Abstract
Toxinogenic endobacteria were isolated from a collection of Rhizopus spp. representing highly diverse geographic origins and ecological niches. All endosymbionts belonged to the Burkholderia rhizoxinica complex according to matrix-assisted laser desorption ionization-time of flight biotyping and multilocus sequence typing, suggesting a common ancestor. Comparison of host and symbiont phylogenies provides insights into possible cospeciation and horizontal-transmission events.
Bacterial symbionts and their metabolic potential play essential roles for many organisms. They may benefit from improved fitness, survival, and even acquired virulence (7, 12, 22). In the course of our studies of the biosynthesis of rhizoxin, the causative agent of rice seedling blight (10), we found that the phytotoxin is produced not by the fungus Rhizopus microsporus but by symbiotic bacteria (Burkholderia rhizoxinica) that reside within the fungus cytosol (13, 15, 23). Furthermore, cloning and sequencing of the rhizoxin biosynthesis gene cluster revealed the molecular basis of bacterial toxin production (14). In sum, this represents an unparalleled example for a symbiosis in which a fungus harbors bacteria for the production of a virulence factor. In analogy, we found that the first reported “mycotoxins” from lower fungi, the highly toxic cyclopeptides rhizonin A and B (25, 28), are also produced by symbiotic bacteria (Burkholderia endofungorum) and not by the fungus (16). While both rhizoxins and rhizonins have been believed to promote zygomycoses (21), there is no indication for toxin-producing endosymbiotic bacteria in clinical isolates (18).
In nature, toxin production plays a pivotal role in the development of the fungus-bacterium association. Studies of the evolution of host resistance indicate that the association resulted from a pathogenicity mutualism shift in insensitive zygomycetes (24). The fungus lost its ability to sporulate independently and became totally dependent on endobacteria for reproduction through spores, thus warranting the persistence of the symbiosis and its efficient distribution through vegetative spores (17).
To gain a broader view of the occurrence, biosynthetic potential, and relationship of toxinogenic endofungal bacteria, we investigated a collection of Rhizopus spp. consisting of 20 isolates classified as R. microsporus (of which 13 belong to R. microsporus var. microsporus, four to R. microsporus var. chinensis, two to R. microsporus var. oligosporus, and one to R. microsporus var. rhizopodiformis), one isolate classified as Rhizopus sp., and one Rhizopus oryzae strain. We initially monitored the presence of bacterial symbionts by PCR using universal primers (16S rRNA genes) and rhizoxin production in all available Rhizopus strains. Liquid cultivation of fungi in production medium with and without antibiotic followed by organic solvent extraction yielded crude extracts that were analyzed by high-performance liquid chromatography (HPLC) and mass spectrometry (MS). In total, eight fungal strains were identified or confirmed as rhizoxin positive and thus expected to harbor endosymbionts. In all cases, this assumption was verified by PCR and confocal scanning microscopy. By means of an optimized protocol, we finally succeeded in the isolation and cultivation of all eight bacterial symbiont strains in pure cultures (isolates B1 to B8) (Table 1).
TABLE 1.
Fungal strains and their bacterial endosymbionts
| Taxon | Strain designationa | Origin | Bacterial endosymbiont (isolate) |
|---|---|---|---|
| Rhizopus microsporus van Tieghem | ATCC 62417 | Rice seedlings, Japan | Burkholderia rhizoxinica HKI-0454 (B1) |
| Rhizopus sp. strain F-1360 | ATCC 20577 | Soil, Japan | Burkholderia sp. strain HKI-0512 (B2) |
| Rhizopus microsporus Tieghem var. microsporus | CBS 111563 | Sufu starter culture, rice wine tablet, Vietnam | Burkholderia sp. strain HKI-0455 (B3) |
| Rhizopus microsporus Tieghem var. microsporus | CBS 699.68 | Soil, Ukraine | Burkholderia sp. strain HKI-402 (B4) |
| Rhizopus microsporus Tieghem | CBS 112285 | Ground nuts, Mozambique | Burkholderia endofungorum HKI-0456 (B5) |
| Rhizopus microsporus var. chinensis (Saito) Schipper & Stalpers | CBS 261.28 | Not specified, United States of America | Burkholderia sp. strain HKI-0513 (B6) |
| Rhizopus microsporus Tieghem var. microsporus | CBS 700.68 | Forest soil, Georgia | Burkholderia sp. strain HKI-0403 (B7) |
| Rhizopus microsporus Tieghem var. microsporus | CBS 308.87 | Man, from deep necrotic tissue within the hand following a spider bite, Australia | Burkholderia sp. strain HKI-0404 (B8) |
ATCC, American Type Culture Collection, Manassas, VA; CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands.
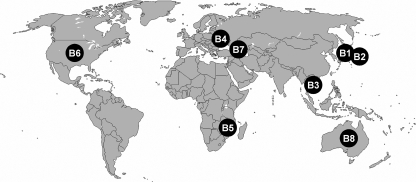
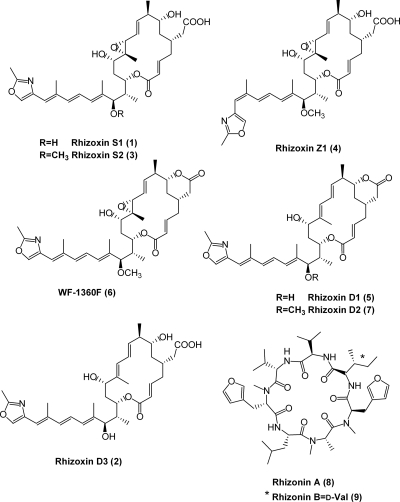
Notably, the eight Rhizopus isolates are from geographically distinct collection sites, covering all five continents (Africa, America, Asia, Australia, and Europe) and representing diverse ecological niches of the host (plants, soil, food, and necrotic tissue) (Fig. 1; Table 1). HPLC and MS analyses of the metabolic profiles and comparison with authentic references revealed that all endofungal bacterial strains are capable of producing considerable amounts of rhizoxin derivatives 1 to 7 (23) (Fig. 2). Among the rhizoxin derivatives, rhizoxin S2 (derivative 3) is the main product formed by all isolates, followed by compounds WF-1360F (derivative 6) (11), rhizoxin Z1 (derivative 4), and rhizoxin S1 (derivative 1) (23), while derivatives 2, 5, and 7 are formed only in minor amounts. Significant differences in production of rhizoxins were not found among the isolates (see Fig. S1 in the supplemental material). Only one isolate, Burkholderia endofungorum HKI-0456 (isolate B5), also produces the hepatotoxic cyclopeptides rhizonin A (derivative 8) and B (derivative 9) under laboratory conditions (16).
FIG. 1.
Survey of collection sites of toxinogenic R. microsporus strains used in this study.
FIG. 2.
Structures of the main rhizoxin derivatives (derivatives 1 to 7) produced by all eight fungal endosymbionts (isolates B1 to B8) and structures of rhizonin A and B (derivatives 8 and 9), produced by the symbiont B. endofungorum HKI-0456 (isolate B5).
A preceding phylogenetic analysis of the 16S rRNA gene of the type strains B. rhizoxinica HKI-0454 (B1) and B. endofungorum HKI-0456 (B5) showed that both isolates belong to the genus Burkholderia (13). Although the two strains resemble each other in terms of endofungal lifestyle and physiology, DNA-DNA hybridization experiments enforced the division of the two isolates into two distinct species. To establish the taxonomic positions of all eight bacterial symbionts, we isolated genomic DNA from the recovered strains and obtained full-length 16S rRNA gene sequences by PCR using 16S universal primers (15). Sequence comparisons revealed that all isolated endosymbiotic bacteria are closely related to species of the genus Burkholderia.
However, the close relationship of the symbionts is particularly intriguing considering the highly diverse collection localities of the host strains (Table 1). Despite the clear grouping of the bacteria associated with Rhizopus, the phylogenetic relationship within the endofungal symbiont complex could not be resolved by 16S rRNA gene data alone (see Fig. S2 in the supplemental material). Several computational methods failed to infer a statistically meaningful phylogeny. To overcome uncertainties in the 16S rRNA gene and biotyping analyses and to further characterize the genotypes of the eight isolates of the B. rhizoxinica complex, we performed a multilocus sequence typing (MLST) analysis. Seven conserved gene loci from all isolates were amplified by PCR, sequenced, and phylogenetically analyzed. To facilitate the comparison with an MLST study of the related bacterium Burkholderia pseudomallei (8), fragments of the following genes were chosen: ace (acetoacetyl-coenzyme A reductase), gltB (glutamate synthase, large subunit), gmhD (ADP-l-glycero-d-manno-heptose-6-epimerase), lepA (GTP-binding protein), lipA (lipoate synthase), and ndh (NADH:ubiquinone oxidoreductase). Shotgun sequencing of the genomes of two symbiont isolates indicated that the narK locus used in the B. pseudomallei study is obviously not present in the endofungal Burkholderia strains (G. Lackner, L. P. Partida-Martinez, and C. Hertweck, unpublished results). Therefore, as a characteristic feature of the ecotype, a locus from the rhizoxin biosynthesis gene cluster, rhiE (14), was sequenced in all isolates. The rhiE locus codes for a part of the polyketide synthase involved in rhizoxin biosynthesis in endofungal bacteria (14). It should be mentioned that a homologous rhizoxin biosynthesis gene cluster has been identified in the phylogenetically distant strain Pseudomonas fluorescens Pf-5 (3, 19).
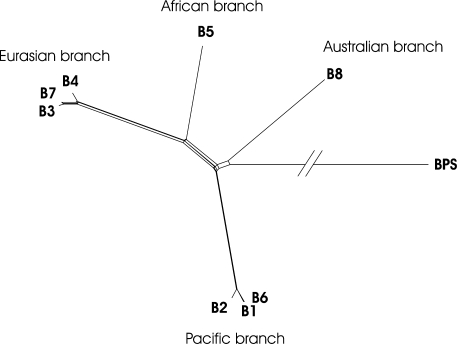
All loci were analyzed independently to test for incongruence between the data sets. The majority of the single-locus trees yielded a topology similar to that shown in Fig. S3 in the supplemental material. Only the ace locus resulted in a different tree: it showed a split (Eurasian, B8) (B5, others) different from the split (Eurasian, B5) (B8, others) found in the majority of single-locus trees (gltB, gmhD, and lipA) (see Fig. S3 in the supplemental material). The remaining loci (lepA, ndh, and rhiE) failed to infer statistically supported clades containing the strain B5 or B8. The phi test for recombination implemented in the program SplitsTree4 (5, 9a) indicated evidence for recombination, if the ace locus was included in the data set (P = 0.044). The removal of the ace locus abolished the signal. We conducted concatenated analyses including gltB, gmhD, lipA, lepA, ndh, and rhiE in the presence and absence of the ace locus. The resulting phylogenetic trees were recovered using distance matrix, maximum-parsimony, and Bayesian methods (see Fig. S3 and S4 in the supplemental material). We found that the tree topology is independent from the presence or absence of the ace locus. An alternative way to illustrate the phylogenetic groupings is in a network (9) (Fig. 3). The type strain B. rhizoxinica HKI-0454 (isolate B1) and isolate B6 share identical alleles in all sequenced loci and thus could be considered the same species. This high degree of similarity is supported by matrix-assisted laser desorption ionization protein profiling (see Fig. S5 in the supplemental material). However, it is remarkable that the geographic origins of isolates B1 and B6 (Japan and the United States, respectively) are different. Another member of this “Pacific group,” isolate B2 from Japan, is the closest relative. This observation strongly suggests that the Japanese and U.S. isolates have a common ancestor. Another highly supported clade, the “Eurasian group,” consists of the isolates B3 (Vietnam), B4 (Ukraine), and B7 (Georgia). Again, strain B3 and B7 are highly similar at the nucleotide level despite their geographic distance. Although related to this clade, the B. endofungorum type strain HKI-0456 (isolate B5), isolated from ground nuts in Mozambique, is unique in both genotypic and phenotypic aspects. Isolate B8 from Australia is related even more remotely to all other strains. The phylogenetic data obtained in this study suggest that all Burkholderia symbiont strains found in Rhizopus have a common ancestor.
FIG. 3.
Phylogenetic network of the endofungal symbiont complex (isolates B1 to B8) and B. pseudomallei (BPS), based on MLST. The graph was obtained by the neighbor-net method, implemented with the SplitsTree4 program. Uncertainties in the data are visualized by the network structure in the center.
To test whether the phylogenetic data obtained from the MLST analysis contain further information about the evolution of the endofungal symbiosis, we analyzed the extent of detectable recombination in the data set. The presence of recombination could be explained by the exchange of genetic material between bacterial lineages that might have occurred during horizontal transmission of endosymbionts. Less likely, recombination could mean that the mutualistic association was established several times in some of the lineages.
The number of incompatible splits in the center of the phylogenetic-network structure (Fig. 3) leaves some uncertainty about the correct placement of the strains B8 and B5, and the phi test for recombination indicated evidence for recombination if the ace locus was included. However, recombination does not appear to be a dominant factor in the evolution of the core genome of the known endosymbionts. Notably, the strong congruence between the trees retrieved from the symbiont data is in stark contrast to the recently reported high rate of recombination in free-living Burkholderia spp. (2, 6). Presumably, this is not due to a lack of recombination machinery, as homologous recombination works fine in at least three of the strains under laboratory conditions. Rather, we assume that the mainly vertically transmitted and geographically separated symbionts have evolved primarily separately from each other. Remaining traces of recombination might be hints of coinfection events in the early history of the symbiosis.
Another genetic feature of the endosymbionts presented here is their relatively low GC content compared to that for related, but free-living Burkholderia species. The bacterial endosymbiont of aphids, Buchnera aphidicola, is known to have some mutational bias toward low GC content (26). Indeed, all of the endosymbionts have significantly lower GC contents than their sequenced free-living relatives, e.g., B. pseudomallei, Burkholderia thailandensis, and Burkholderia cenocepacia, in all conserved loci (see Fig. S6 in the supplemental material). Although the data obtained in this study are only preliminary evidence and future studies at the whole-genome level could certainly provide more insights into the nucleotide evolution of fungal endosymbionts, it is possible that mechanisms similar to those for Buchnera species are responsible for the reduced GC content in the fungal endosymbionts. The observation that the reduction in GC content is not as striking as that in Buchnera species is then well in accordance with the expectation that the fungus-bacterium endosymbiosis is young compared to the Buchnera-aphid mutualistic relationship.
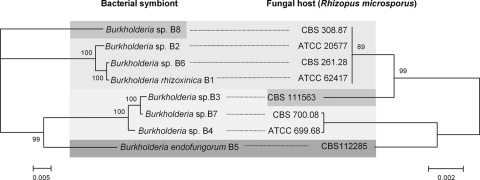
To obtain hints about possible cospeciation or horizontal-transmission events, we compared the phylogenetic relationships between the endobacteria and their fungal hosts. Nucleotide sequences of the 18S ribosomal DNA (rDNA), 28S rDNA, and internal transcribed spacer (ITS) regions were chosen to elucidate phylogenetic relationships between fungal hosts (1). These attempts were hampered since 18S rDNA sequences were highly conserved among the fungal isolates. The only variable site distinguished symbiotic from nonsymbiotic R. microsporus strains (see Fig. S7 in the supplemental material). 28S rDNA sequences are known to accumulate single nucleotide changes at a relatively low rate as well (27). Our 28S rDNA data set, with a total length of 604 nucleotides, contained only eight variable sites. With three of them being parsimony informative, no meaningful phylogram could be inferred from the 28S rDNA sequences. The ITS region is known to evolve more rapidly and is used to provide discrimination within species (27). The curated ITS alignments consisted of 621 sites, 14 being variable and 10 being parsimony informative. Four short insertions or deletions, which are mostly ignored by phylogeny inference software, were found. The phylogram based on both ITS data sets was juxtaposed with the endosymbiont tree determined by MLST (Fig. 4). The host strains of the Burkholderia sp. isolates B1, B2, and B6 (ATCC 62417, ATCC 20577, and CBS 261.28, respectively) were identical and reproduced the Pacific group of endobacteria. Strikingly, in contrast to their bacterial partners, the fungal host strains CBS 308.87 (Burkholderia sp. isolate B8) from Australia and CBS 111563 (Burkholderia sp. isolate B3) from Vietnam are members of the Pacific group as well. The strains CBS 700.08 (Burkholderia sp. isolate B7) from Georgia and ATCC 699.68 (Burkholderia sp. isolate B4) from Ukraine appeared to represent the Eurasian branch. This group was known from the bacterial phylogeny, but the fungal clade missed the close relationship to CBS 111563 (Burkholderia sp. isolate B3). Again, the African branch, consisting exclusively of CBS 112285 (Burkholderia sp. isolate B5), shared a common ancestor with the Eurasian group. These results are in accordance with cospeciation of some fungal hosts and their endosymbionts, especially for Burkholderia sp. isolates B1, B2, and B6 and B5, B4, and B7 (Fig. 4). However, there might be first evidence for some host switching events in the history of the endofungal bacteria (Burkholderia sp. isolates B3 and B8). Although this hypothesis is based mainly on a few informative sites within the ITS data set, three insertion or deletion events within the alignment support the extended Pacific group (Burkholderia sp. isolates B1, B2, B6, B3, and B8), indicating the horizontal transfer of symbionts and/or genetic material between strains. While it is possible that multiple events led to this unusual symbiosis, a scenario in which all symbiont strains are derived from an ancestral association seems to be more likely. The endosymbiont-dependent sporulation of the host strain indicates that the fungus-bacterium interaction is highly specialized. Furthermore, vertical transmission of the symbionts through spores is an efficient strategy for rapid distribution (4, 20). Nonetheless, our data suggest that the horizontal transmission of symbionts might also have played a role during the evolution of the endofungal bacteria.
FIG. 4.
Juxtaposition of phylogenetic trees derived from the MLST data of the endofungal symbiont complex (isolates B1 to B8) and the ITS sequence data of the fungal host (strain designations of fungal isolates of the genus Rhizopus are given). Dashed lines are representative of a symbiotic relationship. The numbers on top of the branches indicate the clade probability values. Shading designates similar clade affiliations for the bacterial symbiont and the fungal host.
In conclusion, we have investigated eight bacterial endosymbiont strains isolated from toxinogenic R. microsporus strains in pure culture. All isolates are representatives of the same unique “endofungal” ecotype, albeit the hosts' origins cover all five continents and occur in highly diverse niches. The bacterial endosymbionts share characteristic phenotypic traits, like secondary metabolite production and protein profile, as demonstrated by HPLC-MS and matrix-assisted laser desorption ionization-time of flight biotyping, respectively. Phylogenetic analyses (16S rRNA genes) provide strong evidence that all symbiont strains originate from a common ancestor and form a new complex within the genus Burkholderia. This observation is strongly supported by MLST, according to which all eight symbiont isolates can be grouped into continental branches. Results revealing both similar and deviating geographical groupings of fungal isolates in comparison to bacterial endosymbionts allow hypothesizing about the possible cospeciation of fungal and bacterial symbionts and some extent of horizontal-transmission events. All bacterial strains investigated seem to have evolved mainly separately from each other, not showing extensive recombination. In addition, we present preliminary evidence that there might be a mutational bias toward high AT contents, as is known for other endosymbiotic bacteria.
Supplementary Material
Acknowledgments
This project was financially supported by the International Leibniz Research School for Microbial and Biomolecular Interactions (ILRS) and the Jena School for Microbial Communication (JSMC).
We thank G.-M. Schwinger for fungal-strain maintenance.
Footnotes
Published ahead of print on 13 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abe, A., Y. Oda, K. Asano, and T. Sone. 2006. The molecular phylogeny of the genus Rhizopus based on rDNA sequences. Biosci. Biotechnol. Biochem. 70:2387-2393. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, A., E. Mahenthiralingam, P. Drevinek, C. Pope, D. Waine, D. Henry, et al. 2008. Elucidating global epidemiology of Burkholderia multivorans in cases of cystic fibrosis by multilocus sequence typing. J. Clin. Microbiol. 46:290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brendel, N., L. P. Partida-Martinez, K. Scherlach, and C. Hertweck. 2007. A cryptic PKS/NRPS gene cluster in the plant commensal Pseudomonas fluorescens Pf-5 codes for the biosynthesis of an antimitotic rhizoxin complex. Org. Biomol. Chem. 5:2211-2213. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. K., and M. S. Hovmoller. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297:537-541. [DOI] [PubMed] [Google Scholar]
- 5.Bruen, T. C., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, A., L. Ward, D. Godoy, R. Norton, M. Mayo, D. Gal, et al. 2008. Genetic diversity of Burkholderia pseudomallei isolates in Australia. J. Clin. Microbiol. 46:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudler, R., and L. Eberl. 2006. Interactions between bacteria and eukaryotes via small molecules. Curr. Opin. Biotechnol. 17:268-273. [DOI] [PubMed] [Google Scholar]
- 8.Godoy, D. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huson, D., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 9a.Huson, D. H. 1998. SplitsTree: a program for analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki, S., H. Kobayashi, J. Furukawa, M. Namikoshi, S. Okuda, Z. Sato, I. Matsuda, and T. Noda. 1984. Studies on macrocyclic lactone antibiotics. VII. Structure of a phytotoxin “rhizoxin” produced by Rhizopus chinensis. J. Antibiot. 37:354-362. [DOI] [PubMed] [Google Scholar]
- 11.Kiyoto, S., Y. Kawai, T. Kawakita, E. Kino, M. Okuhara, I. Uchida, H. Tanaka, M. Hashimoto, and H. Terano. 1986. A new antitumor complex, WF-1360, WF-1360A, B, C, D, E and F. J. Antibiot. 39:762-772. [DOI] [PubMed] [Google Scholar]
- 12.Moran, N. A. 2006. Symbiosis. Curr. Biol. 16:R866-R871. [DOI] [PubMed] [Google Scholar]
- 13.Partida-Martinez, L. P., I. Groth, M. Roth, I. Schmitt, K. Buder, and C. Hertweck. 2007. Burkholderia rhizoxinica and Burkholderia endofungorum, bacterial endosymbionts of the rice pathogenic fungus Rhizopus microsporus. Int. J. Syst. Evol. Microbiol. 57:2583-2590. [DOI] [PubMed] [Google Scholar]
- 14.Partida-Martinez, L. P., and C. Hertweck. 2007. A gene cluster encoding rhizoxin biosynthesis in Burkholderia rhizoxina, the bacterial endosymbiont of the fungus Rhizopus microsporus. Chembiochem 8:41-45. [DOI] [PubMed] [Google Scholar]
- 15.Partida-Martinez, L. P., and C. Hertweck. 2005. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437:884-888. [DOI] [PubMed] [Google Scholar]
- 16.Partida-Martinez, L. P., C. Looß, K. Ishida, M. Ishida, and C. Hertweck. 2007. Rhizonin, the first mycotoxin isolated from lower fungi, is not a fungal metabolite but is produced by bacterial endosymbionts. Appl. Environ. Microbiol. 73:793-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partida-Martinez, L. P., S. Monajembashi, K. O. Greulich, and C. Hertweck. 2007. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr. Biol. 17:773-777. [DOI] [PubMed] [Google Scholar]
- 18.Partida-Martinez, L. P., R. Rüchel, E. Dannoui, and C. Hertweck. 2008. Lack of evidence for endofungal bacteria in the development of Rhizopus-borne zygomycoses in humans. Mycoses 51:266-269. [DOI] [PubMed] [Google Scholar]
- 19.Paulsen, I. T., C. M. Press, J. Ravel, D. Y. Kobayashi, G. S. Myers, D. V. Mavrodi, R. T. DeBoy, R. Seshadri, Q. Ren, R. Madupu, R. J. Dodson, A. S. Durkin, L. M. Brinkac, S. C. Daugherty, S. A. Sullivan, M. J. Rosovitz, M. L. Gwinn, L. Zhou, D. J. Schneider, S. W. Cartinhour, W. C. Nelson, J. Weidman, K. Watkins, K. Tran, H. Khouri, E. A. Pierson, L. S. Pierson III, L. S. Thomashow, and J. E. Loper. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-05. Nat. Biotechnol. 23:873-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raboin, L. M., A. Selvi, K. M. Olivieira, F. Paulet, C. Calatayud, M. F. Zapater, P. Brottier, R. Lazaran, O. Garsmeur, J. Carlier, and A. D'Hont. 2006. Evidence for the dispersal of a unique lineage from Asia to America and Africa in the sugarcane fungal pathogen Ustilago scitaminea. Fungal Genet. Biol. 44:64-76. [DOI] [PubMed] [Google Scholar]
- 21.Ribes, J. A., C. L. Vanover-Sams, and D. J. Baker. 2000. Zygomycetes in human disease. Clin. Microbiol. Rev. 13:236-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenblueth, M., and E. Martinez-Romero. 2006. Bacterial endophytes and their interactions with hosts. Mol. Plant-Microbe Interact. 19:827-837. [DOI] [PubMed] [Google Scholar]
- 23.Scherlach, K., L. P. Partida-Martinez, H.-M. Dahse, and C. Hertweck. 2006. Antimitotic rhizoxin derivatives from cultured symbionts of the rice pathogenic fungus Rhizopus microsporus. J. Am. Chem. Soc. 128:11529-11536. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt, I., L. P. Partida-Martinez, R. Winkler, K. Voigt, A. Einax, J. Wöstemeyer, and C. Hertweck. 2008. Evolution of host-resistance in a toxin-producing fungal-bacterial mutualism. ISME J. 2:632-641. [DOI] [PubMed] [Google Scholar]
- 25.Steyn, P. S., A. A. Tuinman, F. R. van Heerden, P. H. van Rooyen, P. L. Wessels, and C. J. Rabie. 1983. The isolation, structure, and absolute configuration of the mycotoxin rhizonin A, a novel cyclic heptapeptide containing N-methyl-3-(furyl)alanine, produced by Rhizopus microsporus. J. Chem. Soc. Chem. Commun. 1983:47-49. [Google Scholar]
- 26.Wernegreen, J. J., and D. J. Funk. 2004. Mutation exposed: a neutral explanation for extreme base composition of an endosymbiont genome. J. Mol. Evol. 59:849-858. [DOI] [PubMed] [Google Scholar]
- 27.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis et al. (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.
- 28.Wilson, T., C. J. Rabie, J. E. Fincham, P. S. Steyn, and M. A. Schipper. 1984. Toxicity of rhizonin A, isolated from Rhizopus microsporus, in laboratory animals. Food Chem. Toxicol. 22:275-281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.