Abstract
Comparative genomic hybridization analysis of 32 Nordic group I Clostridium botulinum type B strains isolated from various sources revealed two homogeneous clusters, clusters BI and BII. The type B strains differed from reference strain ATCC 3502 by 413 coding sequence (CDS) probes, sharing 88% of all the ATCC 3502 genes represented on the microarray. The two Nordic type B clusters differed from each other by their response to 145 CDS probes related mainly to transport and binding, adaptive mechanisms, fatty acid biosynthesis, the cell membranes, bacteriophages, and transposon-related elements. The most prominent differences between the two clusters were related to resistance to toxic compounds frequently found in the environment, such as arsenic and cadmium, reflecting different adaptive responses in the evolution of the two clusters. Other relatively variable CDS groups were related to surface structures and the gram-positive cell wall, suggesting that the two clusters possess different antigenic properties. All the type B strains carried CDSs putatively related to capsule formation, which may play a role in adaptation to different environmental and clinical niches. Sequencing showed that representative strains of the two type B clusters both carried subtype B2 neurotoxin genes. As many of the type B strains studied have been isolated from foods or associated with botulism, it is expected that the two group I C. botulinum type B clusters present a public health hazard in Nordic countries. Knowing the genetic and physiological markers of these clusters will assist in targeting control measures against these pathogens.
Clostridium botulinum produces a potent neurotoxin during its growth. The toxin causes a potentially lethal paralytic disease, botulism, in humans and animals. The classical food-borne botulism follows the consumption of toxin-containing food or drink, while infant and adult intestinal botulism results from in vivo spore germination, outgrowth, and toxin production in the gut. Apart from attenuated intestinal microbial population, other factors affecting the colonization of C. botulinum in the intestinal forms of botulism are not known.
Based on their physiology and genetic background, C. botulinum strains are divided into groups I to IV (13). Strains of groups I and II are associated with human disease. Group I strains produce neurotoxin serotypes A, B, and/or F, while the group II strains produce type B, E, or F toxin. Physiologically, groups I and II differ markedly from each other as well as from groups III and IV. Genomic analysis of group I and II C. botulinum strains by 16S rrn sequencing (13), ribotyping (10), and amplified fragment length polymorphism (11, 15, 16) is consistent with the divergent physiologies of the two groups (18).
Nordic C. botulinum group I strains show a remarkable homogeneity (15, 20, 21, 23). In a large pulsed-field gel electrophoresis (PFGE) analysis, the majority of group I strains isolated from various sources from Finland, Norway, and Denmark formed type B neurotoxin and clustered into two large groups, with the members of each group sharing identical or nearly identical restriction patterns (20, 23). Many of these strains were recovered from honey for human consumption (23), and one strain was related to an infant botulism case (22). Apart from a recent study showing that strains of the two type B clusters, further referred to as clusters BI and BII, differ in their abilities to grow at extreme temperatures (12), the physiological, epidemiological, and genetic markers of the two clusters are not known. An understanding of such traits will assist in designing control measures against these potential food- and environment-borne pathogens.
The availability of group I C. botulinum genome sequences has enabled the construction of whole-genome DNA microarrays and a comprehensive genomic analysis of C. botulinum strains (26, 27). In this paper, we describe a comparative genomic hybridization (CGH) analysis of 32 Nordic group I C. botulinum type B cluster BI or BII strains with a DNA microarray based on the protein-coding sequences (CDS) in the ATCC 3502 genome. Strains within each cluster showed no substantial variation. Furthermore, strains belonging to the two clusters differed by their responses to 145 CDS probes, suggesting differential resistance to toxic compounds and a relatively large antigenic variability. Sequencing of botB in a representative cluster BI strain and a representative cluster BII strain revealed subtype B2 neurotoxin genes in both strains.
MATERIALS AND METHODS
Clostridium botulinum strains.
A total of 32 group I C. botulinum type B strains isolated from various geographical sources and ecological niches in the Nordic countries in 2000 to 2003 were analyzed (Table 1 and Fig. 1). All strains were previously shown to carry a type B neurotoxin gene, botB (21, 24). PFGE analysis showed that these strains fell into two large genetically homogeneous clusters (20, 23). The genome-sequenced group I C. botulinum type A1 strain ATCC 3502 (Hall A) (28) was used as a hybridization reference.
TABLE 1.
Strains of group I Clostridium botulinum included in the genomic analysis
| Strain | Origin | Place and yr of isolationa | Cluster |
|---|---|---|---|
| ATCC 3502 | United States | NK, 1920s | |
| B-129/1 | Bees, Finland | DFEH, 2002 | BI |
| KV-16/4 | Beeswax, Finland | DFEH, 2001 | BI |
| KV-8/5 | Beeswax, Finland | DFEH, 2001 | BI |
| KV-41/3 | Beeswax, Finland | DFEH, 2001 | BI |
| 90-K/5 | Beeswax, Finland | DFEH, 2000 | BI |
| 515-K/2 | Beeswax, Finland | DFEH, 2000 | BI |
| KS-3/5 | Pollen from bee bread, Finland | DFEH, 2001 | BI |
| KS-57/7 | Pollen from bee bread, Finland | DFEH, 2001 | BI |
| KS-102/10 | Pollen from bee bread, Finland | DFEH, 2002 | BI |
| 525-S/8 | Pollen from bee bread, Finland | DFEH, 2000 | BI |
| KH-64/2 | Comb honey, Finland | DFEH, 2001 | BI |
| KH-51/1 | Comb honey, Finland | DFEH, 2001 | BI |
| He-3323 | Horse feces, Finland | DFEH, 2002 | BI |
| He-3396 | Horse feces, Finland | DFEH, 2002 | BI |
| M-153/1 | Soil, Finland | DFEH, 2002 | BI |
| M-197/17 | Soil, Finland | DFEH, 2002 | BI |
| M-1/3 | Soil, Finland | DFEH, 2001 | BI |
| M-18/3 | Soil, Finland | DFEH, 2001 | BI |
| M-170/4 | Soil, Finland | DFEH, 2002 | BI |
| 3213/5 | Soil, Finland | DFEH, 2000 | BI |
| P-120/6 | Vacuum cleaner dust, Finland | DFEH, 2002 | BI |
| NKH-50/1 | Comb honey, Norway | DFEH, 2003 | BI |
| DA-32/1 | Honey, Denmark | DFEH, 2003 | BI |
| DA-58/1 | Honey, Denmark | DFEH, 2003 | BI |
| KV-39/1 | Beeswax, Finland | DFEH, 2001 | BII |
| 518-K/3 | Beeswax, Finland | DFEH, 2000 | BII |
| KH-318/1 | Comb honey, Finland | DFEH, 2003 | BII |
| KS-44/10 | Pollen from bee bread, Finland | DFEH, 2001 | BII |
| M-43/15 | Soil, Finland | DFEH, 2001 | BII |
| M-193/15 | Soil, Finland | DFEH, 2002 | BII |
| M-171/14 | Soil, Finland | DFEH, 2002 | BII |
| M-195/20 | Soil, Finland | DFEH, 2002 | BII |
NK, not known (isolated by I. C. Hall in the United States in the 1920s); DFEH, Department of Food and Environmental Hygiene, University of Helsinki, Helsinki, Finland.
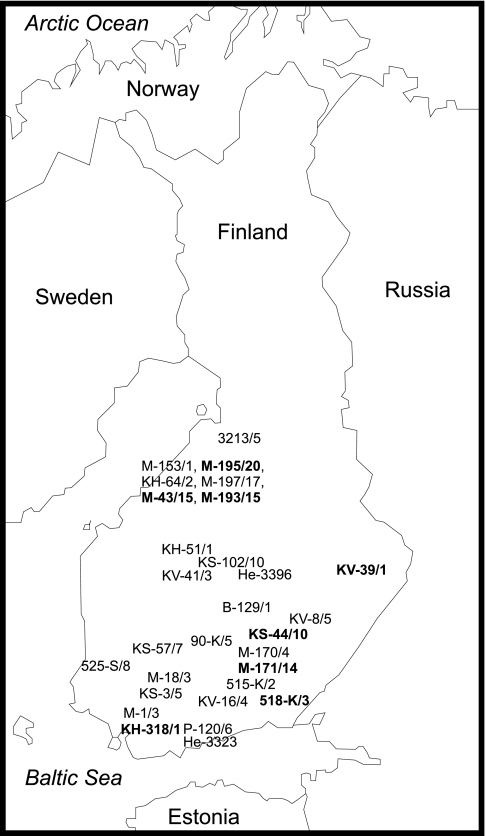
FIG. 1.
Sites of isolation of Finnish group I C. botulinum type B strains. Cluster BI strains have been marked with normal type, and cluster BII strains have been marked with boldface type.
DNA extraction.
Each strain was streaked from a spore suspension onto an anaerobic blood agar plate and incubated anaerobically at 37°C for 48 h. A single colony was further inoculated into 10 ml of anaerobic tryptose-peptone-glucose-yeast extract (TPGY) medium and incubated overnight at 37°C. Genomic DNA was extracted from the remaining culture as previously described (15), dissolved in 100 μl of sterile distilled water, and stored at −70°C until use. For biological replicates, the DNA of each strain was extracted twice.
DNA microarrays.
The DNA microarrays based on the C. botulinum ATCC 3502 genome (27) were constructed at the Institute of Food Research. The ATCC 3502 genome contains 3,648 chromosomal CDSs and 18 plasmid CDSs. These CDSs cover 82% of the entire genome of the ATCC 3502 strain, leaving 18% of the genome for intergenic space. Each microarray slide contained two arrays, one array consisting of 3,421 PCR amplicon probes of 100 to 500 bp, representing 3,403 (93.3%) chromosomal CDSs and 18 (100%) plasmid CDSs in the ATCC 3502 genome. Specific probes could not be designed to or amplification products were not obtained for the remaining 245 (6.7%) chromosomal CDSs in the ATCC 3502 genome; hence, they were excluded from the array. In addition, each array contained 31 amplicons representing the C and N terminals of type A1, A3, B, C, D, E, F, and G botulinum neurotoxin genes and genes encoding the nontoxic neurotoxin-associated proteins.
Probe preparation.
A total of 6 μg of genomic DNA of each C. botulinum strain was randomly labeled with fluorescent dye (BioPrime DNA labeling system; Invitrogen Corporation, Carlsbad, CA). The 50-μl labeling reaction mixtures contained 0.3 μg/μl random primers (Invitrogen); 0.2 mM dATP, dGTP, and dTTP (Promega Corporation, Madison, WI); 0.1 mM dCTP (Promega); 40 μM Cy3-dCTP (GE Healthcare, Buckinghamshire, United Kingdom) (type A control strain) or Cy5-dCTP fluorescent dye (GE Healthcare) (type B test strains); and 0.8 U/μl Klenow fragment (Invitrogen). The reaction mixtures were incubated at 37°C for 1 to 2 h, and the reaction was stopped by adding 5 μl of 0.5 M EDTA (pH 8.0; Invitrogen). For each hybridization, the Cy5-labeled DNA from each type B strain was combined with similarly prepared, Cy3-labeled reference DNA from ATCC 3502, and the mixture was purified with a DNA purification kit (QIAquick PCR purification kit; Qiagen, Hilden, Germany) and finally eluted with 42 μl of elution buffer (Qiagen). A volume of 24 μl of hybridization buffer (10× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] [Invitrogen], 0.08 M HEPES buffer [Sigma-Aldrich Chemie, Steinheim, Germany], 16.5× Denhardt's solution [Invitrogen], and 0.75% sodium dodecyl sulfate [SDS] [Invitrogen]), 70 μg/ml human Cot-1 DNA (Invitrogen), and 1.4 μg/μl yeast tRNA (Invitrogen) were added to a final volume of 70 μl. The probe mixtures were denatured in a vigorously boiling water bath for 5 min and cooled down.
Hybridization and washes.
Before hybridization, UV-cross-linked array slides were incubated in prewarmed prehybridization buffer (1% bovine serum albumin [Sigma-Aldrich], 6× SSC, and 0.5% SDS) at 65°C for 1 to 2 h. After prehybridization, the slides were washed three times for 5 min with sterile distilled water and dried by centrifugation for 15 s in a minicentrifuge. A coverslip (LifterSlip 25× 60I-2-4789; Erie Scientific Company, Portsmouth, NH) was placed on top of each microarray slide covering both arrays, and 70 μl of each probe mixture was pipetted under the coverslip, avoiding air bubble formation. The slides were hybridized overnight at 54°C. The hybridized slides were washed twice for 15 min with 2× SSC-0.2% SDS prewarmed to 50°C, twice for 10 min with 0.5× SSC at room temperature, twice for 8 min with 0.1× SSC at room temperature, and once for 3 min with 0.01× SSC and 0.05% Tween 20 at room temperature, followed by drying (as described above).
The two DNA extracts of each strain were hybridized onto separate slides, with each slide containing two arrays, yielding four replicate hybridizations per strain.
Scanning, image processing, and data normalization.
The slides were scanned (Axon GenePix Autoloader 4200 AL; Westburg, Leusden, The Netherlands) at 532 and 635 nm using a 10-μm resolution. Image processing was done with GenePix Pro 6.0 software. Foreground and local background intensities of each spot were characterized by the mean and median pixel values of the spot, respectively. Spots with segmented foreground areas of less than 50 pixels and spots whose foreground intensities in the control channel (Cy3) were less than 1,000 raw intensity units above the background were excluded from the analysis (11.4% of all spots). Local background was subtracted from the foreground signal, and the intensity values of two replicate spots for each CDS on a slide were averaged. Ratios between Cy5 and Cy3 intensities were converted into a logarithmic scale, and the slide-dependent median was subtracted. Based on self-self-hybridizations with ATCC 3502 DNA, the amount of noise was estimated to be small; the standard error of signal log ratios was less than 0.13, indicating good hybridization quality.
Thresholding signal intensities and hierarchical clustering.
In order to set a threshold for the presence and absence of CDSs, GACK software was used (17). The method is based on estimating the spread of signal log ratios of present CDSs by fitting a normal curve to the histogram of all signal log ratio values of a slide. The threshold is slide dependent. To select the CDSs most reliably differentiating between strains, the GACK trinary categorization was used. This method estimates if a CDS is present, absent, or highly divergent or if it belongs to the middle category representing no reliable labeling (slightly divergent CDSs). The thresholds for the estimated probability of presence (EPP) were set to 100% for present CDSs and 0% for absent or highly divergent CDSs. These settings give the most stringent conditions for a CDS to be declared in the two categories. Hierarchical clustering of strains was determined using the statistical computing package R (14). A dendrogram was created based on averaged GACK values of replicate slides (Fig. 2). Euclidean distances were computed between strains, and hierarchical clustering was created using an average linkage method.
FIG. 2.
Dendrogram of 32 Nordic group I C. botulinum type B strains and ATCC 3502 based on CGH analysis with ATCC 3502 microarrays. The two clusters have been marked as clusters BI and BII. The scale describes the Euclidean distance between two strains, i.e., the square root of a sum of squared gene-wise differences. Since the trinary GACK method gives values of −1, 0, and 1 for gene absent/highly divergent, unreliable labeling, and gene present, respectively, the squared difference of one gene present in one strain and absent in another strain equals [1 − (−1)]2̂ = 4. Thus, a Euclidean distance of 20 corresponds to 100 genes missing in one strain and present in another strain, and a Euclidean distance of 40 corresponds to 400 genes missing in one strain and present in another strain.
CDSs discriminating between clusters BI and BII.
Based on GACK trinary categorization, the numbers of “CDS present” and “CDS absent/highly divergent” calls were calculated within each cluster for each CDS. When finding CDSs that were present in cluster BI and absent in cluster BII, no “CDS absent/highly divergent” call was allowed for any cluster BI strain, and no “CDS present” call was allowed for any cluster BII strain. The opposite requirement was applied when finding the CDSs present in cluster BII and absent in cluster BI. The statistical significance of the difference between the clusters for having or lacking the CDS was obtained by Fisher's exact test. A P value was calculated for each CDS, and the list of P values was then converted into false-discovery-rate (FDR) values. There were 149 CDSs with FDR values of <0.01 and 145 CDSs with FDR values of <0.001. Since the list of 145 CDSs is based on very stringent criteria, it contains only highly reliable CDSs for differentiating between the two clusters.
Validation of DNA microarray results with PCR.
To verify the DNA microarray results, two CDSs (CBO2590 and CBO3250) with a wide range of signal log ratios for all 32 type B strains were selected for PCR. CBO2590 probe spots gave low intensity values and CBO3250 spots gave high intensity values for the control channel in all slides. PCR was carried out in a 25-μl reaction mixture volume containing 1× buffer, 200 μM of each deoxynucleotide triphosphate, 0.5 μM of each primer (CBO2590-f [5′-TGCTGAGCATAACCAAACTG-3′] and CBO2590-r [5′-CTGAAGTCCCAGGACATGTAGA-3′], or CBO3250-f [5′-CCATTGTTCCACCTCCTCAT-3′] and CBO3250-r [5′-TGTGTCCAGAAGCAGCTAAAA-3′]), and 1 IU of DNA polymerase (DyNAZyme; Finnzymes, Espoo, Finland). A total of 10 to 20 ng of DNA from each C. botulinum type B strain was used as a template. The reaction conditions included an initial denaturation step at 95°C for 5 min and 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min, followed by a final extension step at 72°C for 3 min. The amplification products were visualized under UV light in 2% agarose gels following staining with ethidium bromide.
Validation of DNA microarray results with DNA sequencing.
In addition to PCR, amplification fragments representing 12 CDSs predicted to discriminate between strains within cluster BI or BII based on signal log ratios or GACK results were sequenced using primers described in Table S1 in the supplemental material. Sequencing was carried out using an Applied Biosystems ABI Prism 3130xl genetic analyzer (1). Sequence data were analyzed with Staden package release 1.6.0 (32), where base calling was done using Phred (7). Sequences were aligned with ClustalW, and they were cut to have equal lengths. Pairwise similarities were computed between all sequences.
Sequencing of botB.
The botB genes of cluster I strain M-18/3 and cluster II strain KV-39/1 were sequenced (5) using primers described in Table S2 in the supplemental material.
Growth in the presence of sodium arsenite.
The arsenic resistance of three BI strains (M-1/3, P-120/6, and DA-58/1) and three BII strains (KH-318/1, KS-44/10, and M-43/15) was tested in TPGY medium in the absence and presence of sodium arsenite. In brief, 100 μl of a spore suspension of each strain was inoculated into 10 ml of anaerobic TPGY broth and incubated overnight at 37°C under anaerobic conditions, followed by subculture into 10 ml of fresh TPGY broth and overnight incubation under the same conditions. A volume of 40 μl of each culture was further transferred into 4 ml of fresh anaerobic TPGY broth containing 0, 0.05, 0.1, 0.5, or 1 mM sodium arsenite (As[III]; Sigma-Aldrich). Five replicate 350-μl aliquots of each suspension were pipetted into wells of sealable 100-well microtiter plates and incubated at 37°C for 24 h in an automated turbidity reader (Bioscreen C microbiology reader; Growth Curves, Helsinki, Finland) placed in an anaerobic workstation. The optical density in each well at 600 nm was read at 2-h intervals. Means of all replicates and all individual strains of a cluster were calculated and plotted on a time-turbidity curve.
Growth in the presence of cadmium chloride.
The cadmium resistance of cluster BI and BII strains was studied as described above for arsenic using four replicates and an 18-h total incubation period without and with 5 mM cadmium chloride (Sigma-Aldrich). The difference between cluster BI and BII strains was evaluated by calculating maximum growth rates from the exponential phase of growth curves fitted with DMFit (2). The median values of the four replicates for all individual strains were calculated. The statistical significance of the difference between the two clusters was tested using a Student's t test.
Microarray data accession number.
The microarray data reported here have been deposited in the Array Express database under accession number E-TABM-309.
Nucleotide sequence accession numbers.
The sequence data for the amplification fragments representing the 12 CDSs predicted to discriminate between strains within cluster BI or BII reported here were submitted to the EMBL database under accession numbers AM774033 to AM774040, AM774049 to AM774099, and AM774101 to AM774126 (see Table S1 in the supplemental material). The sequence data for the botB genes of cluster I strain M-18/3 and cluster II strain KV-39/1 have been deposited in the EMBL database under accession numbers FM865704 and FM865705, respectively.
RESULTS
Overall genetic diversity.
A total of 413 CDSs (11.3% of all ATCC 3502 CDSs and 12.1% of ATCC 3502 CDSs represented on the microarray) were variable among the 32 group I C. botulinum type B strains and the ATCC 3502 type A reference strain (Table 2). Numbers of CDSs that are different between the type B strains and ATCC 3502 are presented for each functional category in the fourth column of Table 2. In general, all the type B strains studied lacked the plasmid (Cb245.01 to Cb245.18)-, two prophage (CBO1682 to CBO1754 and CBO2312 to CBO2391)-, and the flagellar glycosylation island (CBO2696 to CBO2732)-related CDSs present in ATCC 3502 (27).
TABLE 2.
Variable CDSs in the Nordic group I Clostridium botulinum type B strains in relation to ATCC 3502 chromosomal and plasmid CDSsa
| CDS functional class | No. of CDSs present in cluster BI strains but not cluster BII strains | No. of CDSs present in cluster BII strains but not cluster BI strains | No. of CDSs missing from all type B strains studied | Total no. of CDSs present in ATCC 3502 chromosome and plasmid | No. (%) of discriminating CDSs in each functional classb |
|---|---|---|---|---|---|
| Cell processes | |||||
| Chemotaxis and mobility | 1 | 1 | 3 | 68 | 5 (7) |
| Chaperones | 15 | 0 (0) | |||
| Protection responses | 1 | 1 | 5 | 2 (40) | |
| Cell killing | 1 | 4 | 10 | 5 (50)* | |
| Detoxification | 2 | 11 | 2 (18) | ||
| Drug/analog sensitivity | 1 | 14 | 1 (7) | ||
| Radiation sensitivity | 1 | 0 (0) | |||
| Transport/binding proteins | 4 | 10 | 6 | 156 | 20 (13) |
| Amino acids and amines | 1 | 38 | 1 (3) | ||
| Cations | 3 | 36 | 3 (8) | ||
| Carbohydrates, organic acids, and alcohols | 1 | 35 | 1 (3) | ||
| Anions | 8 | 0 (0) | |||
| Other substances | 4 | 44 | 4 (10) | ||
| Adaptation | |||||
| Adaptations, atypical conditions | 8 | 0 (0) | |||
| Osmotic adaptation | 1 | 4 | 1 (25) | ||
| Fe storage | 5 | 36 | 5 (14) | ||
| Cell division | 23 | 0 (0) | |||
| Differentiation/sporulation | 5 | 2 | 81 | 7 (9) | |
| Macromolecule degradation | |||||
| Degradation of DNA | 10 | 0 (0) | |||
| Degradation of RNA | 1 | 9 | 1 (11) | ||
| Degradation of polysaccharides | 10 | 0 (0) | |||
| Degradation of proteins, peptides, glycoproteins | 1 | 5 | 50 | 6 (12) | |
| Macromolecule synthesis | 12 | 211 | 12 (6) | ||
| Amino acid biosynthesis | 54 | 0 (0) | |||
| Biosynthesis of cofactors, carriers | 1 | 3 | 1 | 86 | 5 (6) |
| Central intermediary metabolism | 1 | 1 | 109 | 2 (2) | |
| Degradation of small molecules | 86 | 0 (0) | |||
| Energy metabolism, carbon | 1 | 2 | 113 | 3 (3) | |
| Fatty acid biosynthesis | 6 | 18 | 6 (33) | ||
| Nucleotide biosynthesis | 24 | 0 (0) | |||
| Periplasmic/exported/lipoproteins | 1 | 8 | 1 (13) | ||
| Inner membrane | 2 | 4 | 2 (50) | ||
| Outer membrane constituents | 2 | 0 (0) | |||
| Surface polysaccharides and antigens | 1 | 13 | 40 | 14 (35)* | |
| Surface structures | 1 | 7 | 1 (14) | ||
| Gram-positive membrane | 1 | 14 | 11 | 281 | 26 (9) |
| Gram-positive exported protein/lipoprotein | 2 | 5 | 5 | 91 | 12 (13) |
| Gram-positive surface-anchored protein | 4 | 0 (0) | |||
| Gram-positive peptidoglycan, teichoic acid | 1 | 1 | 43 | 2 (5) | |
| Ribosome constituents | 1 | 2 | 63 | 3 (5) | |
| Horizontally acquired elements | |||||
| Phage-related functions and prophages | 1 | 4 | 111 | 166 | 116 (70)* |
| Plasmid-related functions | 4 | 5 | 4 (80)* | ||
| Transposon/IS element-related functions | 3 | 4 | 12 | 7 (58)* | |
| Pathogenicity | 1 | 1 | 16 | 2 (13) | |
| Regulation | 5 | 0 (0) | |||
| Two-component system | 2 | 1 | 74 | 3 (4) | |
| RNA polymerase core enzyme binding | 1 | 15 | 1 (7) | ||
| Defined regulatory families | |||||
| AsnC | 2 | 0 (0) | |||
| AraC | 2 | 4 | 24 | 6 (25)* | |
| GntR | 2 | 18 | 2 (11) | ||
| LacI | 2 | 0 (0) | |||
| LysR | 6 | 0 (0) | |||
| MarR | 1 | 12 | 1 (8) | ||
| TetR | 1 | 1 | 12 | 2 (17) | |
| DeoR | 5 | 0 (0) | |||
| LuxR (GerR) | 1 | 0 (0) | |||
| MerR | 1 | 10 | 1 (10) | ||
| ArsR | 1 | 5 | 1 (20) | ||
| PadR | 3 | 4 | 3 (75)* | ||
| Other regulatory proteins | 3 | 1 | 85 | 4 (5) | |
| Unknown function, conserved proteins | 3 | 26 | 48 | 642 | 77 (12) |
| Not classified | 1 | 14 | 15 | 383 | 30 (8) |
| Total | 17 | 128 | 268 | 3,421 | 413 (12) |
*, significant (P < 0.05) overrepresentation of a functional class as tested with one-sided Fisher's exact test.
The CDS functional classification is done according to data presented previously by Sebaihia et al. (27).
CGH analysis of group I C. botulinum type B clusters BI and BII.
The CGH analysis of the Nordic type B strains clustered the 32 strains into two homogeneous clusters, called clusters BI and BII, distinct from ATCC 3502. No substantial strain-to-strain variation was observed within each cluster (Fig. 2). This was confirmed with sequencing of DNA fragments representing 12 CDSs showing the greatest variation in signal log ratios or predicted by GACK to be variably present within cluster BI or within cluster BII. Strains representing the same cluster showed 99.3 to 100% identity in the sequenced DNA fragments (see Table S1 in the supplemental material).
A total of 145 gene probes discriminated between clusters BI and BII, with 128 CDSs being present exclusively in cluster BII strains and 17 CDSs being present exclusively in cluster BI strains. Numbers of CDSs that are different between clusters BI and BII are presented for each functional category in Table 2. Some of the differential CDSs and related physiological phenomena are discussed in more detail in Discussion.
To verify the DNA microarray results, CBO2590, giving low-intensity values on the array, and CBO3250, giving high-intensity values on the array, were selected for PCR. The array signal log ratios of all strains for the two probe spots ranged from 0 to −3.5, with no clear cutoff for the presence and absence of CDSs. All the eight cluster BII strains yielded a positive PCR result (i.e., a DNA fragment of the expected size with the intensity of a control DNA) for both CBO2590 and CBO3250 CDSs. In addition, two cluster BI strains (515-K/2 and M-1/3) yielded a weak positive result (i.e., a very faint fragment of the expected size visualized on an agarose gel when 10 μl of the reaction mixture was loaded within the agarose gel) for CBO2590, and two cluster BI strains (515-K/2 and He-3323) yielded a weak positive result for CBO3250. The cutoff hybridization signal log ratios of −0.4 and −0.25 for CBO2590 and CBO3250, respectively, corresponded to a 100% PCR-positive result. The cutoff log ratios for the GACK EPP of 100% for all genes were in the range of −0.6 to −0.15 (median, −0.23). As for the missing or divergent CDSs, the cutoff signal log ratios for the GACK EPP of 0% varied between −1.1 and −0.5 between slides (median, −0.77). A total of 22 out of 24 (92%) cases with a 0% EPP prediction with microarrays gave a negative PCR result.
Nucleotide sequence of botB.
The botB genes in cluster I strain M-18/3 and cluster II strain KV-39/1 shared 99.6% sequence identity, corresponding to 99.7% similarity at the amino acid level. Most differences were observed in the C terminal of the heavy chain. Compared to 27 botB gene sequences available in the public domain, botB of both of our strains clustered together with subtype B2 (Fig. 3).
FIG. 3.
Phylogram of the botB sequences from Nordic cluster BI strain M-18/3 and cluster BII strain KV-39/1 and 27 previously published botB sequences. Both Nordic strains clustered together with strains previously identified as belonging to subtype B2 (11).
Growth in the presence of sodium arsenite.
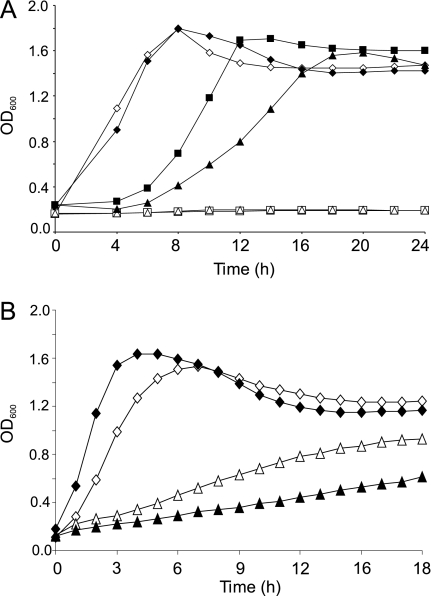
Cluster BII strains KH-318/1, KS-44/10, and M-43/15 showed growth in the presence of 0.05 and 0.1 mM (3.7 and 7.5 mg/liter) sodium arsenite, while growth was not observed with cluster BI strains M-1/3, P-120/6, and DA-58/1 under the same conditions (Fig. 4A). The growth of all cluster BI and BII strains was prevented by the addition of 0.5 mM sodium arsenite (Fig. 4A).
FIG. 4.
Growth of cluster BI strains (M-1/3, P-120/6, and DA-58/1) (marked with open symbols) and cluster BII strains (KH-318/1, KS-44/10, and M-43/15) (marked with solid symbols) in the presence of 0 to 0.1 mM sodium arsenite (diamonds, 0 mM; squares, 0.05 mM; triangles, 0.1 mM) (A) or 0 to 5 mM cadmium chloride (diamonds, 0 mM; triangles, 5 mM) (B). Each curve represents the means of five (A) or four (B) replicate measurements of the three individual strains. OD600, optical density at 600 nm.
Growth in the presence of cadmium chloride.
In the absence of cadmium chloride, cluster BII strains KH-318/1, KS-44/10, and M-43/15 grew at a 1.2-fold-higher rate than did cluster BI strains M-1/3, P-120/6, and DA-58/1 (P < 0.005) (Fig. 4B). However, in the presence of 5 mM cadmium chloride, the same cluster BI strains grew at a twofold-higher rate than did cluster BII strains (P < 0.01) (Fig. 4B).
DISCUSSION
CGH analysis of 33 group I C. botulinum strains revealed that a total of 413 CDSs, thus, 12.1% of all ATCC 3502 CDSs (11.3% of ATCC 3502 CDSs represented on the microarray), were variable among these strains, suggesting that the “core gene set” of all the group I C. botulinum type B strains examined in the present study were 88 to 89% of the ATCC 3502 CDSs. Sebaihia et al. (27) previously reported that nine group I strains, representing a wider range of toxinotypes and geographical locations than in the present study, possessed 87 to 96% of the CDSs of ATCC 3502. While further testing with a more heterogeneous strain collection is required to confirm the true group I C. botulinum “core gene set,” it is apparent that the findings for group I C. botulinum contrast greatly with data from a previous genomic analysis of 75 Clostridium difficile strains, which shared only 20% of CDSs (31). Our results thus support the theory of a stable genome (27) and a limited overall diversity among group I C. botulinum strains (15, 21).
CGH analysis clustered the 32 Nordic group I C. botulinum type B strains into two homogeneous groups similar to those previously observed using PFGE (20, 23). Sequencing of DNA fragments of CDSs representing the greatest strain-to-strain variation within a cluster showed 99.3 to 100% identity between strains of the same cluster, indicating that strains within each cluster were highly similar. However, as the array probes represent only protein coding sequences, covering only 28% of the ATCC 3502 genome, genome sequencing or hybridization onto tiled genome arrays are needed to finally judge whether the two clusters consist of genetically identical strains or whether diversity exists.
Of the 145 CDS probes discriminating between clusters BI and BII, 128 CDSs were present exclusively in BII strains (Table 2), suggesting that cluster BII strains are more closely related to ATCC 3502 than are cluster BI strains. Reflecting the environmental origin of group I C. botulinum, the most apparent differences between the two clusters were related to resistance to toxic compounds such as arsenic and cadmium, which are frequently detected in the environment. While the CDS for the arsenate reductase gene arsC (CBO0751) was present in all strains studied, arsA and arsB (CBO0755 and CBO0756, respectively), encoding the ATP-dependent arsenite efflux pump, and arsR (CBO0753) and arsD (CBO0754), encoding trans-acting repressors, were present exclusively in the cluster BII strains. Therefore, the cluster BII strains were anticipated to tolerate a higher arsenic concentration in their growth medium than were cluster BI strains. Further laboratory tests on three cluster BI strains and three cluster BII strains confirmed that the cluster BII strains (KH-318/1, KS-44/10, and M-43/15) were able to grow in the presence of up to 0.1 mM sodium arsenite, while no growth was observed for the cluster BI strains (M-1/3, P-120/6, and DA-58/1) under the same conditions. The ars resistance system was originally described for bacteria as being plasmid-borne but was later found to be chromosomally located in many bacterial species (28, 33). The ars operon in ATCC 3502 is flanked by mobile elements such as transposase and phage-related CDSs. While our cluster BI strains lacked many such CDSs (CBO0744 to CBO0747 and CBO0749), the presence of several other associated CDSs (CBO0732 to CBO0735, CBO0738 to CBO0742, CBO0762, and CBO0768) in both cluster BI and BII strains suggests a phage-mediated background of the arsenic resistance genes. The acquisition of arsenic resistance in Nordic C. botulinum cluster BII strains is probably an evolutionary response to high levels of environmental arsenic contamination; ground waters and soil in certain areas in Finland have been reported to have high arsenic concentrations in excess of 0.01 mM (http://www.ymparisto.fi/default.asp?contentid=291469&lan=EN), while typical concentrations in European rivers are 100-fold lower (29).
All the C. botulinum type B strains studied carried the cadA operon, related to cadmium resistance, consisting of cadA (CBO0478), which encodes a cadmium exporter, and cadC, which codes for a transcriptional regulator (CBO0477). In Staphylococcus aureus, a plasmid carrying cadAC was associated with resistance to 50 to 100 μM of Cd2+, while strains lacking this gene tolerated only 1/10 of this cadmium concentration (24). All the 32 Nordic type B strains were able to grow in the presence of 5 mM cadmium chloride, suggesting the presence of an active cadAC operon in both cluster BI and BII strains. However, all the three tested BI strains (M-1/3, P-120/6, and DA-58/1) showed significantly higher growth rates in the presence of 5 mM cadmium chloride than did the cluster BII strains tested (KH-318/1, KS-44/10, and M-43/15), while their growth in the absence of cadmium was significantly slower than that of the cluster BII strains. The difference may be explained by the presence of cadmium resistance mechanisms other than those that are efflux related. All cluster BI strains carried exclusively CBO0841, which shares 50 to 60% amino acid similarity to cadmium transporters in several other prokaryotes and 52% similarity to cadD in S. aureus. This gene resembles cadB, which has been suggested to confer cadmium resistance by binding Cd2+ in the membrane rather than by promoting cation efflux (25). In S. aureus, cadD conferred resistance to cadmium sulfate at a lower level than did cadAC (6).
The surface structures of C. botulinum vegetative cells have not been widely investigated, probably because this bacterium is generally considered to cause disease through exogenous neurotoxin production rather than being infectious or invasive. Considering the generally small genetic homogeneity of group I C. botulinum strains, a marked variation in the compositions of cell wall and surface structure-related CDSs between the two type B clusters and also between the type B strains and ATCC 3502 was observed. A total of 24 CDSs related to gram-positive cell wall or surface antigens were differential between cluster BII and BI strains, and 34 CDSs were missing from all the type B strains. In the absence of further genomic information, these data indicate that the two type B clusters studied may represent antigenically distinct lineages, which also differ from ATCC 3502. A recent insight into the available clostridial genomes confirmed the vast variety of membrane protein- and surface antigen-encoding genes between different clostridial species (4).
The ATCC 3502 genome contains two loci putatively encoding polysaccharide capsule formation (locus I contains CBO2678 to CBO2729, and locus II contains CBO3092 to CBO3114). While all our type B strains lacked 31 of the 51 CDSs of locus I, they possessed 20 out of 21 CDSs of locus II. The CDSs present in all strains included several putative glycosyltransferase CDSs as well as those encoding the suggested core capsule synthesis proteins, namely, O-antigen polymerase (CBO3098), chain length determinant protein (CBO2689), and UDP-N-acetylglucosamine 2-epimerase (CBO3100 and CBO0148) (4). These results strongly suggest C. botulinum cells to be capable of capsule formation under some circumstances. Capsule formation in other bacteria is considered to protect the bacteria against phagocytosis or assist in adhesion and virulence. As an environmental organism, C. botulinum may benefit from features assisting in adhesion to plant material or dead epithelial tissues. More importantly, considering that infant botulism, which is the most common form of human botulism in the United States today, and adult infectious botulism are due to colonization and subsequent in vivo toxin production by group I C. botulinum strains in the gut, the proposed ability to form a capsule and large antigenic variability may play an important role in the pathogenesis of the intestinal forms of human botulism.
Apart from the two regulators controlling the arsenic resistance operon, the cluster BII strains exclusively contained 10 other regulatory CDSs, of which the PadR family members have been related to phenolic acid metabolism (3) and the AraC/XylS members have been associated with carbon metabolism, virulence, and the stress response (9). In addition to PadR and AraC/XylS family regulators, CDSs present exclusively in cluster BII strains were related to signal transduction systems. Although the specific role of these regulators in the physiology of the cluster BII C. botulinum strains has yet to be elucidated, they probably contribute to the ability of cluster BII strains to better adapt to environmental stresses than cluster BI strains. Accordingly, the cluster BII strains together with ATCC 3502 were previously reported to grow at lower temperatures than were cluster BI strains (12).
All the Nordic group I C. botulinum type B strains carried botB and the CDSs encoding the nontoxic nonhemagglutinin component, three hemagglutinins, and the transcriptional neurotoxin regulator. No additional entire or partial neurotoxin-related CDSs were present in the type B strains. The botB sequences of both cluster BI strain M-18/3 and cluster BII strain KV-39/1 clustered together with the subtype B2 genes detected in group I C. botulinum strains (11). Subtype B2 seems to be the most common type B subtype equally distributed around the world (11): subtype B2 strains have been isolated from North America, Europe, and East Asia. However, although the narrow genetic diversity among the 32 type B strains studied suggests that all the strains may represent toxin subtype B2, further sequencing efforts are required to confirm their neurotoxin subtype.
The fact that the cluster BI and BII strains had nearly identical botB sequences but seemed to be genetically and antigenically different suggests that the neurotoxin gene cassettes have generally an independent evolutionary background distinct from that of the C. botulinum organism. Two of the transposase-related CDSs (CBO0800 and CBO0809) flanking the neurotoxin gene cluster in ATCC 3502 (27) were absent or highly divergent in the Nordic type B strains, and the other two CDSs (CBO0807 and CBO0810) gave only a weak positive result. In accordance with the recent report on the group I type B strain Okra and the bivalent type Ba strain 657 carrying insertion sequence (IS) elements different from those in ATCC 3502 (30), our CGH findings suggest that the IS elements related to the type B neurotoxin gene cassette in general are distinct from those related to the type A toxin gene. In light of current knowledge, a mobile background of both type A and type B neurotoxin gene cassettes seems highly likely, as several C. botulinum strains with type A and/or type B toxin genes on plasmids of various sizes have been reported (8, 19).
The reliability of DNA microarray hybridization signals was verified by PCR targeted to CBO2590 and CBO3250. These probes gave the widest range of signal log ratios between all slides, with CBO2590 consistently giving weaker signals for the reference DNA (ATCC 3502) than did CBO3250. The signal log ratios of all strains for these two probe spots ranged from 0 to −3.5, with no clear cutoff for the presence and absence of CDSs. The cutoff signal log ratios for the GACK EPP of 100% were in accordance with those for a 100% PCR-positive result, indicating that the most stringent GACK prediction for a CDS to be present was correct. As for the missing or divergent CDSs, most (92%) PCR results, being negative, were in accordance with GACK 0% EPP predictions. Only three strains gave a weak PCR-positive result for CBO2590 and/or CBO3250, while the corresponding signal log ratios were in the range of the 0% EPP prediction for an absent or highly divergent CDS. A weak PCR-positive result is probably due to sequence divergence at a primer annealing site, decreasing the reaction efficiency. Since the cutoff DNA identity for a positive hybridization signal is perhaps 80% and that for a 100% EPP prediction is over 90% (16), whereas a PCR may tolerate up to 50 to 60% mismatch at the primer 5′ end but could fail due to only one mismatch at the primer 3′ end, some discrepancy between PCR and hybridization is expected to occur. Our results are in accordance with those reported previously by Kim et al. (17), indicating that the GACK predictions were generally reliable.
In conclusion, the CGH analysis with ATCC 3502 microarrays revealed that the two predominating Nordic group I C. botulinum type B clusters have different resistance to toxic compounds, reflecting variable evolutionary responses to environmental stress. Evidence of an ability to form a capsule and the apparently large variation in the cell surface structures of C. botulinum strains indicate a different adaptation to various environmental or clinical niches, which may be of considerable importance when defining the pathogenesis and epidemiology of C. botulinum and the infectious forms of human botulism.
Supplementary Material
Acknowledgments
This work was funded by the Academy of Finland (grants 1115133, 1118602, 1120180, and 206319), the Finnish Ministry of Agriculture and Forestry (grant 4655/501/2003), the Finnish Funding Agency for Technology and Innovation (grant 2431/31/04), the Walter Ehrström Foundation, and the Competitive Strategic Grant of the BBSRC.
We thank Hanna Korpunen for excellent laboratory assistance.
Footnotes
Published ahead of print on 6 March 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ala-Poikela, M., E. Svensson, A. Rojas, T. Horko, L. Paulin, J. Valkonen, and A. Kvarnheden. 2005. Genetic diversity and mixed infections of begomoviruses infecting tomato, pepper and cucurbit crops in Nicaragua. Plant Pathol. 54:448-459. [Google Scholar]
- 2.Baranyi, J., and T. A. Roberts. 1994. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 23:277-294. [DOI] [PubMed] [Google Scholar]
- 3.Barthelmebs, L., B. Lecomte, C. Divies, and J. F. Cavin. 2000. Inducible metabolism of phenolic acids in Pediococcus pentosaceus is encoded by an autoregulated operon which involves a new class of negative transcriptional regulator. J. Bacteriol. 182:6724-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brüggemann, H., and G. Gottschalk. 2008. Comparative genomics of clostridia: link between the ecological niche and cell surface properties. Ann. N. Y. Acad. Sci. 1125:73-81. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., H. Korkeala, J. Aarnikunnas, and M. Lindström. 2007. Sequencing the botulinum neurotoxin gene and related genes in Clostridium botulinum type E strains reveals orfx3 and a novel type E neurotoxin subtype. J. Bacteriol. 189:8643-8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crupper, S. S., V. Worrell, G. C. Stewart, and J. J. Iandolo. 1999. Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J. Bacteriol. 181:4071-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewing, B., L. Hillier, M. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 8.Franciosa, G., A. Maugliani, C. Scalfaro, and P. Aureli. 2008. Subtypes of the type B botulinum neurotoxin gene are widely distributed on extrachromosomal elements, p. 27. In Abstr. Clostridium botulinum Epidemiol. Diagn. Genet. Control Prev., Helsinki, Finland, 16 to 19 June 2008. http://www.clostridia.net/helsinki/c-diff-abstractbook.pdf.
- 9.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hielm, S., J. Björkroth, E. Hyytiä, and H. Korkeala. 1999. Ribotyping as an identification tool for Clostridium botulinum strains causing human botulism. Int. J. Food Microbiol. 47:121-131. [DOI] [PubMed] [Google Scholar]
- 11.Hill, K. K., T. J. Smith, C. H. Helma, L. O. Ticknor, B. T. Foley, R. T. Svensson, J. L. Brown, E. A. Johnson, L. A. Smith, R. T. Okinaka, P. J. Jackson, and J. D. Marks. 2007. Genetic diversity among botulinum neurotoxin-producing clostridial strains. J. Bacteriol. 189:818-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinderink, K., M. Lindström, and H. Korkeala. 2009. Group I Clostridium botulinum strains show significant variation in growth at low and high temperatures. J. Food Prot. 72:375-383. [DOI] [PubMed] [Google Scholar]
- 13.Hutson, R. A., D. E. Thompson, P. A. Lawson, R. P. Schocken-Itturino, E. C. Bottger, and M. D. Collins. 1993. Genetic interrelationships of proteolytic Clostridium botulinum types A, B, and F and other members of the Clostridium botulinum complex as revealed by small-subunit rRNA gene sequences. Antonie van Leeuwenhoek 64:273-283. [DOI] [PubMed] [Google Scholar]
- 14.Ihaka, R., and R. Gentleman. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299-314. [Google Scholar]
- 15.Keto-Timonen, R., M. Nevas, and H. Korkeala. 2005. Efficient DNA fingerprinting of Clostridium botulinum types A, B, E, and F by amplified fragment length polymorphism analysis. Appl. Environ. Microbiol. 71:1148-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keto-Timonen, R., A. Heikinheimo, E. Eerola, and H. Korkeala. 2006. Identification of Clostridium species and DNA fingerprinting of Clostridium perfringens by amplified fragment length polymorphism analysis. J. Clin. Microbiol. 44:4057-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:RESEARCH0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindström, M., and H. Korkeala. 2006. Laboratory diagnostics of Clostridium botulinum. Clin. Microbiol. Rev. 19:298-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall, K. M., M. Bradshaw, S. Pellett, and E. A. Johnson. 2007. Plasmid encoded neurotoxin genes in Clostridium botulinum serotype A subtypes. Biophys. Res. Commun. 361:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nevas, M., M. Lindström, K. Hautamäki, S. Puoskari, and H. Korkeala. 2005. Prevalence and diversity of Clostridium botulinum types A, B, E and F in honey produced in the Nordic countries. Int. J. Food Microbiol. 105:145-151. [DOI] [PubMed] [Google Scholar]
- 21.Nevas, M., M. Lindström, S. Hielm, K. J. Björkroth, M. W. Peck, and H. Korkeala. 2005. Diversity of proteolytic Clostridium botulinum strains, determined by a pulsed-field gel electrophoresis approach. Appl. Environ. Microbiol. 71:1311-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevas, M., M. Lindström, A. Virtanen, S. Hielm, M. Kuusi, S. S. Arnon, E. Vuori, and H. Korkeala. 2005. Infant botulism acquired from household dust presenting as sudden infant death syndrome. J. Clin. Microbiol. 43:511-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nevas, M., M. Lindström, A. Hörman, R. Keto-Timonen, and H. Korkeala. 2006. Clostridium botulinum in the honey production environment. Environ. Microbiol. 8:1085-1094. [DOI] [PubMed] [Google Scholar]
- 24.Nucifora, G., L. Chu, T. K. Misra, and S. Silver. 1989. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc. Natl. Acad. Sci. USA 86:3544-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry, R. D., and S. Silver. 1982. Cadmium and manganese transport in Staphylococcus aureus membrane vesicles. J. Bacteriol. 150:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raphael, B. H., C. Luquez, L. M. McCroskey, L. A. Joseph, M. J. Jacobson, E. A. Johnson, S. E. Maslanka, and J. D. Andreadis. 2008. Genetic homogeneity of Clostridium botulinum type A1 strains with unique toxin gene clusters. Appl. Environ. Microbiol. 74:4390-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sebaihia, M., M. W. Peck, N. P. Minton, N. R. Thomson, M. T. Holden, W. J. Mitchell, A. T. Carter, S. D. Bentley, D. R. Mason, L. Crossman, C. J. Paul, A. Ivens, M. H. J. Wells-Bennik, I. J. Davis, A. M. Cerdeno-Tarraga, C. Churcher, M. A. Quail, T. Chillingworth, T. Feltwell, A. Fraser, I. Goodhead, Z. Hance, K. Jagels, N. Larke, M. Maddison, S. Moule, K. Mungall, H. Nobertczak, E. Rabbinowitsch, M. Standeres, M. Simmonds, B. White, S. Whitehead, and J. Parkhill. 2007. Genome sequence of a proteolytic (group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res. 17:1082-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silver, S., and L. T. Phung. 2005. Genes and enzymes involved in the bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smedley, P. L., and D. G. Kinniburgh. 2001. Source and behaviour of arsenic in natural waters, p. 1-61. In United Nations synthesis report on arsenic in drinking water. World Health Organization, Geneva, Switzerland. http://www.who.int/water_sanitation_health/dwq/arsenicun1.pdf.
- 30.Smith, T. J., K. K. Hill, B. T. Foley, J. C. Detter, A. C. Munk, D. C. Bruce, N. A. Doggett, L. A. Smith, J. D. Marks, G. Xie, and T. S. Brettin. 2007. Analysis of the neurotoxin complex genes in Clostridium botulinum A1-A4 and B1 strains: BoNT/A3, /Ba4 and /B1 clusters are located within plasmids. PLoS ONE 2:e1271. doi: 10.1371/journal.pone.0001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stabler, R. A., D. N. Gerding, J. G. Songer, D. Drudy, J. S. Brazier, H. T. Trinh, A. A. Witney, J. Hinds, and B. W. Wren. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 33.Stolz, J. F., P. Basu, J. M. Santini, and R. S. Oremland. 2006. Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60:107-130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.