Abstract
Here, we report a fluorescence in situ hybridization (FISH) method for rapid detection of Cronobacter strains in powdered infant formula (PIF) using a novel peptide nucleic acid (PNA) probe. Laboratory tests with several Enterobacteriaceae species showed that the specificity and sensitivity of the method were 100%. FISH using PNA could detect as few as 1 CFU per 10 g of Cronobacter in PIF after an 8-h enrichment step, even in a mixed population containing bacterial contaminants.
Cronobacter strains were originally described as Enterobacter sakazakii (12), but they are now known to comprise a novel genus consisting of six separate genomospecies (20, 21). These opportunistic pathogens are ubiquitous in the environment and various types of food and are occasionally found in the normal human flora (11, 12, 16, 32, 47). Based on case reports, Cronobacter infections in adults are generally less severe than Cronobacter infections in newborn infants, with which a high fatality rate is associated (24).
The ability to detect Cronobacter and trace possible sources of infection is essential as a means of limiting the impact of these organisms on neonatal health and maintaining consumer confidence in powdered infant formula (PIF). Conventional methods, involving isolation of individual colonies followed by biochemical identification, are more time-consuming than molecular methods, and the reliability of some currently proposed culture-based methods has been questioned (28). Recently, several PCR-based techniques have been described (23, 26, 28-31, 38). These techniques are reported to be efficient even when low levels of Cronobacter cells are found in a sample (0.36 to 66 CFU/100 g). However, PCR requires DNA extraction and does not allow direct, in situ visualization of the bacterium in a sample.
Fluorescence in situ hybridization (FISH) is a method that is commonly used for bacterial identification and localization in samples. This method is based on specific binding of nucleic acid probes to particular DNA or RNA target regions (1, 2). rRNA has been regarded as the most suitable target for bacterial FISH, allowing differentiation of potentially viable cells. Traditionally, FISH methods are based on the use of conventional DNA oligonucleotide probes, and a commercial system, VIT-E sakazakii (Vermicon A.G., Munich, Germany), has been developed based on this technology (25). However, a recently developed synthetic DNA analogue, peptide nucleic acid (PNA), has been shown to provide improved hybridization performance compared to DNA probes, making FISH procedures easier and more efficient (41). Taking advantage of the PNA properties, FISH using PNA has been successfully used for detection of several clinically relevant microorganisms (5, 15, 17, 27, 34-36).
Cronobacter probe design and theoretical evaluation.
The Primrose program (http://www.cf.ac.uk/biosi/research/biosoft/Primrose/index.html) coupled with the 16S rRNA databases of Ribosomal Database Project II (version 9.55; http://rdp.cme.msu.edu/html) (3, 8) was employed to identify oligonucleotides that potentially could be used as probes to detect Cronobacter. Two oligonucleotides capable of detecting all Cronobacter strains with no nontarget sequence matches with Primrose were identified. The oligonucleotide that had a lower number of nontarget sequences when one nucleotide mismatch was allowed and also had a higher G+C content (and thus higher predicted melting temperature) was selected for further study. The PNA oligomer sequence obtained was 5′-TGC AGG ATT CTC TGG-3′. This probe hybridizes between positions 971 and 985 of the Cronobacter sp. strain ATCC 29544 16S rRNA sequence and was designated SakPNA971. The sequence was synthesized (Panagene, Daejeon, South Korea), and the oligonucleotide N terminus was attached to an Alexa Fluor 594 molecule via a double AEEA linker.
The theoretical specificity and sensitivity of the probe were evaluated further with the updated and comprehensive databases available at Ribosomal Database Project II and at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). The values were determined as previously reported by Guimarães and coworkers (15). Searches confirmed that SakPNA971 detected not only all 99 E. sakazakii or Cronobacter sp. sequences but also 11 nontarget sequences with a total of 110 matches (last accession date, April 2008). Therefore, the estimated theoretical specificity and sensitivity were 90 and 100%, respectively.
The 11 non-Cronobacter strains detected by SakPNA971 were identified as 7 Acidithiobacillus sp. strains (accession numbers S000342376, S000392292, S000467258, S000712736, S000712737, S000722390, and S000859142), 3 uncultured Pseudomonadales strains (accession numbers S000668021, S000869002, and S000970044), and 1 Mangroveibacter plantisponsor strain (accession number S000893171). Considering that the numbers of Acidithiobacillus sp. and Pseudomonadales strains detected represent just 1.15% and 0.04%, respectively, of the total numbers of sequences in the database for these species, the risk of misidentification in PIF is very low. For M. plantisponsor, a salt-tolerant nitrogen-fixing bacterium (N. Ramesh Kumar and S. Nair, unpublished data), there is a lack of information about its morphology and its ecological reservoirs. Moreover, none of these species, with the possible exception of some Pseudomonadales species, are expected to contaminate infant formula (10, 13, 45).
Protocol optimization and autofluorescence-related factors.
Protocols were developed both for hybridization on slides, based on the method described by Guimarães et al. (15), and for hybridization in suspension, as reported by Perry-O' Keefe et al. (35). However, the following modifications of the hybridization and fixation steps were necessary to obtain successful hybridization. Inclusion of a 10-min paraformaldehyde immersion step before ethanol fixation as a means of increasing the signal-to-noise ratio was critical and also appeared to result in a reduction in sample autofluorescence. No differences in signal intensity were found with various ethanol concentrations (50 and 80%) in the fixation step. Hybridization and washing temperatures were also modified as the highest signal-to-noise ratio was obtained at 57°C (see Fig. S1 in the supplemental material). Hybridization times of 30, 45, 60, and 90 min were found to be equally efficient. However, as autofluorescence appeared to increase with hybridization time, the 30-min assay was used preferentially. All these observations were applicable for hybridizations performed on slides and for hybridizations performed in suspension.
After hybridization, samples were stored at 4°C in the dark for a maximum of 24 h before microscopy. Visualization by microscopy was performed using an Olympus BX51 (Olympus Portugal SA, Porto, Portugal) epifluorescence microscope equipped with a filter sensitive to the Alexa Fluor 594 signaling molecule attached to PNA probe (excitation wavelengths, 530 to 550 nm; barrier wavelength, 570 nm; emission long-pass wavelength, 591 nm). To ensure that the signal obtained was not related to autofluorescence, all samples were visualized with other available filters. For every experiment, a negative control was performed simultaneously, using all the steps for standard hybridization but without addition of the probe to the hybridization solution.
Cronobacter probe specificity test.
Following optimization of the hybridization conditions, the specificity of the PNA probe was tested using 51 Cronobacter strains (representative of the phenotypic and genetic diversity in this genus), 23 Enterobacter strains, and 31 strains of other related bacteria or expected contaminants of milk and milk products (Table 1). The bacterial strains used were kept at −80°C and subcultured once on tryptic soy agar (VWR, Portugal) at 37°C for 24 h before each experiment. As shown in Table 1 and as expected from the predicted sensitivity value, SakPNA971 hybridized with all the Cronobacter strains, whereas no hybridization was observed for the other species used, so the practical levels of sensitivity and specificity were 100%.
TABLE 1.
Results of the Cronobacter probe specificity test
| Microorganism(s) (no. of strains tested)a | Results of FISH with PNA |
|---|---|
| Cronobacter sakazakii (36), including ATCC 29544T and 274b | + |
| Cronobacter dublinensis subsp. lactaridi (2), Cronobacter dublinensis subsp. lausannensis, and Cronobacter dublinensis subsp. dublinensis | + |
| Cronobacter muytjensii (3), including ATCC 51329T | + |
| Cronobacer malonaticus (6) | + |
| Cronobacter genomospecies 1 | + |
| Cronobacter turicensis | + |
| Enterobacter helveticus (8) | − |
| Enterobacter pulveris (6) | − |
| Enterobacter asburiae (2) | − |
| Enterobacter hormaechei (2) | − |
| Enterobacter turicensis (2) | − |
| Enterobacter cloacae | − |
| Enterobacter aerogenes ATCC 12048T | − |
| Enterobacter amnigenus ATCC 33072T | − |
| Shigella flexneri ATCC 12022T | − |
| Staphylococcus aureus ATCC 12600T, ATCC 6538T, and ATCC 13565T | − |
| Staphylococcus epidermidis ATCC 35983T, ATCC 35984T, ATCC 1798T, and ATCC 14990T | − |
| Escherichia coli ATCC 25922T, N5,b and N9b | − |
| Salmonella enterica serotype Enteritidis ATCC 13076T, 349,b 355,b 357,b A1,b CC,b Sal4,b and Sal6,bSalmonella enterica serotype Heidelberg 354,b and Salmonella enterica serotype Typhimurium NCTC 12416T | − |
| Pseudomonas fluorescens ATCC 13525T and N3b | − |
| Pseudomonas aeruginosa ATCC 10145T | − |
| Serratia plymuthica F4b | − |
| Listeria monocytogenes 747,b 925,b 930,b 994,b and 1562b | − |
| Bacillus cereusb | − |
T = type strain.
Isolate.
Detection of Cronobacter in PIF.
The FISH procedure using PNA was then adapted to detect Cronobacter directly in a commercially available PIF, based on the previously work of Mohan Nair and Venkitanarayanan (31). Initially, we performed an experiment to assess the experimental detection limit of FISH with PNA. For this, Cronobacter muytjensii ATCC 51329 was resuspended in reconstituted PIF (10% [wt/wt]; NAN 1 Premium; Nestle) at concentrations ranging from 1 × 108 to 1 × 102 CFU/ml. The PIF was reconstituted in water at ca. 60°C, a temperature commonly used for rehydration (13). One-milliliter aliquots of each dilution were concentrated by centrifugation as described above, and hybridization was performed in suspension or on glass slides. Microscopic visualization showed that this procedure could detect the pathogen at a concentration of 1 × 106 CFU/ml.
The theoretical detection limit was also calculated based on the assumption that for trustworthy analysis each microscopy field should contain at least five cells. This calculation considered the microscopic field area (0.0158 mm2), the slide well area (200.96 mm2), and the dilution used during the procedure. The value obtained, 2 × 105 cells/ml, was lower than the value determined by laboratory testing. The difference could be explained by the fact that the theoretical value is between the 10-fold dilutions analyzed, 1 × 102 to 1 × 108 CFU/ml. Moreover, during laboratory testing some cells might be lost in the centrifugation steps, decreasing the real cellular concentration.
After determination of the detection limit, we used the FISH procedure with PNA to detect Cronobacter in PIF samples with different levels of contamination. Serial 10-fold dilutions of Cronobacter sp. strain ATCC 51329 were prepared using reconstituted formula to obtain final concentrations of Cronobacter ranging from 1 × 10−4 to 1 × 107 CFU/ml (corresponding to 1 × 10−2 to 1 × 109 CFU/10 g). Given the short lag time and the growth rate of Cronobacter in PIF, it was expected that a single cell could produce a concentration of more than 1 × 107 CFU/ml in an 8-h enrichment period (19, 22). For this reason, after 8 h of enrichment at 37°C, 1-ml samples were taken and diluted 1:10, and hybridization was performed in suspension or on glass slides, as described above. A noncontaminated culture was prepared in parallel and exposed to the same conditions. This experiment was performed three times and was repeated with two other Cronobacter strains (ATCC 29544 and 274). The bacterial concentration in each enriched culture was determined by conventional counting and by FISH with PNA (see Table S1 in the supplemental material). Quantification by FISH with PNA was obtained by epifluorescence microscopy by counting a total of 15 fields. Approximately 57 to 87% of the PNA-FISH-labeled cells were detected by culture methods, and the final concentrations were always higher than the theoretical or experimental detection limit previously determined.
The procedure was able to detect Cronobacter in PIF samples with an initial concentration of 1 × 10−2 CFU/ml (and even with an initial concentration of 1 × 10−3 CFU/ml for 4 of 27 replicates) after an 8-h preenrichment step. The previously reported levels of Cronobacter in PIF samples are very low. For instance, Muytjens et al. examined 141 different powdered formulas and isolated Cronobacter at levels ranging from 0.36 to 66 CFU per 100 g (33). In other work, Simmons et al. isolated 8 Cronobacter CFU per 100 g from PIF associated with an outbreak of clinical illness in Tennessee (39). Based on our findings, our assay is capable of detecting less than 1 CFU per 10 g of PIF, which compares well with previous reports.
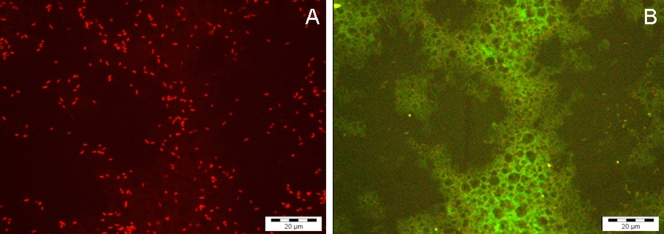
Autofluorescence of infant formula proteins remained detectable, but, as the sample was diluted at least 1:10 before hybridization, autofluorescence did not interfere with bacterial detection (Fig. 1). Moreover, when hybridization was performed in suspension, autofluorescence was almost undetectable.
FIG. 1.
(A) Detection of C. sakazakii 274 on a glass slide using the SakPNA971 probe, a preenriched culture (10% PIF), and an initial concentration of 1 to 3 CFU/100 ml reconstituted PIF. (B) Visualization of the same microscopic field with the green channel (negative control), showing autofluorescence of infant formula proteins and the absence of fluorescent cells.
The experiments with PIF were repeated with the same strains using freeze-dried Cronobacter cells, as in the experiments performed previously by Iversen and Forsythe (18). With this approach we intended to investigate if the physiological state of the cells and contamination in powdered or reconstituted infant formula could affect the outcome of FISH with PNA or the concentration determined. The results showed that there was no difference between the two physiological conditions tested.
Detection of Cronobacter in a mixed population.
SakPNA971 was also employed with a mixed population containing Cronobacter cells together with other species. For this experiment, serial 10-fold dilutions of Cronobacter sp. strain ATCC 51329 were made in reconstituted formula, as described above, resulting in concentrations ranging from 103 to 108 CFU/ml, and mixed with Bacillus cereus at 108 CFU/ml. One-milliliter samples were concentrated by centrifugation (10,000 × g for 5 min), and hybridization was performed as described above. The sample was then counterstained with 20 μl (10 μg/ml) of 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) and incubated for 10 min in the dark. The excess DAPI was removed, and the sample was allowed to air dry, mounted with nonfluorescent immersion oil (Merck), and covered with a coverslip. The same procedure was repeated with other species, including Salmonella enterica serotype Enteritidis ATCC 13076 and Pseudomonas aeruginosa ATCC 10145, and also with all four species simultaneously. Pseudomonas and Bacillus species were chosen because they are common contaminants of milk products and infant foods (4, 10, 13, 14, 37, 43), and S. enterica was chosen because it is a pathogen that is phylogenetically more closely related to Cronobacter that has also been reported to be an occasional contaminant of PIF (6, 7, 42, 44).
DAPI counterstaining facilitated visualization of the total bacterial population and allowed determination of the percentage of Cronobacter cells in the heterotrophic mixture of cells.
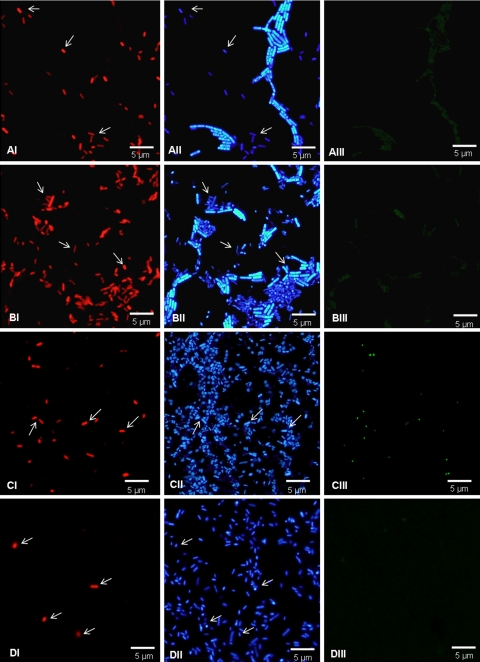
It was observed that FISH with PNA allows easy detection of the target microorganism in mixed populations, even when the concentration of the target microorganism is clearly lower (Fig. 2). In mixed samples containing Cronobacter cells that were 10- to 100-fold diluted compared to the P. aeruginosa, B. cereus, and S. enterica cells, Cronobacter cells were easily detected using the SakPNA971 probe. Even when the proportion of Cronobacter cells was 1:300 (ratio of Cronobacter cells to P. aeruginosa cells to B. cereus cells to S. enterica cells, 1:100:100:100), Cronobacter cells were easily detectable. For samples containing Cronobacter cells diluted more than 1,000-fold, detection was not possible as the number of cells was below the detection limit of FISH with PNA established previously.
FIG. 2.
Detection of C. sakazakii ATCC 29544 in suspension with a mixed population using SakPNA971. (A) Reconstituted PIF with C. sakazakii and B. cereus. (B) Reconstituted PIF with C. sakazakii, B. cereus, P. aeruginosa ATCC 10145, and S. enterica serotype Enteritidis ATCC 13076. (C) Reconstituted PIF with C. sakazakii, S. enterica serotype Enteritidis ATCC 13076 (100-fold concentrated), and P. aeruginosa ATCC 10145 (100-fold concentrated). (D) Reconstituted PIF with C. sakazakii and S. enterica serotype Enteritidis ATCC 13076 (100-fold concentrated). (Panels I) Detection of C. sakazakii using the red fluorescent SakPNA971 probe. (Panels II) Counterstaining with DAPI (total population). (Panels III) Visualization of the same microscopic field with the green channel (negative control). The arrows indicate C. sakazakii cells that could easily be visualized in the DAPI channel with the red-labeled SakPNA971 probe. All images were obtained using the same exposure time.
In conclusion, FISH procedure using the SakPNA971 probe has been shown to be a sensitive and specific method for detection of Cronobacter in PIF. Using this approach, a qualitative assay detected Cronobacter in less than 12 h, and the detection limit was less than 1 CFU per 10 g of PIF.
It is also important to note that most previously described protocols that use a molecular approach to detect E. sakazakii are PCR-based protocols that target DNA (23, 26, 28, 29, 31). The previous studies reported development of several primers and DNA probes, but a DNA extraction step is required and, when low levels of Cronobacter are present, an additional enrichment step is also needed. The total time required for the FISH assay using PNA is similar to or less than the times reported for PCR-based methods (9, 28, 29, 31), while culture-based techniques and biochemical tests, which are currently used in hospitals, take about 5 to 7 days to confirm the presence of the organism (9, 31). FISH using PNA is likely to be easily adapted for identification and quantification of Cronobacter in several other types of samples, such as cerebrospinal fluid and blood (17, 40, 46), although this was not tested in this work.
Future work could take advantage of suitable PNA probes that already have been developed and the very narrow emission band of the Alexa fluorophore attached to SakPNA971 to develop multiplex assays that detect a large number of different pathogens. Moreover, combination with flow cytometry, as reported by Hartmann and coworkers, could result in easier, faster, and accurate distinction of several species simultaneously (17).
Supplementary Material
Acknowledgments
This work was supported by the Portuguese Institute Fundação para a Ciência e Tecnologia (Ph.D. fellowship SFRH/BD/29297/2006 and postdoctoral fellowship SFRH/BPD/42208/2007).
Footnotes
Published ahead of print on 6 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R., and B. M. Fuchs. 2008. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6:339-348. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R., F. O. Glockner, and A. Neef. 1997. Modern methods in subsurface microbiology: in situ identification of microorganisms with nucleic acid probes. FEMS Microbiol. Rev. 20:191-200. [Google Scholar]
- 3.Ashelford, K. E., A. J. Weightman, and J. C. Fry. 2002. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 30:3481-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, H., G. Schaller, W. von Wiese, and G. Terplan. 1994. Bacillus cereus in infant foods and dried milk products. Int. J. Food Microbiol. 23:1-15. [DOI] [PubMed] [Google Scholar]
- 5.Brehm-Stecher, B. F., J. J. Hyldig-Nielsen, and E. A. Johnson. 2005. Design and evaluation of 16S rRNA-targeted peptide nucleic acid probes for whole-cell detection of members of the genus Listeria. Appl. Environ. Microbiol. 71:5451-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brouard, C., E. Espie, F. X. Weill, A. Kerouanton, A. Brisabois, A. M. Forgue, V. Vaillant, and H. de Valk. 2007. Two consecutive large outbreaks of Salmonella enterica serotype Agona infections in infants linked to the consumption of powdered infant formula. Pediatr. Infect. Dis. J. 26:148-152. [DOI] [PubMed] [Google Scholar]
- 7.Cahill, S. M., I. K. Wachsmuth, L. Costarrica Mde, and P. K. Ben Embarek. 2008. Powdered infant formula as a source of Salmonella infection in infants. Clin. Infect. Dis. 46:268-273. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derzelle, S., and F. Dilasser. 2006. A robotic DNA purification protocol and real-time PCR for the detection of Enterobacter sakazakii in powdered infant formulae. BMC Microbiol. 6:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dogan, B., and K. J. Boor. 2003. Genetic diversity and spoilage potentials among Pseudomonas spp. isolated from fluid milk products and dairy processing plants. Appl. Environ. Microbiol. 69:130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drudy, D., M. O'Rourke, M. Murphy, N. R. Mullane, R. O'Mahony, L. Kelly, M. Fischer, S. Sanjaq, P. Shannon, P. Wall, M. O'Mahony, P. Whyte, and S. Fanning. 2006. Characterization of a collection of Enterobacter sakazakii isolates from environmental and food sources. Int. J. Food Microbiol. 110:127-134. [DOI] [PubMed] [Google Scholar]
- 12.Farmer, J. J., III, M. A. Asbury, F. W. Hickman, D. J. Brenner, and the Enterobacteriaceae Study Group. 1980. Enterobacter sakazakii: a new species of “Enterobacteriaceae” isolated from clinical specimens. Int. J. Syst. Bacteriol. 30:569-584. [Google Scholar]
- 13.Forsythe, S. J. 2005. Enterobacter sakazakii and other bacteria in powdered infant milk formula. Matern. Child. Nutr. 1:44-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gras-Le Guen, C., D. Lepelletier, T. Debillon, V. Gournay, E. Espaze, and J. C. Roze. 2003. Contamination of a milk bank pasteuriser causing a Pseudomonas aeruginosa outbreak in a neonatal intensive care unit. Arch. Dis. Child. Fetal Neonatal Ed. 88:F434-F435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guimarães, N., N. F. Azevedo, C. Figueiredo, C. W. Keevil, and M. J. Vieira. 2007. Development and application of a novel peptide nucleic acid probe for the specific detection of Helicobacter pylori in gastric biopsy specimens. J. Clin. Microbiol. 45:3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurtler, J. B., J. L. Kornacki, and L. R. Beuchat. 2005. Enterobacter sakazakii: a coliform of increased concern to infant health. Int. J. Food Microbiol. 104:1-34. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann, H., H. Stender, A. Schafer, I. B. Autenrieth, and V. A. Kempf. 2005. Rapid identification of Staphylococcus aureus in blood cultures by a combination of fluorescence in situ hybridization using peptide nucleic acid probes and flow cytometry. J. Clin. Microbiol. 43:4855-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iversen, C., and S. J. Forsythe. 2007. Comparison of media for the isolation of Enterobacter sakazakii. Appl. Environ. Microbiol. 73:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iversen, C., M. Lane, and S. J. Forsythe. 2004. The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula milk. Lett. Appl. Microbiol. 38:378-382. [DOI] [PubMed] [Google Scholar]
- 20.Iversen, C., A. Lehner, N. Mullane, E. Bidlas, I. Cleenwerck, J. Marugg, S. Fanning, R. Stephan, and H. Joosten. 2007. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov., Cronobacter sakazakii subsp. sakazakii comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies I. BMC Evol. Biol. 7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iversen, C., N. Mullane, B. McCardell, B. D. Tall, A. Lehner, S. Fanning, R. Stephan, and H. Joosten. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 58:1442-1447. [DOI] [PubMed] [Google Scholar]
- 22.Kandhai, M. C., M. W. Reij, C. Grognou, M. van Schothorst, L. G. Gorris, and M. H. Zwietering. 2006. Effects of preculturing conditions on lag time and specific growth rate of Enterobacter sakazakii in reconstituted powdered infant formula. Appl. Environ. Microbiol. 72:2721-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keyser, M., R. C. Witthuhn, L. C. Ronquest, and T. J. Britz. 2003. Treatment of winery effluent with upflow anaerobic sludge blanket (UASB)-granular sludges enriched with Enterobacter sakazakii. Biotechnol. Lett. 25:1893-1898. [DOI] [PubMed] [Google Scholar]
- 24.Lai, K. K. 2001. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore) 80:113-122. [DOI] [PubMed] [Google Scholar]
- 25.Lehner, A., S. Nitzsche, P. Breeuwer, B. Diep, K. Thelen, and R. Stephan. 2006. Comparison of two chromogenic media and evaluation of two molecular based identification systems for Enterobacter sakazakii detection. BMC Microbiol. 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehner, A., T. Tasara, and R. Stephan. 2004. 16S rRNA gene based analysis of Enterobacter sakazakii strains from different sources and development of a PCR assay for identification. BMC Microbiol. 4:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehtola, M. J., C. J. Loades, and C. W. Keevil. 2005. Advantages of peptide nucleic acid oligonucleotides for sensitive site directed 16S rRNA fluorescence in situ hybridization (FISH) detection of Campylobacter jejuni, Campylobacter coli and Campylobacter lari. J. Microbiol. Methods 62:211-219. [DOI] [PubMed] [Google Scholar]
- 28.Liu, Y., X. Cai, X. Zhang, Q. Gao, X. Yang, Z. Zheng, M. Luo, and X. Huang. 2006. Real time PCR using TaqMan and SYBR Green for detection of Enterobacter sakazakii in infant formula. J. Microbiol. Methods 65:21-31. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y., Q. Gao, X. Zhang, Y. Hou, J. Yang, and X. Huang. 2006. PCR and oligonucleotide array for detection of Enterobacter sakazakii in infant formula. Mol. Cell. Probes 20:11-17. [DOI] [PubMed] [Google Scholar]
- 30.Malorny, B., and M. Wagner. 2005. Detection of Enterobacter sakazakii strains by real-time PCR. J. Food Prot. 68:1623-1627. [DOI] [PubMed] [Google Scholar]
- 31.Mohan Nair, M. K., and K. S. Venkitanarayanan. 2006. Cloning and sequencing of the ompA gene of Enterobacter sakazakii and development of an ompA-targeted PCR for rapid detection of Enterobacter sakazakii in infant formula. Appl. Environ. Microbiol. 72:2539-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullane, N. R., C. Iversen, B. Healy, C. Walsh, P. Whyte, P. G. Wall, T. Quinn, and S. Fanning. 2007. Enterobacter sakazakii, an emerging bacterial pathogen with implications for infant health. Minerva Pediatr. 59:137-148. [PubMed] [Google Scholar]
- 33.Muytjens, H. L., H. Roelofswillemse, and G. H. J. Jaspar. 1988. Quality of powdered substitutes for breast milk with regard to members of the family Enterobacteriaceae. J. Clin. Microbiol. 26:743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira, K., G. W. Procop, D. Wilson, J. Coull, and H. Stender. 2002. Rapid identification of Staphylococcus aureus directly from blood cultures by fluorescence in situ hybridization with peptide nucleic acid probes. J. Clin. Microbiol. 40:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry-O'Keefe, H., S. Rigby, K. Oliveira, D. Sorensen, H. Stender, J. Coull, and J. J. Hyldig-Nielsen. 2001. Identification of indicator microorganisms using a standardized PNA FISH method. J. Microbiol. Methods 47:281-292. [DOI] [PubMed] [Google Scholar]
- 36.Prescott, A. M., and C. R. Fricker. 1999. Use of PNA oligonucleotides for the in situ detection of Escherichia coli in water. Mol. Cell. Probes 13:261-268. [DOI] [PubMed] [Google Scholar]
- 37.Reyes, J. E., J. M. Bastias, M. R. Gutierrez, and L. Rodriguez Mde. 2007. Prevalence of Bacillus cereus in dried milk products used by Chilean School Feeding Program. Food Microbiol. 24:1-6. [DOI] [PubMed] [Google Scholar]
- 38.Seo, K. H., and R. E. Brackett. 2005. Rapid, specific detection of Enterobacter sakazakii in infant formula using a real-time PCR assay. J. Food Prot. 68:59-63. [DOI] [PubMed] [Google Scholar]
- 39.Simmons, B. P., M. S. Gelfand, M. Haas, L. Metts, and J. Ferguson. 1989. Enterobacter sakazakii infections in neonates associated with intrinsic contamination of a powdered infant formula. Infect. Control Hosp. Epidemiol. 10:398-401. [DOI] [PubMed] [Google Scholar]
- 40.Sogaard, M., H. Stender, and H. C. Schonheyder. 2005. Direct identification of major blood culture pathogens, including Pseudomonas aeruginosa and Escherichia coli, by a panel of fluorescence in situ hybridization assays using peptide nucleic acid probes. J. Clin. Microbiol. 43:1947-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stender, H., M. Fiandaca, J. J. Hyldig-Nielsen, and J. Coull. 2002. PNA for rapid microbiology. J. Microbiol. Methods 48:1-17. [DOI] [PubMed] [Google Scholar]
- 42.Toyofuku, H., K. Kubota, and K. Morikawa. 2006. Outbreaks of Salmonella in infants associated with powdered infant formula. Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku 124:74-79. (In Japanese.) [PubMed] [Google Scholar]
- 43.Tudela, E., J. Croize, A. Lagier, and M. R. Mallaret. 2008. Microbiological monitoring of milk samples and surface samples in a hospital infant formula room. Pathol. Biol. (Paris) 56:272-278. (In French.) [DOI] [PubMed] [Google Scholar]
- 44.Usera, M. A., A. Echeita, A. Aladuena, M. C. Blanco, R. Reymundo, M. I. Prieto, O. Tello, R. Cano, D. Herrera, and F. Martinez-Navarro. 1996. Interregional foodborne salmonellosis outbreak due to powdered infant formula contaminated with lactose-fermenting Salmonella virchow. Eur. J. Epidemiol. 12:377-381. [DOI] [PubMed] [Google Scholar]
- 45.Wiedmann, M., D. Weilmeier, S. S. Dineen, R. Ralyea, and K. J. Boor. 2000. Molecular and phenotypic characterization of Pseudomonas spp. isolated from milk. Appl. Environ. Microbiol. 66:2085-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, D. A., M. J. Joyce, L. S. Hall, L. B. Reller, G. D. Roberts, G. S. Hall, B. D. Alexander, and G. W. Procop. 2005. Multicenter evaluation of a Candida albicans peptide nucleic acid fluorescent in situ hybridization probe for characterization of yeast isolates from blood cultures. J. Clin. Microbiol. 43:2909-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zogaj, X., W. Bokranz, M. Nimtz, and U. Romling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.