Abstract
Insect intestinal tracts harbor several novel, deep-rooting clades of as-yet-uncultivated bacteria whose biology is typically completely unknown. Here, we report the isolation of the first representative of the termite group 1 (TG1) phylum from sterile-filtered gut homogenates of a humivorous scarab beetle larva. Strain Pei191T is a mesophilic, obligately anaerobic ultramicrobacterium with a gram-negative cell envelope. Cells are typically rod shaped, but cultures are pleomorphic in all growth phases (0.3 to 2.5 μm long and 0.17 to 0.3 μm wide). The isolate grows heterotrophically on sugars and ferments d-galactose, d-glucose, d-fructose, d-glucosamine, and N-acetyl-d-glucosamine to acetate, ethanol, hydrogen, and alanine as major products but only if amino acids are present in the medium. PCR-based screening and comparative 16S rRNA gene sequence analysis revealed that strain Pei191T belongs to the “intestinal cluster,” a lineage of hitherto uncultivated bacteria present in arthropod and mammalian gut systems. It is only distantly related to the previously described so-called “endomicrobia” lineage, which comprises mainly uncultivated endosymbionts of termite gut flagellates. We propose the name “Elusimicrobium minutum” gen. nov., sp. nov. (type strain, Pei191T = ATCC BAA-1559T = JCM 14958T) for the first isolate of this deep-branching lineage and the name “Elusimicrobia” phyl. nov. for the former TG1 phylum.
Insect intestinal tracts harbor an enormous diversity of as-yet-uncultivated bacteria that are characterized only by their 16S rRNA gene sequences and whose biology is typically completely obscure (9, 17, 49). As in other environments (45), many of these sequences form deep-branching phylogenetic lineages that do not contain a single isolate (18, 28). One of these lineages is the termite group 1 (TG1), which was originally discovered by Ohkuma and Kudo (37) and recognized as a phylum-level group (candidate division) by Hugenholtz et al. (20). TG1 bacteria form a major proportion of the microbial community in the hindgut of lower termites (17, 69), where they inhabit the cytoplasm of the intestinal flagellates (38, 53). These so-called “endomicrobia” are specific for the respective flagellate species (21) and, at least in the case of “Candidatus Endomicrobium trichonymphae,” are cospeciating with their flagellate host (22).
However, the TG1 phylum also comprises several other deep-rooting lineages (>15% 16S rRNA gene sequence divergence). They are present in a variety of environments, including soils, sediments, and intestinal tracts (14). One of these lineages, the “intestinal cluster,” comprises sequences originating exclusively from intestinal habitats, including the termite gut, but is only distantly related to the lineage comprising the “endomicrobia” (14). Here, we report the isolation of a member of the intestinal cluster from the hindgut of a humivorous scarab beetle larva and its physiological and ultrastructural characterization. We propose a new species, “Elusimicrobium minutum” gen. nov. sp. nov., and define the phylogenetic framework for the first cultivated representative of the TG1 phylum.
MATERIALS AND METHODS
Cultivation and inoculum.
Pachnoda ephippiata (Coleoptera: Scarabaeidae) larvae were from a laboratory population raised on organic soil (30). Hindgut homogenates of four third-instar larvae were prepared under anoxic conditions (30). The homogenates (2 ml) were diluted in 10 ml of phosphate buffer solution (58) and centrifuged at 5,000 × g. The supernatants—originally intended as a medium supplement to promote growth of fastidious gut bacteria—were passed twice through 0.2-μm-pore-size cellulose acetate membrane filters (FP 30; Schleicher & Schüll), and the filtrate was added to a series of rubber-stoppered cultivation tubes containing basal medium amended with d-glucose, d-fructose, and N-acetyl-d-glucosamine (each, 2 mM). From culture tubes that showed visible growth, pure cultures were isolated by two consecutive agar dilution series in basal medium amended only with glucose (5 mM), following the protocols of Pfennig and Trüper (43).
AM5 basal medium was (3) reduced with 2 mM dithiothreitol and kept under a headspace of N2-CO2 (80:20, vol/vol). For enrichments, the medium was supplemented with Casamino Acids and yeast extract (0.4% each; Becton, Dickinson and Company, Le Pont de Claix, France). For isolation and growth experiments, each supplement was at 0.02%. Glucose (5 mM) was used as a substrate unless indicated otherwise. Cultures were routinely grown in 16-ml rubber-stoppered culture tubes with 5 ml of medium at 30°C and pH 7. To assay pH dependence of growth, the pH of the medium was adjusted by adding sterile solutions of NaOH or HCl (each 1 M). To assay the salt tolerance, the medium was mixed at different ratios with medium amended with 3.5% NaCl plus MgCl2 (10:1, by weight). Growth was determined photometrically by following the increase in optical density at 578 nm (OD578). Growth yields were estimated by using an OD-to-cell mass conversion factor that was determined with cultures (200 ml). Cell densities were estimated by differential interference contrast (DIC) microscopy (see below) using a Thoma counting chamber.
Metabolic profile.
Culture tubes with basal medium AM5 were amended with the respective substrates, inoculated with 1% preculture, and incubated as described above. Supernatants of fully grown cultures were analyzed for substrate utilization and product formation by high-performance liquid chromatography using an ion exclusion column (Resin-GPZH10812S2508; Grom, Herrenberg, Germany), a UV detector, and a refractive index detector (57). Metabolites were identified by comparing the retention times and the signal ratios of the UV and refractive index detectors to those of authentic standards. Pyruvate was also verified via an enzymatic assay using l-lactate dehydrogenase (EC 1.1.1.27) and standard procedures (6). Amino acids were quantified by precolumn derivatization with o-phthaldialdehyde and 3-mercaptopropionic acid followed by reverse-phase high-performance liquid chromatography and fluorimetric analysis (330/450 nm) (12) on a SIL OPA-3 column (Grom) using gradient elution according to the manufacturer's instructions. Ammonia was measured colorimetrically (2). Hydrogen was analyzed by gas chromatography using a thermal conductivity detector (57). For computation of electron balances, all metabolites were formally oxidized to CO2, and the number of electrons theoretically released from the respective amounts of products was compared with that of the amount of substrate consumed. Expressed on a percent basis, this calculation yielded the electron recovery as previously described (58).
Substrate spectra.
Utilization of a wide range of substrates was tested with API-50CH test kits (Biomerieux, Marcy-l'Etoile, France) according to the manufacturer's instructions, with the following exceptions. The test strips were incubated in a 4-liter anaerobic jar under a N2/CO2 atmosphere; the recommended medium was replaced with basal medium containing only 10 mM bicarbonate buffer, 2 mM glucose, and 18 mg liter−1 phenol red; and the results were read after 4 weeks of incubation.
MALDI-TOF analysis of amino acids.
Strain Pei191T was incubated with 13C-labeled glucose (99 at%; Cambridge Isotope Laboratories, Andover, MA). The supernatant of a fully grown culture was mixed (1:2:1, by volume) with an internal standard solution (1 mM aminoadipic acid) and a matrix solution containing 0.4% (wt/vol) α-cyano-4-hydroxycinnamic acid, 0.1% tetrafluoroacetic acid, and 70% acetonitrile. A small drop of the mixture (1 μl) was dried on a gold target and examined in an ABI 4800 Plus matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analyzer using a molecular mass window from 55 to 160 (25). Cultures with unlabeled glucose, the uninoculated medium, and the uninoculated medium containing 20 proteinogenic amino acids (1 mM each) were treated the same way and served as controls or standards on the same target.
Light microscopy.
Light microscopy was done with a Zeiss Axiophot epifluorescence microscope equipped with a cooled charge-coupled-device camera; filter sets for fluorescence microscopy were described by Schmitt-Wagner et al. (50). Nonstained cultures were routinely examined at a magnification of ×1,250 using DIC microscopy.
Gram staining was done according to Süßmuth et al. (54); Acetobacterium woodii (DSM1030T) and Desulfovibrio vulgaris (DSM2119) were used as controls. For 4′,6-diamidino-2-phenylindol (DAPI) staining, 3-ml samples were acidified with HCl (40 mM), fixed with formaldehyde (3%), mixed with 1 ml of DAPI solution (100 μg/ml), and incubated for 20 min at room temperature. The samples were filtered onto a black 0.2-μm-pore-size polycarbonate membrane (Cyclopore Track-Etched Membrane; Whatman). Color images were recorded using a Chroma 31000 filter set (Chroma, Fürstenfeldbruck, Germany), inverted, and enhanced in contrast with image-editing software, using only the green and blue channels of the image.
Fluorescence in situ hybridization was performed according to Pernthaler et al. (42) with a specific Cy3-labeled probe (Elm1034, 5′-GCA GCA CCT CGG CTG GCT TT-3′) and the fluorescein-labeled general eubacterial probe EUB338 (1). Desulfurella acetivorans (DSM5264) served as a negative control. Optimal hybridization conditions were determined by increasing the formamide concentration from 0 to 50% in steps of 10%.
TEM.
For transmission electron microscopy (TEM), cells were fixed directly in the growth medium with 3% (wt/vol) glutaraldehyde for 1 h, gently centrifuged, and washed twice in phosphate buffer, followed by 2% (wt/vol) osmium tetroxide fixation. Cells were dehydrated and embedded in Epon 812 resin using standard procedures (66).
Negative-contrast electron microscopy was done according to Valentine et al. (64), except that the cells were fixed with 2% (wt/vol) glutaraldehyde directly in the growth medium. After gentle centrifugation, cells were resuspended and placed on hydrophilized or nonhydrophilized carbon-coated 400-mesh grids. After preparation with 2% (wt/vol) uranyl acetate, specimens were immediately examined by electron microscopy.
Photomicrographs were taken in a Philips 301G electron microscope using negative film and then digitally scanned and enhanced in contrast with image-editing software.
PCR-based screening.
16S rRNA gene sequences of the intestinal cluster were amplified with primer set 3 (14), using DNA extracted from the following samples: horse feces; rabbit feces; cow rumen; and gut homogenates of lower termites (Hodotermopsis sjoestedti, Kalotermes flavicollis, and Zootermopsis nevadensis; pseudergates), higher termites (Nasutitermes corniger and Cubitermes ugandensis; worker caste), cockroaches (Blaberus giganteus and Nauphoeta cinnerea; adults), scarab beetle larvae (P. ephippiata), lepidopteran larvae (Manduca sexta), crickets (Achaeta domestica), and locusts (Schistocerca gregaria). PCR products were cloned and sequenced as previously described (14).
16S rRNA genes of the isolates were amplified by PCR using 27f (8) and 1492r (29) bacterial primers and sequenced on both strands.
Phylogenetic analysis.
The 16S rRNA gene sequence of strain Pei191T was fitted into an alignment of about 270,000 bacterial sequences in the ARB-Silva database (44) (version 96; http://www.arb-silva.de), using the automated tools of the ARB software package (33) (version 08.03.14org; http://www.arb-home.de). Putative chimeras in the Silva database were removed. Highly variable base positions were removed from the alignment using a frequency-based filter (50% criterion) calculated for all high-quality near-full-length sequences in the TG1 phylum (190); only 20 selected sequences from the “Candidatus Endomicrobia” (cluster I) were included to avoid bias toward this highly overrepresented group. A maximum-likelihood tree was constructed using fastDNAml (39); tree topology was tested using RAxML-III (52) and maximum-parsimony analysis (1,000 bootstraps each); these tools were also implemented in the ARB software package. Tree topologies were challenged by using different data sets varying with respect to positional masking and outgroup. Insufficiently resolved nodes are reported as a multifurcation. The sequences in the intestinal cluster were analyzed in the same way, except that the filter accommodated only the base positions covered by primer set 3 (14).
Other analyses.
Whole-cell fatty acids, respiratory lipoquinones, and polar lipids were analyzed by the Identification Service and B. J. Tindall, Deutsche Sammlung für Mikroorganismen und Zellkulturen, Braunschweig, Germany, using 0.5 g of freeze-dried cell material washed with distilled water. Fatty acids were analyzed according to Kämpfer and Kroppenstedt (24). Lipoquinones and polar lipids were extracted and analyzed by two-dimensional thin-layer chromatography (60, 61, 62).
RESULTS
Isolation and morphological characterization.
During the course of a cultivation-based characterization of the larval gut microbiota of P. ephippiata, we observed that uninoculated culture tubes amended with clarified gut homogenate showed microbial growth after 3 to 6 months of storage. Since nonsupplemented culture tubes from the same batch had remained sterile, we suspected that the contamination was introduced with the sterile-filtered gut homogenate. 16S rRNA gene sequencing of the DNA extracted from the tube cultures indicated that the contaminating cells were members of TG1, a bacterial candidate phylum without any cultured representatives.
We isolated the contaminating bacterium using a deep-agar dilution series. Small, white, lentil-shaped colonies appeared after 4 weeks. A pure culture, strain Pei191T, was obtained from the highest dilution. The 16S rRNA gene sequence of strain Pei191T was identical to the sequence obtained from the original culture. Repetition of the procedure with sterile-filtered gut homogenates obtained from a different batch of P. ephippiata larvae led to the isolation of strain Pei192, which had a 16S rRNA gene sequence identical to that of Pei191T. A third strain, Pei193, again with an identical sequence, was obtained from a liquid dilution series out of a tube that was inoculated with an equivalent of 10−10 ml of unfiltered gut homogenate of P. ephippiata.
Owing to their extremely small size, the isolates were very difficult to observe by light microscopy. DIC light microscopy revealed that the growing cultures consisted of rod-shaped cells of variable lengths (0.3 to 2.5 μm), but images were poorly resolved. This was confirmed by TEM (Fig. 1A to E), which additionally revealed that the cells had an extremely small diameter (0.17 to 0.30 μm). However, larger coccoid cells and cells with undefined shapes were also observed in all growth stages (see below).
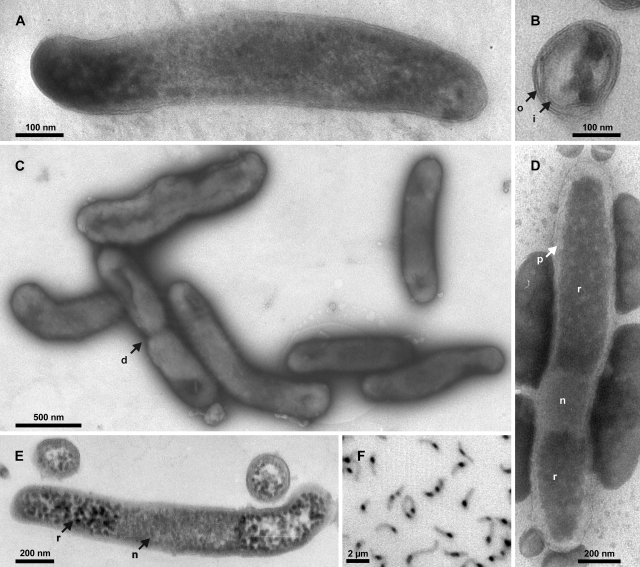
FIG. 1.
Images of strain Pei191T. (A) TEM image of a longitudinal section of a smaller cell, showing both cell poles delimited by the outer membrane. (B) TEM image of a radial section resolving both inner and outer membranes and several ribosomes. (C) Negative-contrast TEM image of cells in the mid-exponential growth phase prepared on a hydrophilized grid. (D) Negative-contrast TEM image of a cell prepared on a nonhydrophilized grid. (E) TEM image of longitudinal and radial sections of larger cells. (F) Fluorescence photomicrograph (negative image) of DAPI-stained cells in the mid-exponential growth phase. Arrows denote cell division (d), inner membrane (i), nucleoid (n), outer membrane (o), periplasmic space (p), and riboplasm (r).
To exclude the possibility that this apparent pleomorphism was caused by the presence of a second bacterium, we confirmed the purity of the cultures by simultaneous fluorescence in situ hybridization with the oligonucleotide probe EUB338, targeting almost all Bacteria, and the probe Elm1034, specifically designed to target the 16S rRNA of strain Pei191T. A formamide series showed that probe ELM1034 discriminated Desulfurella acetivorans, the isolate with the least number of mismatches, in the range of 10 to 40% formamide (see Table S1 in the supplemental material); the optimal condition for the simultaneous hybridization of strain Pei191T with both probes was at a 30% formamide concentration. Subsequent examination of various cultures of strain Pei191T in different growth stages revealed that all morphotypes hybridized with both probes, corroborating that—irrespective of their shape—all cells belonged to the same phylotype.
Strain Pei191T was gram negative in the classical Gram stain. TEM of ultrathin sections of Pei191 cultures revealed that the cell envelope consists of two membranes with a thickness of 6 nm and a periplasmic space with a thickness of 6 to 8 nm (Fig. 1B); a thin structure possibly representing a peptidoglycan layer was only poorly resolved in some negative stains (Fig. 1D). Sometimes, the outer membrane showed protuberances, which were scarce in exponentially growing cultures but numerous in the late stationary or death phase (details not shown). Structures resembling flagella or spores were never observed. Both TEM and negative-contrast electron microscopy showed that the poles of the rod-shaped cells were densely filled with ribosomes, whereas the centers of the cells, probably representing the nucleoids, were invariably free of ribosomes (Fig. 1A, D, and E). The central location of the DNA is confirmed by the bright fluorescence of this region in DAPI-stained wet mounts (Fig. 1F), where DNA gives a much stronger signal than RNA (56). Based on the negative-contrast images obtained with nonhydrophilized grids (Fig. 1D), the average volume of the nucleoid, obtained by geometric approximation, was only 0.010 μm3 (ranging from 0.006 to 0.020 μm3).
Growth and nutrition.
In the API test, strain Pei191T showed moderate acid production from d-glucose, d-fructose, d-galactose, and N-acetyl-d-glucosamine and only weak acid production from d-ribose, methyl-α-d-mannopyranoside, d-turanose, potassium 2-ketogluconate, and potassium 5-ketogluconate. No acid was produced from d-adonitol, amygdalin, d-arabinose, l-arabinose, d-arabitol, l-arabitol, arbutin, d-cellobiose, dulcitol, erythritol, d-fucose, l-fucose, gentiobiose, potassium gluconate, methyl-β-d-glucopyranoside, glycerol, glycogen, inositol, inulin, d-lactose, d-lyxose, d-maltose, d-mannitol, d-mannose, d-melicitose, d-melibiose, d-raffinose, l-rhamnose, salicin, d-sorbitol, l-sorbose, starch, d-sucrose, d-tagatose, d-threhalose, xylitol, methyl-β-d-xylopyranoside, d-xylose, or l-xylose. In liquid culture, strain Pei191T grew on d-glucose, d-fructose, d-galactose, d-glucosamine, and N-acetyl-d-glucosamine, which were fermented to acetate, ethanol, lactate, hydrogen, and alanine (Table 1). No growth was observed on lactate, pyruvate, formate, or amino acids. Casein, xylan, arabinogalactan, microcrystalline cellulose (Avicel), and filter paper were not used. Fumarate was not reduced. Oxygen, nitrate, and sulfate were not used as electron acceptors. Thiosulfate (5 mM) inhibited growth.
TABLE 1.
Growth yields and fermentation products of strain Pei191T on basal medium with different substratesa
| Substrate | Casamino Acids (0.02%)h | Substrate consumed (mmol) | Cell biomass (mg)b | Product formed (mmol)c
|
Electron recovery (%)d | Growth yield (g mol−1) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Acetate | Ethanol | Lactate | Pyruvatee | H2 | Amino acidsf | ||||||
| Glucose | + | 5.0 | 149 | 3.0 | 3.1 | 0.3 | 0.1 | 7.8 | 0.8 | 101 | 29.8 |
| Glucose | − | 4.1g | 99 | 2.2 | 1.9 | 0.3 | 0.1 | 5.7 | 1.2 | 97 | 24.4 |
| Fructose | + | 5.0 | 90 | 3.3 | 5.0 | 0.4 | 0.1 | 6.5 | ND | 100 | 18.0 |
| N-Acetyl-d-glucosamine | + | 5.0 | 82 | 8.6 | 4.1 | 0.3 | 0.1 | 8.2 | ND | 96 | 16.5 |
Basal medium contains 0.02% yeast extract. Values are normalized to 1 liter of medium.
Calculated from the increase in optical density using the OD-to-dry mass ratio of 276 mg l−1 at an OD578 of 1, experimentally determined for glucose-grown cells.
Formate, propionate, and succinate were below the detection limit (0.01 mM). Butyrate, 2-methylprionate, 2- or 3-methylbutyrate, phenylacetate, and 3-hydroyphenylacetate were detected in minor amounts.
Electron recovery was calculated assuming the formula <C4H8O2N> for cell biomass. Electron recovery in amino acids (produced − consumed) was calculated separately for each amino acid, and the result was added to that obtained for the other metabolites. The contribution of yeast extract to electron recovery was estimated with <3% (details not shown).
The identity of pyruvate was confirmed by enzymatic assay.
Net increase of total amino acids. ND, not determined.
Slow growth and incomplete use of glucose in the absence of Casamino Acids.
+, with Casamino Acids; −, without Casamino Acids.
Growth on glucose (5 mM) required both yeast extract and Casamino Acids at a minimum concentration of 0.02% each. No growth occurred in the absence of yeast extract, and at lower concentrations of yeast extract, glucose was not completely consumed. In the absence of Casamino Acids, glucose consumption was incomplete. Casamino Acids could be replaced by the addition of individual amino acids, which were consumed during growth, albeit to different extents (Table 2), but not by an equivalent amount of casein. Additives that stimulated growth but were apparently not consumed were l-arginine, l-β-alanine, l-2-aminobutyrate, dl-3-aminobutyrate, l-5-aminovalerate, l-6-aminocaproate, 11-aminoundecanoic acid, dl-threo-β-methylaspartate, l-citrulline, l-ornithine, putrescine, cadaverine, 2-aminopropanol, 3-aminopropanol, and 4-aminobutanol. Growth was also stimulated by the addition of l-cysteine, l-cystine, l-proline, 3-aminobenzoic acid, 2-aminophenol, 3-aminophenol, urea, and uric acid (all at 2 mM).
TABLE 2.
Consumption of amino acids added individually to cultures of strain Pei191T growing on glucose (5 mM) and yeast extract (0.02%)a
| Amino acid (2 mM)b | Consumed (mM) | Product(s) observedc |
|---|---|---|
| l-Asparagine | 2.0 | ND |
| l-Serine | 2.0 | Lactate, pyruvated |
| l-Glutamine | 1.8 | Unidentified |
| l-Threonine | 1.4 | Glycine |
| l-Methionine | 1.4 | Unidentified |
| l-Leucine | 1.4 | Butyrate, 3-methylbutyrate |
| l-Glycine | 1.3 | Pyruvate |
| l-Lysine | 1.3 | Unidentified |
| l-Isoleucine | 1.2 | 2-Methylbutyrate |
| l-Tryptophan | 1.1 | ND |
| l-Phenylalanine | 1.0 | Phenylacetic acid, unidentified |
| l-Valine | 1.0 | 2-Methylpropionate |
| Diaminopimelate (1 mM) | 1.0 | Lysine |
| l -Ornithine | 0.9 | ND |
| l-Glutamic acid | 0.7 | Unidentified |
| l-Histidine | 0.5 | ND |
| l-4-Aminobutyrate | 0.4 | Lactate, pyruvate |
| l-Aspartic acid | 0.4 | ND |
| l-Tyrosine | 0.3 | 4-Hydroxyphenylacetate |
The yeast extract contained no free amino acids. For amino acids that were not consumed, see text.
Initial concentration.
In addition to the products formed from glucose. ND, not detected.
The identity of pyruvate was confirmed by enzymatic assay.
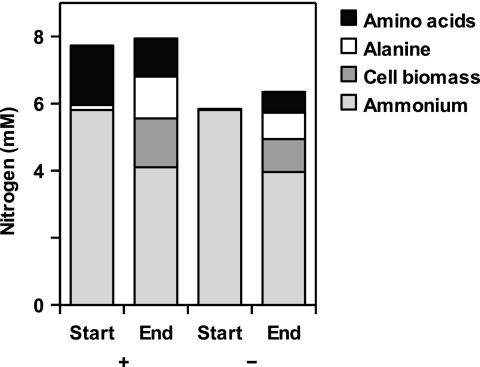
For several amino acids, it was possible to identify the corresponding oxidative decarboxylation products among the fermentation products formed during growth (Table 2). A nitrogen balance of glucose-grown cultures indicated a net formation of amino acids (0.5 to 1.2 mM), mostly alanine, and a net consumption of ammonia (0.5 to 1 mM) (Fig. 2). Alanine was also the only amino acid that did not stimulate growth on glucose.
FIG. 2.
Nitrogen content in different metabolites of strain Pei191T cultures grown with (+) or without (−) Casamino Acids (0.02%), immediately after inoculation (Start) and at the end of the growth phase.
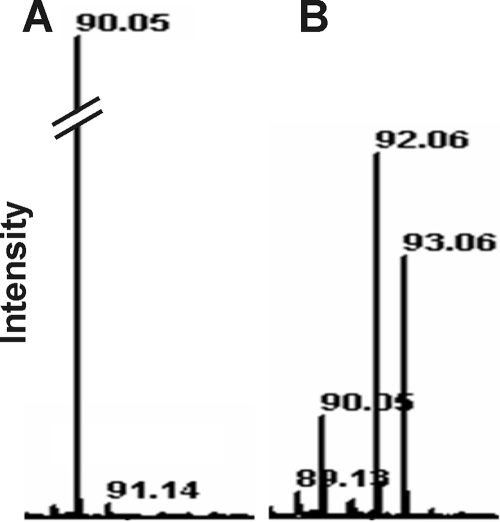
MALDI-TOF analysis of culture supernatants revealed that cultures grown on uniformly 13C-labeled glucose formed alanine labeled in two and three C atoms (Fig. 3). Less than 15% of the total alanine was unlabeled, indicating that most of the alanine was formed from the carbon skeleton of glucose.
FIG. 3.
MALDI-TOF spectra of the supernatants of cultures of strain Pei191T grown with [12C]glucose (A) or [13C]glucose (B). Only the mass ranges relevant for [12C]alanine (90) and [13C]alanine (92 and 93) are shown.
On basal medium with glucose, the cells grew within a pH range of 6.2 to 8.2. Growth was possible at a temperature range of 20 to 32°C but not at 15 or 37°C. Highest growth rates were obtained at 30°C and pH 7.5. Growth was best in freshwater medium, but strain Pei191T also grew at 3.5% salt concentration; the lag phase increased greatly (20 to 40 days) at concentrations above 1.5%.
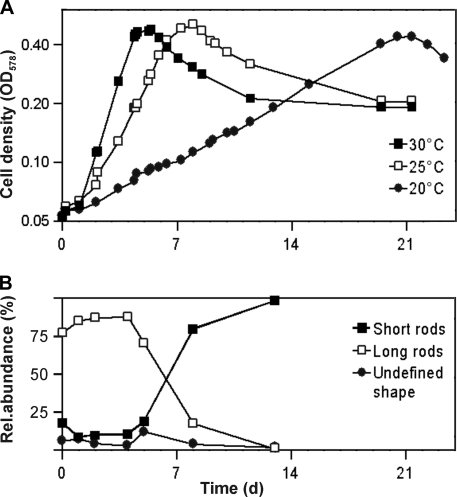
The growth yield on glucose was 29.8 g (dry weight) mol−1; substrate-free controls showed that growth on basal medium was negligible. Cultures grown under this condition showed cell densities (normalized to an OD578 of 1) of 1010 cells ml−1 or 75.1 μg (dry weight) ml−1. The doubling time during growth on glucose was 19 h (Fig. 4A); higher growth rates (doubling time, 11 h) were observed when Casamino Acids and yeast extract were increased to 0.1% (wt/vol) each. When cultures of strain Pei191T were filtered through a 0.2-μm-pore-size membrane filter, subcultures on fresh medium grew very slowly. Even after the second transfer, doubling times were still >80 h but decreased again with subsequent transfers.
FIG. 4.
(A) Growth curves of strain Pei191T at different temperatures (semilogarithmic plot). (B) Relative abundance of the major cell shapes during the different growth phases, illustrating the increase of short forms in the stationary phase (growth temperature, 30°C). d, days.
Strain Pei191T required a reduced medium. Although the bacterium did not grow in the presence of oxygen (0.5%) even in static cultures, the redox zonation (visualized by resorufin) showed that cells pregrown in deep-agar tubes retarded the diffusive influx of oxygen into the tubes considerably longer than uninoculated controls, indicating the principal capacity to reduce molecular oxygen if it does not accumulate to high concentrations.
Chemotaxonomic analysis.
Analysis of the whole-cell fatty acid composition of strain Pei191T revealed an unusual pattern dominated by iso- and anteiso-fatty acids (see Table S2 in the supplemental material). There were no similarities to the pattern of any species included in the Microbial Identification System database (MIDI, Newark, DE). Polar lipid analysis showed phosphatidylethanolamine and phosphatidylglycerol as major components; numerous unidentified phospholipids and amino-phospholipids were present (see Fig. S1 in the supplemental material). Quinones were not detected.
Phylogenetic analysis.
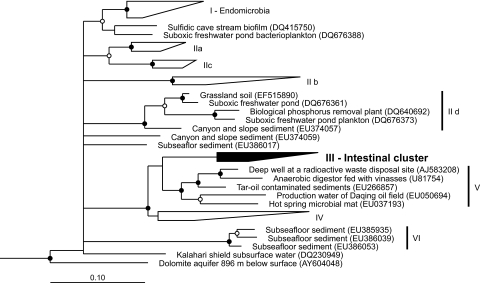
Already a preliminary analysis of the 16S rRNA gene sequence of strain Pei191T had indicated that it belongs to a phylogenetic lineage that so far contains no cultivated representatives, the TG1 phylum (14). Phylogenetic analysis of all near-full-length sequences in the TG1 phylum confirmed that strain Pei191T falls into the so-called intestinal cluster (Fig. 5, cluster III), a stable monophyletic group that comprises clones obtained exclusively from gut-derived samples of termites (14, 35), cows (14, 55), and chimpanzees (31). However, there were also several novel deep-rooting lineages in addition to those already described 2 years ago (14).
FIG. 5.
Maximum-likelihood tree of the TG1 phylum, based on an alignment (1,260 base positions) of all available near-full-length 16S rRNA gene sequences. Representative members of other phyla were used as outgroups. Roman numerals indicate the major phylogenetic lineages in the TG1 phylum. The tree includes 21 novel sequences that were submitted to the GenBank since the first phylogenetic study of the TG1 phylum (14) was published, but details are shown for only those lineages (IId, V, and VI) that were not yet defined in that study.
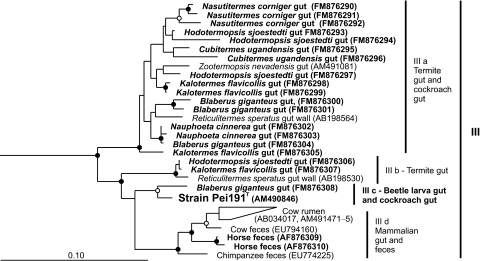
Using a primer set specifically designed to detect members of this cluster (primer set 3) (14), we obtained additional sequences from the guts of numerous wood- and soil-feeding termites and cockroaches and from horse feces. No TG1 sequences were obtained from the guts of several other insects (M. sexta, A. domestica, and S. gregaria) or from rabbit feces. Phylogenetic analysis combining the sequences in cluster III with all partial sequences obtained with primer set 3 showed that the intestinal cluster consists of two major lineages (Fig. 6). One lineage comprises the majority of the clones originating from termites and cockroaches (IIIa). The other lineage comprises several subclusters containing clones from termites (IIIb), the isolates, the sequence-identical clones from P. ephippiata gut homogenate (AM491098), a clone from a cockroach (IIIc), and—well separated from the other subclusters—clones from mammalian samples (IIId).
FIG. 6.
Maximum-likelihood tree of all 16S rRNA gene sequences in the intestinal cluster (III), based on an alignment of 910 base positions. Sequences obtained with primer set 3 (this study) are marked in bold. Sequences in clusters IV and V were included in the analysis and used as outgroups (not shown). In both trees, node support is indicated only if bootstrap values were >70% (open circles) and >95% (filled circles) in both maximum-likelihood and maximum-parsimony analyses.
DISCUSSION
We isolated the first representative of the TG1 phylum, so far predicted only by cultivation-independent methods. Strain Pei191T is a member of a phylogenetic lineage of hitherto uncultivated bacteria occurring in the intestinal tract of insects. It is a strictly anaerobic ultramicrobacterium with a gram-negative cell wall structure and a purely fermentative energy metabolism.
Physiology.
The scope of substrates used by strain Pei191T is quite limited. Only a few sugars and amino sugars are fermented to acetate, ethanol, CO2, and H2—products that are typical of many strict anaerobes (16). Unusual is the production of alanine from glucose. The results of the tracer study indicate that alanine is not formed from other amino acids present in the medium but is, rather, derived from the carbon skeleton of glucose. Alanine is most likely formed by transamination of pyruvate since it carried the 13C label in two or three C atoms, a phenomenon caused by the exchange of the carboxyl group of pyruvate with the unlabeled bicarbonate pool in the medium (67).
Formation of alanine as the end product of anaerobic glucose metabolism has been observed in several eukaryotes, e.g., Saccharomyces cerevisiae (5), the isopod Saduria (Mesidotea) entomon (13), and Giardia lamblia, where alanine production was proposed to be a redox-balancing reaction (41). In prokaryotes, this metabolism has been reported only for a few deep-branching hyperthermophilic archaea (Thermococcales) (26, 27) and bacteria (Thermotogales) (47). It has even been discussed whether this is a remnant of an ancestral metabolism (47). Alanine is the major end product of glucose fermentation in a thermophilic clostridial strain when it is incubated at high ammonium concentrations (40). Remarkably high alanine concentrations were observed already in early studies of the cow rumen (68), an environment characterized by an ample supply of amino acids.
Strain Pei191T did not grow on amino acids, either by fermentation of single amino acids or by Stickland reactions, i.e., the fermentation of amino acid pairs (36). Instead, it fermented amino acids only during growth on glucose, presumably by transamination with pyruvate and subsequent oxidative decarboxylation of the resulting 2-oxoacid, as described for Pyrococcus furiosus (26). This is indicated by the formation of the corresponding oxidative decarboxylation product from most proteinogenic amino acids and the net formation of alanine concomitant to the disappearance of the respective amino acid from the medium. However, transamination of pyruvate may not be the only mechanism of alanine formation. The consumption of ammonium in the growth medium, which significantly exceeds the nitrogen requirements for biomass formation, indicates that part of the alanine is formed by a net amination, as reported for Thermococcus profundus (27). The strong stimulation of growth by the addition of Casamino Acids or individual amino acids and the inhibitory effect of the addition of alanine underscore that alanine formation is an integral component of the energy metabolism of strain Pei191T. However, growth on glucose was also stimulated by other metabolites containing amino groups (e.g., urea or amino alcohols), which indicates that the situation may be even more complex.
Ecology.
Strain Pei191T was isolated from the hindgut of P. ephippiata, a humus-feeding beetle larva. Other members of the intestinal cluster have been retrieved from the intestinal tracts of other insects, including the termite gut, and were also detected in the cow rumen and in the feces of horses and chimpanzees. Common features of these habitats are high concentrations of carbohydrates and amino acids. In the case of P. ephippiata larvae, the hydrolysis products of peptides and other nitrogenous components of soil organic matter (chitin and peptidoglycan) accumulate to millimolar concentrations (2, 32). With its combined metabolism of carbohydrates and amino acids, strain Pei191T seems to be well adapted to the supply of nutrients in its habitat.
Another adaptation to the intestinal habitat may be in the apparent ability of strain Pei191T to remove traces of oxygen. Although it did not grow in liquid medium if the medium was not completely reduced, it was capable of retarding the influx of oxygen into agar tubes containing cells pregrown under anoxic conditions. This feature resembles the situation in Methanobrevibacter cuticularis, which colonizes the gut epithelium of termites. M. cuticularis is extremely sensitive to the presence of oxygen but can reduce it at astonishingly high rates—a trait that may explain why it remains metabolically active despite the continuous influx of oxygen into its natural habitat (59). The exact location of strain Pei191T in the gut of P. ephippiata remains to be established, but other, hitherto uncultivated members of the intestinal cluster have been obtained specifically from the gut wall of the termite gut (35).
In contrast to the distantly related “Candidatus Endomicrobia,” which densely colonizes the cytoplasm of termite gut flagellates (21, 38, 53), members of the other clades in the TG1 phylum are rare in environmental clone libraries (14). This may reflect their low abundance in most other environments and may also be related in part to a low rrn gene copy number among the members of this phylum (15, 19). Also, the isolation of a strain with an identical 16S rRNA gene sequence from a high dilution of P. ephippiata gut homogenate suggests that strain Pei191T is more abundant than indicated by molecular approaches. In view of the small size of strain Pei191T, our failure to detect members of the intestinal cluster by dot blot hybridization (48) of RNA extracted from termite guts (data not shown) may be explained by the small number of ribosomes present in slow-growing ultramicrobacteria (11).
Size.
Strain Pei191T is so small that it passes through a 0.2-μm-pore-size filter membrane typically used for sterilization, a feature that originally led to the concept of ultramicrobacteria (63). Later definitions restricted the term ultramicrobacteria to cells with a volume of <0.1 μm3 (10). Most isolates are slow-growing marine oligotrophs (46, 51), but a few nonoligotrophic ultramicrobacteria have been retrieved from terrestrial habitats (23, 65). Strain Pei191T is the first ultramicrobacterium that has been obtained from a gut environment. The reason for its accidental isolation lies in its extraordinarily small size. Its relatively slow growth on nonselective medium—probably combined with a small population size—are likely explanations of why strain Pei191T and other members of the intestinal cluster have so far eluded isolation (14).
The genome of strain Pei191T, which was recently sequenced, has a size of 1.64 Mbp (15). Since the volume of the nucleoid is extremely small (0.006 to 0.020 μm3, the range probably reflecting the inevitable increase in DNA content during replication), the DNA of strain Pei191T must be tightly packed. Using the volume taken up by a DNA molecule in aqueous solution (34) and assuming the average volume of a single copy of the chromosome to be 0.008 μm3, we estimated that the DNA of strain Pei191T takes up only fourfold the volume of the hydrated DNA molecule even during the exponential growth phase.
Since a small cell can harbor only a limited number of ribosomes, the increase in cell length during the exponential growth phase may reflect the need to accommodate enough ribosomes to achieve a high growth rate. A 10-fold increase in ribosome numbers during the transition from stationary phase to exponential growth phase has been reported for the ultramicrobacterium Sphingomonas sp. strain RB2256 (10), and growth rate and ribosome number are linked also in Escherichia coli (4).
The minimal cell size is limited not only by the cytoplasmic volume required by the ribosomes but also by the DNA. Using the model for the estimation of a theoretical lower size limit of living cells (summarized by De Duve and Osborn [7] in a workshop on size limits of very small microorganisms), corrected for the minimum volume taken up by the hydrated DNA of strain Pei191T (single copy), we estimated the lower size limit for the cytoplasm of Pei191T to be 0.008 μm3. Interestingly, this is in good agreement with the size of the cytoplasm in the smallest cells of Pei191T determined by geometric approximation (Fig. 1A).
Consequently, the reduction of both genome size and ribosome numbers allows bacteria to achieve an extremely small cell size. A small ribosome number results in a low growth rate (4) and would also explain why we found a fourfold longer doubling time in cultures sterile filtered before inoculation: this mechanism would select for small cells, which have fewer ribosomes and hence grow even more slowly than the average culture.
Taxonomy.
Since strain Pei191T is only remotely related to any other bacterial taxon, we propose a new genus and species, Elusimicrobium minutum, with strain Pei191T as the type strain. E. minutum is the first cultivated representative of the candidate phylum TG1. To date, only a putative taxon, “Candidatus Endomicrobium,” has been described; it consists of uncultured endosymbionts in flagellate protozoa in the gut of termites (53).
E. minutum is a member of the intestinal cluster (cluster III), which forms a distinct clade (>15% sequence divergence to other clusters) in the TG1 phylum. Members of this cluster are exclusively found in intestinal habitats, ranging from the guts of termites, cockroaches, and beetle larvae to the mammalian intestinal tract. We propose the family “Elusimicrobiaceae” for all members of cluster III, based on the clear monophyly of this cluster and a within-cluster sequence divergence (0 to 10%) that is typical of the family level in other phyla. In addition, we propose the order “Elusimicrobiales” for the well-supported supercluster formed by clusters III, IV, and V and the phylum “Elusimicrobia” for all members of the TG1 phylum, with E. minutum as the first cultivated representative.
Description of the phylum.
Elusimicrobia phyl. nov. Elusimicrobia (E.lu'si.mi.cro'bi.a. N.L. neut. n. Elusimicrobium, a genus of Bacteria; N.L. neut. pl. n. Elusimicrobia phylum of the genus Elusimicrobium). A lineage of bacteria encompassing “Candidatus Endomicrobium,” the genus Elusimicrobium, and numerous hitherto uncultivated phylotypes that form a stable group according to 16S rRNA gene sequence analysis.
Type order: Elusimicrobiales ord. nov.
Description of Elusimicrobia classis nov.
The description is the same as for the order Elusimicrobiales.
Type order: Elusimicrobiales ord. nov.
Description of Elusimicrobiales ord. nov.
Elusimicrobiales (E.lu'si.mi.cro.bi.al'es. N.L. neut. pl. n. Elusimicrobium type genus of the order; -ales ending to denote an order; N.L. neut. pl. n. Elusimicrobiales the order of the genus Elusimicrobium). It is proposed that the order accommodates bacterial isolates that according to 16S rRNA gene sequence analysis form a stable group within the supercluster formed by clusters III, IV, and V.
Type genus: Elusimicrobium gen. nov.
Description of Elusimicrobiaceae fam. nov.
Elusimicrobiaceae (E.lu'si.mi.cro.bi.a.ce'ae. N.L. neut. n. Elusimicrobium type genus of the family; -aceae ending to denote a family; N.L. neut. n. Elusimicrobiaceae family of the genus Elusimicrobium). The description is the same as for the genus Elusimicrobium. It is proposed that the order accommodates bacterial isolates that according to 16S rRNA gene sequence analysis form a stable group with cluster III.
Type genus: Elusimicrobium gen. nov.
Description of Elusimicrobium gen. nov.
Elusimicrobium (E.lu'si.mi.cro'bi.um. L. part. adj. elusus escaped from capture; N.L. neut. n. microbium, microbe; N.L. neut. n. elusimicrobium an elusive microbe, hard to find, capture, or isolate). Gram-negative staining, nonmotile, rod-shaped cells. Obligately anaerobic and catalase negative. Heterotrophic, purely fermentative metabolism; nitrate, sulfate, and thiosulfate are not reduced.
Type species: Elusimicrobium minutum gen. nov., sp. nov.
Description of Elusimicrobium minutum sp. nov.
Elusimicrobium minutum (mi.nu'tum L. neut. part. adj. minutum, very small, minute). Cells are typically rod shaped, but cultures are pleomorphic in all growth phases (0.3 to 2.5 μm long and 0.17 to 0.3 μm wide); the smallest cells pass through 0.2-μm-pore-size membrane filters. Acid production from d-galactose, d-glucose, d-fructose, d-glucosamine, and N-acetyl-d-glucosamine. Weak acid production from d-ribose, methyl-α-d-mannopyranoside, d-turanose, potassium 2-ketogluconate, and potassium 5-ketogluconate. Growth on glucose requires yeast extract and Casamino Acids (0.02% each); the latter can be replaced by individual amino acids (except alanine). Major end products are acetate, ethanol, lactate, hydrogen, and alanine. Temperature range of growth of 20 to 32°C, optimum at 30°C; no growth at 15 and 37°C. pH range of growth of 6.5 to 8.5; optimum at pH 7.5. The major fatty acids are C15:0 iso, C15:0 anteiso, and C16:0 iso.
Habitat: the intestinal tract of the humus-feeding larva of Pachnoda ephippiata.
Type strain: Pei191T (= ATCC BAA-1559T = JCM 14958T). GenBank accession number: AM490846.
Supplementary Material
Acknowledgments
We thank Jörg Kahnt for MALDI-TOF analysis, Shuning Wang for help with pyruvate measurements, Marianne Johannsen for technical assistance with electron microscopy, and Hans G. Trüper for etymological advice. We are also grateful to Uwe Deggelmann, keeper of the zoological teaching collection of the Biology Department at Konstanz University, for providing P. ephippiata larvae.
Footnotes
Published ahead of print on 6 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andert, J., O. Geissinger, and A. Brune. 2008. Peptidic soil components are a major dietary resource for the humivorous larvae of Pachnoda spp. (Coleoptera: Scarabaeidae). J. Insect Physiol. 54:105-113. [DOI] [PubMed] [Google Scholar]
- 3.Boga, H., and A. Brune. 2003. Hydrogen-dependent oxygen reduction by homoacetogenic bacteria isolated from termite gut. Appl. Environ. Microbiol. 69:779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bremer, H., and P. P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p. 1553-1569. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 5.Chico, E., J. S. Olavarría, and I. Núnez de Castro. 1978. l-Alanine as an end product of glycolysis in Saccharomyces cerevisiae growing under different hypoxic conditions. Antonie van Leeuwenhoek 44:193-201. [DOI] [PubMed] [Google Scholar]
- 6.Czok, R., and W. Lamprecht. 1970. Pyruvat, Phosphoenolpyruvat und d-Glycerat-2-phosphat, p. 1407-1411. In H. U. Bergmeyer (ed.), Methoden der Enzymatischen Analyse, 2nd ed., vol. 2. Verlag Chemie, Weinheim, Germany. [Google Scholar]
- 7.De Duve, C., and M. J. Osborn. 1999. Discussion summary of panel 1, p. 5-9. In A. Knoll, M. J. Osborn, J. Baross, H. C. Berg, N. R. Pace, and M. Sogin (ed.), Size limits of very small microorganisms. Proceedings of a workshop. National Academy Press, Washington, DC. [PubMed]
- 8.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egert, M., B. Wagner, T. Lemke, A. Brune, and M. W. Friedrich. 2003. Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69:6659-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eguchi, M., M. Ostrowski, F. Fegatella, J. Bowman, D. Nichols, T. Nishino, and R. Cavicchioli. 2001. Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl. Environ. Microbiol. 67:4945-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fegatella, F., J. Lim, S. Kjelleberg, and R. Cavicchioli. 1998. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godel, H., T. Graser, P. Földi, P. Pfaender, and P. Fürst. 1984. Measurement of free amino acids in human biological fluids by high-performance liquid chromatography. J. Chromatogr. 297:49-61. [DOI] [PubMed] [Google Scholar]
- 13.Hagerman, L., and A. Szaniawska. 1990. Anaerobic metabolic strategy of the glacial relict isopod Saduria (Mesidotea) entomon. Mar. Ecol. Prog. Ser. 59:91-96. [Google Scholar]
- 14.Herlemann, D. P. R., O. Geissinger, and A. Brune. 2007. The termite group I phylum is highly diverse and widespread in the environment. Appl. Environ. Microbiol. 73:6682-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herlemann, D. P. R., O. Geissinger, W. Ikeda-Ohtsubo, V. Kunin, H. Sun, A. Lapidus, P. Hugenholtz, and A. Brune. 2009. Genomic analysis of “Elusimicrobium minutum,” the first cultivated representative of the phylum “Elusimicrobia” (formerly termite group 1). Appl. Environ. Microbiol. 75:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holdeman, L. V., E. P. Cato, and W. E. C. Moore. 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg.
- 17.Hongoh, Y., M. Ohkuma, and T. Kudo. 2003. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera: Rhinotermitidae). Microb. Ecol. 44:231-242. [DOI] [PubMed] [Google Scholar]
- 18.Hongoh, Y., P. Deevong, S. Hattori, T. Inoue, S. Noda, N. Noparatnaraporn, T. Kudo, and M. Ohkuma. 2006. Phylogenetic diversity, localization, and cell morphologies of members of the candidate phylum TG3 and a subphylum in the phylum Fibrobacteres, recently discovered bacterial groups dominant in termite guts. Appl. Environ. Microbiol. 72:6780-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hongoh, Y., V. K. Sharma, T. Prakash, S. Noda, T. D. Taylor, T. Kudo, Y. Sakaki, A. Toyoda, M. Hattori, and M. Ohkuma. 2008. Complete genome of the uncultured Termite Group 1 bacteria in a single host protist cell. Proc. Natl. Acad. Sci. USA 105:5555-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda-Ohtsubo, W., M. Desai, U. Stingl, and A. Brune. 2007. Phylogenetic diversity of “Endomicrobia” and their specific affiliation with termite gut flagellates. Microbiology 153:3458-3465. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda-Ohtsubo, W., and A. Brune. 2009. Cospeciation of termite gut flagellates and their bacterial endosymbionts: Trichonympha species and “Candidatus Endomicrobium trichonymphae.” Mol. Ecol. 18:332-342. [DOI] [PubMed] [Google Scholar]
- 23.Janssen, P. H., A. Schuhmann, E. Mörschel, and F. A. Rainey. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kämpfer, P., and R. M. Kroppenstedt. 1996. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can. J. Microbiol. 42:989-1005. [Google Scholar]
- 25.Karas, M., D. Bachmann, and F. Hillenkamp. 1985. Influence of the wavelength in high-irradiance ultraviolet laser desorption mass spectrometry of organic molecules. Anal. Chem. 57:2935-2939. [Google Scholar]
- 26.Kengen, S. W. M., and A. J. Stams. 1994. Formation of l-alanine as a reduced end product in carbohydrate fermentation by the hyperthermophilic archaeon Pyrococcus furiosus. Arch. Microbiol. 161:168-175. [Google Scholar]
- 27.Kobayashi, T., S. Higuchi, K. Kimura, T. Kudo, and K. Horikoshi. 1995. Properties of glutamate dehydrogenase and its involvement in alanine production in a hyperthermophilic archaeon, Thermococcus profundus. J. Biochem. 118:587-592. [DOI] [PubMed] [Google Scholar]
- 28.Köhler, T., U. Stingl, K. Meuser, and A. Brune. 2008. Novel lineages of Planctomycetes densely colonize the alkaline gut of soil-feeding termites (Cubitermes spp.). Environ. Microbiol. 10:1260-1270. [DOI] [PubMed] [Google Scholar]
- 29.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemke, T., U. Stingl, M. Egert, M. W. Friedrich, and A. Brune. 2003. Physicochemical conditions and microbial activities in the highly alkaline gut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69:6650-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ley, R. E., M. Hamady, C. Lozupone, P. J. Turnbaugh, R. R. Ramey, J. S. Bircher, M. L. Schlegel, T. A. Tucker, M. D. Schrenzel, R. Knight, and J. I. Gordon. 2008. Evolution of mammals and their gut microbes. Science 320:1647-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, X., and A. Brune. 2007. Transformation and mineralization of soil organic nitrogen by the humivorous larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Plant Soil 301:233-244. [Google Scholar]
- 33.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandelkern, M., J. G. Elias, D. Eden, and D. M. Crothers. 1981. The dimensions of DNA in solution. J. Mol. Biol. 152:153-161. [DOI] [PubMed] [Google Scholar]
- 35.Nakajima, H., Y. Hongoh, R. Usami, T. Kudo, and M. Ohkuma. 2005. Spatial distribution of bacterial phylotypes in the gut of the termite Reticulitermes speratus and the bacterial community colonizing the gut epithelium. FEMS Microbiol. Ecol. 54:247-255. [DOI] [PubMed] [Google Scholar]
- 36.Nisman, B. 1954. The Stickland reaction. Bacteriol. Rev. 18:16-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohkuma, M., and T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohkuma, M., T. Sato, S. Noda, S. Ui, T. Kudo, and Y. Hongoh. 2007. The candidate phylum “Termite Group 1” of bacteria: phylogenetic diversity, distribution, and endosymbiont members of various gut flagellated protists. FEMS Microbiol. Ecol. 60:467-476. [DOI] [PubMed] [Google Scholar]
- 39.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 40.Örlygsson, J., R. Anderson, and B. H. Svensson. 1995. Alanine as an end product during fermentation of monosaccharides by Clostridium strain P2. Antonie van Leeuwenhoek 273:273-280. [DOI] [PubMed] [Google Scholar]
- 41.Paget, T. A., M. H. Raynor, D. W. Shipp, and D. Lloyd. 1990. Giardia lamblia produces alanine anaerobically but not in the presence of oxygen. Mol. Biochem. Parasitol. 42:63-67. [DOI] [PubMed] [Google Scholar]
- 42.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes, p. 207-226. In J. Pernthaler, F. O. Glöckner, W. Schönhuber, and R. Amann (ed.), Methods in microbiology: marine microbiology. Academic Press, London, United Kingdom.
- 43.Pfennig, N., and H. G. Trüper. 1981. Isolation of members of the families Chromatiaceae and Chlorobiaceae, p. 279-289. In M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes, vol. I. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 44.Pruesse, E., C. Quast, K. Knittel, B. Fuchs, W. Ludwig, J. Peplies, and F. O. Glöckner. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 46.Rappé, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 47.Ravot, G., B. Ollivier, M. L. Fardeau, B. K. Patel, K. T. Andrews, M. Magot, and J. L. Garcia. 1996. l-Alanine production from glucose fermentation by hyperthermophilic members of the domains Bacteria and Archaea: a remnant of an ancestral metabolism? Appl. Environ. Microbiol. 62:2657-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahm, K., B. J. MacGregor, B. B. Jørgensen, and D. A. Stahl. 1999. Sulphate reduction and vertical distribution of sulphate-reducing bacteria quantified by rRNA slot-blot hybridization in a coastal marine sediment. Environ. Microbiol. 1:65-74. [DOI] [PubMed] [Google Scholar]
- 49.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69:6007-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Axial dynamics, stability, and interspecies similarity of bacterial community structure in the highly compartmentalized gut of soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 67:6018-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schut, F., E. J. de Vries, J. C. Gottschal, B. R. Robertson, W. Harder, R. A. Prins, and D. K. Button. 1993. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stamatakis, A., T. Ludwig, and H. Maier. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456-463. [DOI] [PubMed] [Google Scholar]
- 53.Stingl, U., R. Radek, H. Yang, and A. Brune. 2005. “Endomicrobia”: cytoplasmic symbionts of termite gut protozoa form a separate phylum of prokaryotes. Appl. Environ. Microbiol. 71:1473-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Süßmuth, R., J. Eberspächer, R. Haag, and W. Springer. 1987. Biochemisch-mikrobiologisches Praktikum. Thieme Verlag, Stuttgart, Germany.
- 55.Tajima, K., S. Arai, K. Ogata, T. Nagamine, H. Matsui, M. Nakamura, R. I. Aminov, and Y. Benno. 2000. Rumen bacterial community transition during adaptation to high-grain diet. Anaerobe 6:273-284. [Google Scholar]
- 56.Tanious, F. A., J. M. Veal, H. Buczak, L. S. Ratmeyer, and W. D. Wilson. 1992. DAPI (4′,6-diamidino-2-phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites. Biochemistry 31:3103-3112. [DOI] [PubMed] [Google Scholar]
- 57.Tholen, A., and A. Brune. 1999. Localization and in situ activities of homoacetogenic bacteria in the highly compartmentalized hindgut of soil-feeding higher termites (Cubitermes spp.). Appl. Environ. Microbiol. 65:4497-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tholen, A., B. Schink, and A. Brune. 1997. The gut microflora of Reticulitermes flavipes, its relation to oxygen, and evidence for oxygen-dependent acetogenesis by the most abundant Enterococcus sp. FEMS Microbiol. Ecol. 24:137-149. [Google Scholar]
- 59.Tholen, A., M. Pester, and A. Brune. 2007. Simultaneous methanogenesis and oxygen reduction by Methanobrevibacter cuticularis at low oxygen fluxes. FEMS Microbiol. Ecol. 62:303-312. [DOI] [PubMed] [Google Scholar]
- 60.Tindall, B. J. 1990. A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst. Appl. Microbiol. 13:128-130. [Google Scholar]
- 61.Tindall, B. J. 1990. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett. 66:199-202. [Google Scholar]
- 62.Tindall, B. J., J. Sikorski, R. M. Smibert, and N. R. Krieg. 2007. Phenotypic characterization and the principles of comparative systematics, p. 330-393. In C. A. Reddy, T. J. Beveridge, J. A. Breznak, G. Marzluf, T. M. Schmidt, and L. R. Snyder (ed.), Methods for general and molecular microbiology, 3rd ed. ASM Press, Washington, DC.
- 63.Torrella, F., and R. Y. Morita. 1981. Microcultural study of bacterial size changes and microcolony and ultramicrocolony formation by heterotrophic bacteria in seawater. Appl. Environ. Microbiol. 41:518-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valentine, R. C., B. M. Shapiro, and E. R. Stadtman. 1968. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry 7:2143-2152. [DOI] [PubMed] [Google Scholar]
- 65.Verbarg, S., H. Rheims, S. Emus, A. Frühling, R. M. Kroppenstedt, E. Stackebrandt, and P. Schumann. 2004. Erysipelothrix inopinata sp. nov., isolated in the course of sterile filtration of vegetable peptone broth, and description of Erysipelotrichaceae fam. nov. Int. J. Syst. Evol. Microbiol. 54:221-225. [DOI] [PubMed] [Google Scholar]
- 66.Wehrmeyer, W., and H. Schneider. 1975. Elektronenmikroskopische Untersuchungen zur reversiblen Veränderung der Chloroplastenfeinstruktur von Rhodella violacea bei Stickstoffmangel. Biochem. Physiol. Pflanz. 7:519-532. [Google Scholar]
- 67.Wilson, J., L. O. Krampitz, and C. H. Werkman. 1948. Reversibility of a phosphoroclastic reaction. Biochem. J. 42:598-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wright, D. E., and R. E. Hungate. 1967. Amino acid concentrations in rumen fluid. Appl. Microbiol. 15:148-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang, H., D. Schmitt-Wagner, U. Stingl, and A. Brune. 2005. Niche heterogeneity determines bacterial community structure in the termite gut (Reticulitermes santonensis). Environ. Microbiol. 7:916-932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.