 1H10D2N | CCTCCTTACTTAATGAAATCGATGATG |
| 1H10D3 | GTGTTTGTTATATCGTCTAAATTGGATG |
| 1H10D3N | TGTATTGTTGTATATGACTGTCAACAC |
| 1H10G | AGTAACGCATCCAGATTTTGCTGGTG |
| 1H10GN | TTCAACCCAAAGGGATTTAGGGAAG |
| 1H10G2 | TCAGCCATTCAAACTAATAAACCCA |
| 1H10G2N | TACGGCAACAAGTTAATTTACGCAA |
| Genome walking, map | |
| GWPO-MAPG | ACAAATATGTTTCGGAAAGCCA |
| GWPO-MAPGN | TTCATCAAATTGATTTCGTGAGG |
| GWPO-MAPD | TGATGGCATGCAAAATATCAGTC |
| GWPO-MAPDN | GAAATCAATTTGATGAAACACGCT |
| GWPO-MAPG2 | AATTCGATTCATCAAATCTTGTGTG |
| GWPO-MAPGN2 | GATAGAATCAGGAACTAACTTCCCTTG |
| vmp1 PCR and sequencing | |
| 1H10F1 | AGGTTGTAAAATCTTTTATGTG |
| 1H10R1 | GCGGATGGCTTTTCATTATTTGAC |
| 1H10F4 | CACAAGCAGAATCTACAAATCC |
| 1H10R4 | AACTGCAGCTTGAGTTCTTGC |
| secY and map PCR and sequencing | |
| POsecF1 | TCTGCTTTGCCTTTGCCTTT |
| POsecR1 | ATTAGTAAACTAGTTCCTCC |
| POadkF1 | GTTGGTCGCAGAATTTGTCC |
| POif1R1 | CCAGAAACATAAGCGGTAATCGT |
FIG. 1.
(A) Genome walking and assembly of the final chromosomal fragment of 4,939 bp. SSH fragments (7) are shown as light gray boxes, and the GenomeWalker PCR fragments are represented by white boxes. The restriction sites indicated in white boxes indicate the type of library that allowed producing the PCR genome walking fragment. (B) Localization of the three coding sequences corresponding to the complete VMP1 protein, the N-terminal part of NAD-dependent DNA ligase, and the beginning of the excinuclease ATPase subunit UVRA. Gray arrows in vmp1 indicate 11-bp direct repeats where duplication (isolate T2_92) or deletion (isolate Moliere) occurred. Black arrows indicate primers 1H10F and 1H10R. (C) Structural domains of the VMP1 protein. SP, N-terminal signal peptide (27 amino acids); TM, transmembrane alpha helix (22 amino acids [aa]); B (84 amino acids) and B′ (80 amino acids), repeated domains.
Initiation of vmp1 translation could proceed at two ATG initiation codons separated by 39 nucleotides. However the first ATG was preceded by a nonclassical ribosome binding sequence situated 14 nucleotides upstream, whereas the second ATG codon was situated 10 nucleotides downstream of a more canonical ribosome binding sequence. This ATG was chosen as a translation start from which the synthesis of a 557-amino-acid protein would be initiated. Following the TAA stop codon, GC-rich short inverted repeats detected by the program MFOLD (47) were followed by a short poly(T) sequence and certainly corresponded to the hairpin sequence (ΔG = −17.4 kJ/mol) of a rho-independent transcription terminator. According to ANTHEPROT 2000 v5.2 (http://antheprot-pbil.ibcp.fr/), the predicted VMP1 sequence possess a signal peptide represented by a N-terminal hydrophobic region of 20 amino acids with a potential cleavage site predicted at glycine 27 and an alpha helix domain of 22 hydrophobic amino acids detected 7 amino acids before the C terminus of VMP1 (Fig. 1C). This alpha helix should permit the anchoring of the protein in the phytoplasma cellular membrane, thus exposing the large hydrophilic mature protein to the cellular surface with only seven amino acids located inside the cell. The mature protein was predicted to have 530 amino acids with a molecular mass of 57.8 kDa and an alkaline pI of 9.03. Repeat searches within the hydrophilic central domain identified two 66% identical repeated domains, called B domains, of 84 and 80 amino acids, preceded by a B′ domain that was only 30% homologous to the B domains (Fig. 1C).
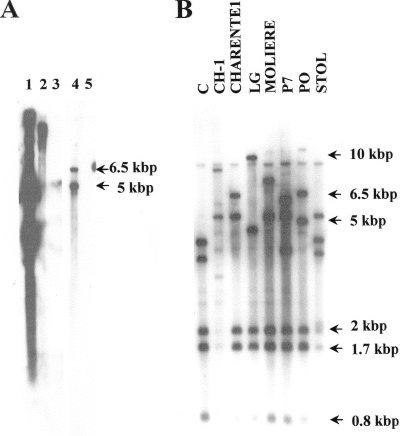
To determine if the vmp1 gene is located on the chromosome or on an extrachromosomal element and to determine the number of vmp1 copies in the genome of the StolP isolate PO, a Southern blot hybridization was performed. The hybridization was first performed at high stringency on native or HindIII-digested total DNA of StolP (PO)-infected periwinkle, using as a probe the digoxigenin-labeled PCR-amplified SR01H10 sequence. No hybridization signal was obtained after CDP-Star chemiluminescent revelation (Roche) from healthy periwinkle DNA, showing that SR01H10 did not bind periwinkle DNA (Fig. 2A, lanes 3 and 5). The probe hybridized to a large DNA band corresponding to the sheared linear genomic DNA which migrated at the top of the gel. No discrete band which could correspond to an extrachromosomal DNA was hybridized by the probe (Fig. 2A, lane 2). Under these high-stringency hybridization conditions (washing steps at 55°C), the probe revealed two HindIII fragments of 5 and 6.5 kbp. Under low-stringency hybridization conditions (washing steps at 50°C), the same probe revealed five DNA fragments of 6.5 kbp, 5 kbp, 2 kbp, 1.7 kbp, and 0.8 kbp in the genome of the StolP isolate PO (Fig. 2B, lane PO). These results suggested that at least two highly homologous copies of the vmp1 gene may be present in the genome of the isolate PO and that other incomplete or poorly homologous copies are also present on its chromosome. Because on the DNA sequence containing vmp1 the first HindIII site is present at position 4330, the HindIII fragment carrying vmp1 should be either the 5-kbp or the 6.5-kbp HindIII fragment detected at high hybridization stringency. Other StolP isolates from various plants in Europe and Lebanon had also at least five HindIII fragments hybridizing the SR1H10 probe. This experiment allowed us to suggest that the vmp1 gene was present as several, possibly divergent, copies on the chromosomes of all StolP isolates analyzed.
FIG. 2.
Southern blots for vmp1 detection in the StolP genome. (A) Hybridization at high stringency with an SR1H10 digoxigenin-labeled PCR-amplified probe. Lanes: 3 and 5, healthy periwinkle total DNA; 2 and 4, StolP PO-infected periwinkle DNA; 2 and 3, undigested; 4 and 5, digested with HindIII; 1, the native pGEMT-Easy plasmid containing the SR1H10 SSH product. (B) Hybridization at low stringency with an SR1H10 digoxigenin-labeled PCR-amplified probe hybridized to HindIII-digested DNA from periwinkle infected by different StolP isolates (indicated above the lanes).
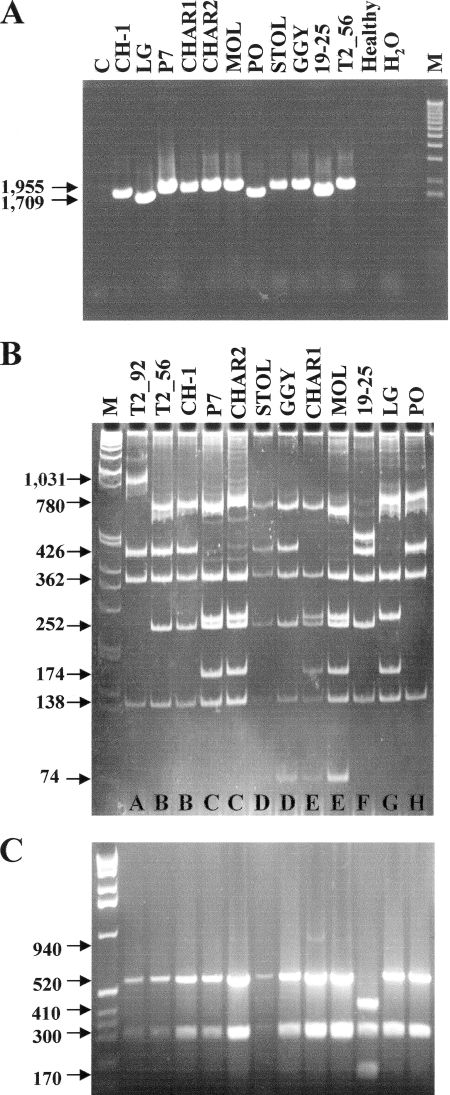
Genetic variability of vmp1 and three housekeeping genes was examined among various StolP isolates isolated from Europe and the Middle East (Table 2) which were maintained and propagated in periwinkle (Catharanthus roseus L.) by graft inoculation. Plant DNA was extracted from 1.5 to 2 g of symptomatic leaf midribs or equivalent noninfected material as described previously (31). PCR amplification using primers 1H10F and 1H10R showed that the vmp1 gene was present in all of the StolP isolates tested except the old stolbur C reference isolate maintained by grafting in periwinkle for 40 years (Fig. 3A). There was a difference in fragments size in the eight StolP isolates tested. StolP PO, 19-25, and LG had a 1.7-kbp vmp1 gene, whereas all the other StolP isolates gave a PCR product of 1.955 kbp (as verified by sequencing all amplicons). Sequences of the larger amplicons revealed that the encoded VMP1 protein contained an additional B domain of 81 amino acids and thus a larger vmp1 gene. Comparison of all vmp1 sequences revealed important sequence variability, and a single restriction of the amplicons using RsaI revealed eight restriction profiles (A to H) among the different StolP isolates (Fig. 3B). The same StolP isolates gave only two different HpaII restriction patterns of the tuf gene amplified using the primer pair tufAYf and tufAYr (38) and treated with HpaII endonuclease according to the published typing protocol (27) (Fig. 3C). The tuf A pattern was found as expected for the tuf A reference isolate StolP 19-25, while all isolates showed the typical tuf B pattern as did the reference tuf B isolate GGY. This comparison demonstrated that vmp1 was much more variable than the tuf housekeeping gene. Similarly, other restriction enzymes revealed five or six different restriction fragment length polymorphism (RFLP) patterns for vmp1. Five different patterns were revealed using DraI and HphI and six patterns with AluI and TaqI (data not shown) (D. Pacifico, A. Cimerman, C. Marzachì, and X. Foissac, presented at the 16th International Congress of the International Organization for Mycoplasmology, Cambridge, United Kingdom, 2006). All of these RFLP data allowed the constitution of 10 RFLP subgroups. Sequencing also revealed that the StolP isolate Moliere had a deletion of 11 bp (repeated motif AAGTAACGCA) downstream of position 1497, whereas the isolate T2_92 had an 11-bp insertion of the same motif at the same position. This deletion and insertion disrupted the C-terminal end of the vmp1 gene in both StolP isolates. A CAC triplet was deleted at position 864 for isolates PO and T2_92, and a GAT and a CAC triplet were inserted at positions 1053 and 1110, respectively, for isolates Moliere, Charente1, GGY, and STOL, without disturbing the translation frame of the corresponding vmp1 genes.
TABLE 2.
StolP isolates propagated on Catharanthus roseus used in this study
| Isolate | Original host | Country, yr | Reference | EMBL accession no. (map/secY/vmp1) |
|---|---|---|---|---|
| C | Tomato | France, 1962 | 8 | AM990987/2083/— |
| CH-1 | Grapevine | Italy, 1991 | 9 | AM990982/2089/2105 |
| Charente1 | H. obsoletus | France, 2000 | J. L. Danet, unpublished data | AM990976/2084/2098 |
| Charente2 | H. obsoletus | France, 2005 | J. L. Danet, unpublished data | AM990977/2085/2099 |
| GGY | Grapevine | Germany, 1995 | 33 | AM990981/2093/2102 |
| LG | Tomato | France, 2000 | S. Eveillard, unpublished data | AM990979/2092/2097 |
| Moliere | Prunus mahaleb | France, 1975 | G. Morvan, unpublished data | AM990984/2090/2096 |
| P7 | Periwinkle | Lebanon, 2001 | 44 | AM990978/2091/2100 |
| PO | H. obsoletus | France, 1996 | 21 | AM990988/2082/2095 |
| STOL | Pepper | Serbia, 1978 | R. Marwitz, unpublished data | AM990983/2086/2103 |
| T2_56 | Tomato | Italy, 1996 | 34 | AM990985/2087/2104 |
| T2_92 | Tomato | Italy, 1996 | 34 | AM990986/2088/2106 |
| 19-25 | Grapevine | Germany | 27 | AM990980/2094/2101 |
FIG. 3.
(A) Size polymorphism of PCR products obtained with primers H10F1 and H10R1 (vmp1) from total DNA of periwinkles infected with different StolP isolates (indicated above the lanes) (Table 2). No amplification was detected for stolbur isolate C, healthy periwinkle DNA, and H2O. Electrophoresis was performed on a 0.8% agarose gel. (B) RsaI RFLP analysis of H10F1/R1 products (vmp1) on an 8% polyacrylamide gel. (C) RFLP analysis of tuf amplicons with HpaII restriction enzyme. Electrophoresis was performed on a 1.5% agarose gel. M, 1-kb ladder from Invitrogen.
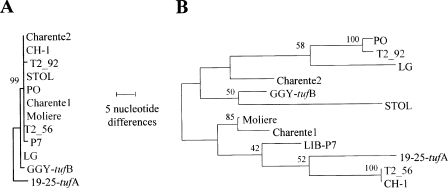
To better compare the variability of vmp1 to that of other genetic loci, the genetic variability of two other housekeeping genes, map and secY, was analyzed. Part of the map gene had been characterized in one of the subtraction libraries produced during a partial genomic survey of the StolP PO genome (7). As secY, a gene frequently used to study phytoplasma genetic diversity, was located upstream from the adk gene and followed by the gene map, the genome walking strategy was continued upward in order to sequence the StolP secY gene. Once the whole secY-adk-map genetic locus was fully amplified and sequenced, primers were designed to amplify map and secY from StolP isolates. Only two single-nucleotide polymorphisms (SNPs) were found in the map gene, one distinguishing the isolate GGY and one specific to the isolate 19-25; all other StolP isolates had a map sequence identical to that of isolate PO. The gene secY appeared to be a bit more variable, with eight SNPs being identified. A phylogenetic analysis performed with MEGA version 4 (41) using the method of maximum of parsimony was obtained after aligning with Clustal W program (42) a concatenation of the map and secY genes (Fig. 4A). When the map-secY results were compared to a similar analysis of vmp1 (Fig. 4B), vmp1 was found to be tremendously more variable. For instance, 94 SNPs affecting 71 codons differentiated the PO and 19-25 vmp1 genes, inducing 68 changes of amino acids between these VMP1 proteins. This very high number of nonsynonymous mutations undoubtedly reflected a strong diversifying selection pressure having been exerted on vmp1.
FIG. 4.
Evolutionary relationships of secY-map and vmp1 genetic loci for 12 StolP isolates. The evolutionary history was inferred using the maximum-parsimony method. (A) One tree out of the 170 most parsimonious trees for the map-secY genetic locus; (B) 1 tree out of the 4 most parsimonious trees for vmp1. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. The trees are drawn to scale, with branch lengths calculated using the average pathway method, and represent the number of nucleotide changes over the whole sequence. All positions containing gaps and missing data were eliminated from the data set. There were a total of 1,888 positions in the final data set for the map-secY genetic locus and 1,522 positions in the final data set for vmp1. Phylogenetic analyses were all conducted with MEGA4.
A potential driving force for phytoplasma evolution is the necessity to adapt to new plant or insect hosts after invasion of new ecological niches resulting from the introduction of new plant hosts or insect vectors (6, 29). However, phytoplasmas, like the other members of the bacterial class Mollicutes, have limited genomes, with size ranging from 530 kbp to 1,350 kbp (32). The first phytoplasma genomes have recently been sequenced, and analysis of their 671 to 839 genes confirmed that phytoplasmas went through reductive evolution but still maintain an important genome plasticity (2, 35, 43). In addition, phytoplasma genes encoding surface proteins involved in the interaction with the insect vector vary much more rapidly than the rest of the genome (24, 40). This diversifying effect is seen as a consequence of a strong positive selection, resulting from the necessary adaptation of phytoplasmas to their complex and changing environment. It is therefore interesting to look for such species-specific and variable genes, as they may constitute discriminant markers for molecular epidemiology as well as relevant candidate genes possibly involved in phytoplasma-insect vector or phytoplasma-plant interactions. In addition, the availability of nonribosomal sequences is essential for a finer molecular differentiation of phytoplasmas within the 16SrXII-A subgroup of phytoplasma classification. As with all Mollicutes, because phytoplasmas lack a cell wall, it is likely that membrane proteins play a central role in the molecular mechanisms governing phytoplasma-host interactions. The structural organization of membrane proteins may reflect biological and ecological properties such as symptom induction in plants and association with different plant species or vector population. Species-specific immunodominant membrane proteins such as spiralin and AMP bind insect actin microfilaments and insect cell glycoproteins, respectively, during the process of insect cell recognition by spiroplasmas or insect cell invasion by phytoplasmas (25, 40). Such mollicute proteins are often characterized by an important variability (15, 24). Therefore, looking for variable membrane proteins might be a relevant strategy to select protein candidates for studying StolP-insect vector interactions and might provide variable markers to survey propagation of StolP isolates from their wild compartment reservoir to vineyards or annual crop fields, where they cause economically damaging diseases and epidemics.
The vmp1 gene was chosen as a candidate for a StolP genetic variability study because part of its sequence had low homology to the gene encoding the surface variable lipoprotein VPMA of Mycoplasma agalactiae, which is encoded by a gene which undergoes site-specific DNA inversion responsible for variation of the corresponding surface protein (14, 19). Such antigenic variation by recombination has never been evidenced either in spiroplasmas or in phytoplasmas. However, we showed that vmp1 is highly variable compared to three housekeeping genes of StolP and that other certainly divergent gene copies could be detected by low-stringency Southern blot hybridization in all StolP isolates from various origins. The noncongruency between vmp1 phylogeny and housekeeping gene phylogeny might indicate recombination between vmp1 gene copies or gene fragments. In addition, it is likely that the VMP1 protein, if expressed, is mainly exposed to the surface of the phytoplasma cell and is subjected to a strong diversifying selection. The presence of repeated domains is a characteristic of many variable surface proteins of mycoplasmas (45), but it is also found in the spiroplasma adhesion-related proteins (3, 36, 46). Repeated domains are often present in proteins promoting eukaryotic cell recognition, such as internalin of Listeria monocytogenes and many other gram-positive surface proteins involved in bacterium-eukaryotic cell interactions (10). VMP1 does not seem to be essential for the propagation of StolP in periwinkle, as the isolate stolbur C lacks the gene and two other isolates (Moliere and T2_92) have incomplete vmp1 genes due to disruptions of the translation frame. We have no indications about the insect transmission properties of these three StolP isolates and therefore cannot link the presence of the full gene to them.
Up to now, three other vmp-like partial CDSs of StolP isolate PO have been characterized, but they represent incomplete CDSs sharing little sequence similarity to vmp1 (unpublished data). They are not organized in clusters as is the case for VPMA (14). As a molecular variable marker, vmp1 is currently being used to survey StolP isolates in the Euro-Mediterranean Basin where various insect vectors or different vector populations of the same insect species have been described (16, 18, 22, 23). Preliminary data indicate that vmp1 RFLP and sequencing represent powerful typing markers to differentiate StolP isolates, but epidemiological studies with a large number of isolates collected from different plants or insect hosts need to be done to determine whether specific genotypes of this marker can be associated with specific insect vector populations, insect vector species, or plant hosts. As a way to progress toward VMP1 function, vmp1 is currently being expressed in heterologous systems and VMP1-derived peptides are being synthesized in order to produce anti-VMP1 antibodies to assess VMP1 expression and to verify its location on the phytoplasma surface. Purification of VMP1 will also be necessary to look for possible interaction between VMP1 and insect vector proteins.
Acknowledgments
A.C. was supported by a Ph.D. fellowship from INRA-SPE and the Conseil Régional d'Aquitaine, and D.P. was supported by a grant from Ministero delle Politiche Agricole e Forestali (GIAVI) and a visiting fellowship from the University of Turin. This work was funded by INRA and the Conseil Régional d'Aquitaine.
Michael Maixner is kindly acknowledged for providing DNA from StolP isolates 19-25 and GGY.
Footnotes
Published ahead of print on 6 March 2009.
REFERENCES
- 1.Arocha, Y., M. Lopez, M. Fernandez, B. Pinol, D. Horta, E. L. Peralta, R. Almeida, O. Carvajal, S. Picornell, M. R. Wilson, and P. Jones. 2005. Transmission of a sugarcane yellow leaf phytoplasma by the delphacid planthopper Saccharosydne saccharivora, a new vector of sugarcane yellow leaf syndrome. Plant Pathol. 54:634-642. [Google Scholar]
- 2.Bai, X. D., J. H. Zhang, A. Ewing, S. A. Miller, A. J. Radek, D. V. Shevchenko, K. Tsukerman, T. Walunas, A. Lapidus, J. W. Campbell, and S. A. Hogenhout. 2006. Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188:3682-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, M., U. Melcher, and J. Fletcher. 2001. Characterization of Spiroplasma citri adhesion related protein SARP1, which contains a domain of a novel family designated sarpin. Gene 275:57-64. [DOI] [PubMed] [Google Scholar]
- 4.Bressan, A., O. Semetey, B. Nusillard, D. Clair, and E. Boudon-Padieu. 2008. Insect vectors (Hemiptera: Cixiidae) and pathogens associated with the disease syndrome “basses richesses” of sugar beet in France. Plant Dis. 92:113-119. [DOI] [PubMed] [Google Scholar]
- 5.Bressan, A., R. Turata, M. Maixner, S. Spiazzi, E. Boudon-Padieu, and V. Girolami. 2007. Vector activity of Hyalesthes obsoletus living on nettles and transmitting a stolbur phytoplasma to grapevines: a case study. Ann. Appl. Biol. 150:331-339. [Google Scholar]
- 6.Caudwell, A. 1983. L'origine des jaunisses à Mycoplasme (MLO) des plantes et l'exemple des jaunisses de la vigne. Agronomie 2:103-111. [Google Scholar]
- 7.Cimerman, A., G. Arnaud, and X. Foissac. 2006. Stolbur phytoplasma genome survey achieved using a suppression subtractive hybridization approach with high specificity. Appl. Environ. Microbiol. 72:3274-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cousin, M. T., P.-L. Maillet, J. P. Gourret, C. Grison, and T. Staron. 1968. Présence de particules de type “jaunisse européenne”. Le stolbur de la tomate, l'aster yellow du glaïeul, la phyllodie du trèfle. Etudes cytologiques et ultrastructurales. Premiers essais de lutte chimique. C. R. Acad. Sci. Ser. D 54:887-895. [Google Scholar]
- 9.Credi, R., and A. Santucci. 1992. Dodder transmission of mycoplasma-like organisms (MLOs) from grapevines affected by a flavescence doree-type disease to periwinkle. Phytopathol. Medit. 31:154-162. [Google Scholar]
- 10.Dramsi, S., P. Dehoux, and P. Cossart. 1993. Common features of Gram-positive bacterial proteins involved in cell recognition. Mol. Microbiol. 9:1119-1122. [DOI] [PubMed] [Google Scholar]
- 11.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 12.Ewing, B., L. Hillier, M. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 13.Firrao, G., M. Andersen, A. Bertaccini, E. Boudon Padieu, J. M. Bové, X. Daire, R. E. Davis, J. Fletcher, M. Garnier, K. Gibb, D. E. Gundersen-Rindal, N. Harrison, C. Hiruki, P. Jones, C. R. Kuske, I. M. Lee, L. Liefting, C. Marcone, S. Namba, B. Schneider, B. B. Sears, E. Seemuller, C. D. Smart, C. Streten, and K. Wang. 2004. ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int. J. Syst. Evol. Microbiol. 54:1243-1255. [DOI] [PubMed] [Google Scholar]
- 14.Flitman-Tene, R., S. Mudahi-Orenstein, S. Levisohn, and D. Yogev. 2003. Variable lipoprotein genes of Mycoplasma agalactiae are activated in vivo by promoter addition via site-specific DNA inversions. Infect. Immun. 71:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foissac, X., C. Saillard, J. Gandar, L. Zreik, and J. M. Bové. 1996. Spiralin polymorphism in strains of Spiroplasma citri is not due to differences in posttranslational palmitoylation. J. Bacteriol. 178:2934-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fos, A., J. L. Danet, L. Zreik, M. Garnier, and J. M. Bové. 1992. Use of a monoclonal-antibody to detect the stolbur mycoplasma-like organism in plants and insects and to identify a vector in France. Plant Dis. 76:1092-1096. [Google Scholar]
- 17.Garnier, M. 2000. The stolbur phytoplasma: an ubiquitous agent. C. R. Acad. Agric. France 86:27-33. [Google Scholar]
- 18.Gatineau, F., J. Larrue, D. Clair, F. Lorton, M. Richard-Molard, and E. Boudon-Padieu. 2001. A new natural planthopper vector of stolbur in the genus Pentastiridius (Hemiptera: Cixiidae). Eur. J. Plant Pathol. 107:263-271. [Google Scholar]
- 19.Glew, M. D., L. Papazisi, F. Poumarat, D. Bergonier, R. Rosengarten, and C. Citti. 2000. Characterization of a multigene family undergoing high-frequency DNA rearrangements and coding for abundant variable surface proteins in Mycoplasma agalactiae. Infect. Immun. 68:4539-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 21.Jarausch, W., J. L. Danet, G. Labonne, F. Dosba, J. M. Broquaire, C. Saillard, and M. Garnier. 2001. Mapping the spread of apricot chlorotic leaf roll (ACLR) in southern France and implication of Cacopsylla pruni as a vector of European stone fruit yellows (ESFY) phytoplasmas. Plant Pathol. 50:782-790. [Google Scholar]
- 22.Johannesen, J., B. Lux, K. Michel, A. Seitz, and M. Maixner. 2008. Invasion biology and host specificity of the grapevine yellows disease vector Hyalesthes obsoletus in Europe. Entomol. Exp. Appl. 126:217-227. [Google Scholar]
- 23.Jovic, J., T. Cvrkovic, M. Mitrovic, S. Krnjajic, M. G. Redinbaugh, R. C. Pratt, R. E. Gingery, S. A. Hogenhout, and I. Toševski. 2007. Roles of Stolbur phytoplasma and Reptalus panzeri (Cixiinae, Auchenorrhyncha) in the epidemiology of Maize redness in Serbia. Eur. J. Plant Pathol. 118:85-89. [Google Scholar]
- 24.Kakizawa, S., K. Oshima, H. Y. Jung, S. Suzuki, H. Nishigawa, R. Arashida, S. Miyata, M. Ugaki, H. Kishino, and S. Namba. 2006. Positive selection acting on a surface membrane protein of the plant-pathogenic phytoplasmas. J. Bacteriol. 188:3424-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Killiny, N., M. Castroviejo, and C. Saillard. 2005. Spiroplasma citri spiralin acts in vitro as a lectin binding to glycoproteins from its insect vector Circulifer haematoceps. Phytopathology 95:541-548. [DOI] [PubMed] [Google Scholar]
- 26.Kostoff, D. 1933. Virus diseases causing sterility. J. Phytopathol. 5:593-602. [Google Scholar]
- 27.Langer, M., and M. Maixner. 2004. Molecular characterisation of grapevine yellows associated phytoplasmas of the stolbur-group based on RFLP-analysis of non-ribosomal DNA. Vitis 43:191-200. [Google Scholar]
- 28.Lee, I. M., R. E. Davis, and D. E. Gundersen-Rindal. 2000. Phytoplasma: phytopathogenic mollicutes. Annu. Rev. Microbiol. 54:221-255. [DOI] [PubMed] [Google Scholar]
- 29.Lee, I. M., D. E. Gundersen-Rindal, and A. Bertaccini. 1998. Phytoplasma: ecology and genomic diversity. Phytopathology 88:1359-1366. [DOI] [PubMed] [Google Scholar]
- 30.Liefting, L. W., R. E. Beever, C. J. Winks, M. N. Pearson, and R. L. S. Forster. 1997. Planthopper transmission of Phormium yellow leaf phytoplasma. Aust. Plant Pathol. 26:148-154. [Google Scholar]
- 31.Maixner, M., U. Ahrens, and E. Seemuller. 1995. Detection of the German grapevine yellows (Vergilbungskrankheit) MLO in grapevine, alternative hosts and a vector by a specific PCR procedure. Eur. J. Plant Pathol. 101:241-250. [Google Scholar]
- 32.Marcone, C., H. C. Neimark, A. Ragozzino, U. Lauer, and E. Seemüller. 1999. Chromosomes size of phytoplasmas composing major phylogenetic groups and subgroups. Phytopathology 89:805-810. [DOI] [PubMed] [Google Scholar]
- 33.Marcone, C., A. Ragozzino, R. Credi, and E. Seemuller. 1996. Detection and characterization of phytoplasmas infecting grapevine in southern Italy and their genetic relatedness to other grapevine yellows phytoplasmas. Phytopathol. Medit. 35:207-213. [Google Scholar]
- 34.Minucci, C., and G. Boccardo. 1997. Genetic diversity in the stolbur phytoplasma group. Phytopathol. Medit. 36:45-49. [Google Scholar]
- 35.Oshima, K., S. Kakizawa, H. Nishigawa, H. Y. Jung, W. Wei, S. Suzuki, R. Arashida, D. Nakata, S. Miyata, M. Ugaki, and S. Namba. 2004. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 36:27-29. [DOI] [PubMed] [Google Scholar]
- 36.Saillard, C., P. Carle, S. Duret-Nurbel, R. Henri, N. Killiny, S. Carrère, J. Gouzy, J. M. Bové, J. Renaudin, and X. Foissac. 2008. The abundant extrachromosomal DNA content of the Spiroplasma citri GII3-3X genome. BMC Genomics 9:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiex, T., J. Gouzy, A. Moisan, and Y. de Oliveira. 2003. FrameD: a flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences. Nucleic Acids Res. 31:3738-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider, B., K. S. Gibb, and E. Seemüller. 1997. Sequence and RFLP analysis of the elongation factor Tu gene used in differentiation and classification of phytoplasmas. Microbiology 143:3381-3389. [DOI] [PubMed] [Google Scholar]
- 39.Sforza, R., D. Clair, X. Daire, J. Larrue, and E. Boudon-Padieu. 1998. The role of Hyalesthes obsoletus (Hemiptera: Cixiidae) in the occurence of bois noir of grapevines in France. J. Phytopathol. 146:549-556. [Google Scholar]
- 40.Suzuki, S., K. Oshima, S. Kakizawa, R. Arashida, H. Y. Jung, Y. Yamaji, H. Nishigawa, M. Ugaki, and S. Namba. 2006. Interaction between the membrane protein of a pathogen and insect microfilament complex determines insect-vector specificity. Proc. Natl. Acad. Sci. USA 103:4252-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran-Nguyen, L. T. T., M. Kube, B. Schneider, R. Reinhardt, and K. S. Gibb. 2008. Comparative genome analysis of “Candidatus Phytoplasma australiense” (subgroup tuf-Australia I; rp-A) and “Ca. Phytoplasma asteris” strains OY-M and AY-WB. J. Bacteriol. 190:3979-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verdin, E., P. Salar, J. L. Danet, B. Gelie, J. M. Bové, M. Garnier, E. Choueiri, F. Jreijiri, and S. El-Zammar. 2004. Phylogenetical characterization and PCR detection of a new phytoplasma in almond (Prunus amygdalus) and peach (Prunus persicae) in the Mediterranean area. Acta Hort. 657:527-532. [Google Scholar]
- 45.Yogev, D., G. F. Browing, and K. S. Wise. 2002. Genetic mechanisms of surface variation, p. 417-4444. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic, Dordrecht, The Netherlands.
- 46.Yu, J., A. C. Wayadande, and J. Fletcher. 2000. Spiroplasma citri surface protein P89 implicated in adhesion to cells of the vector Circulifer tenellus. Phytopathology 90:716-722. [DOI] [PubMed] [Google Scholar]
- 47.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]