Abstract
To further unravel the mechanisms responsible for attenuation of the tuberculosis vaccine Mycobacterium bovis BCG, comparative genomics was used to identify single nucleotide polymorphisms (SNPs) that differed between sequenced strains of Mycobacterium bovis and M. bovis BCG. SNPs were assayed in M. bovis isolates from France and the United Kingdom and from different BCG vaccines in order to identify those that arose during the attenuation process which gave rise to BCG. Informative data sets were obtained for 658 SNPs from 21 virulent M. bovis strains and 13 BCG strains; these SNPs showed phylogenetic clustering that was consistent with the geographical origin of the strains and previous schemes for BCG genealogies. The data revealed a closer relationship between BCG Tice and BCG Pasteur than was previously appreciated, while we were able to position BCG Beijing within a grouping of BCG Denmark-derived strains. Only 186 SNPs were identified between virulent M. bovis strains and all BCG strains, with 115 nonsynonymous SNPs affecting important functions such as global regulators, transcriptional factors, and central metabolism, which might impact on virulence. We therefore refine previous genealogies of BCG vaccines and define a minimal set of SNPs between virulent M. bovis strains and the attenuated BCG strain that will underpin future functional analyses.
Mycobacterium bovis bacillus Calmette-Guérin (BCG) is the only vaccine available against tuberculosis and is the most widely used vaccine in the world. It was derived by the repeated subculture of a strain of Mycobacterium bovis on potato slices soaked in glycerol and ox bile (10), leading to the in vitro accumulation of mutations and ultimately attenuation. Despite the widespread use of BCG, the precise genetic lesions that led to attenuation are not defined. Furthermore, the success of BCG led to its distribution from the Institut Pasteur to laboratories around the world, each of which continued the subculturing process, thereby leading to the generation of a number of daughter strains named after their geographical origin (hence BCG Tokyo, BCG Russia, etc.). The protective efficacy of these strains has been shown to vary in both laboratory models and epidemiological studies (6, 18, 36).
As BCG is the only vaccine currently available against tuberculosis, there is a clear need to understand the molecular basis of attenuation and variable efficacy afforded by BCG. The first study that attempted to identify mutations linked to attenuation was performed by Mahairas and colleagues, who identified three deletions, RD1 to RD3, from the genome of BCG strain Connaught (39). The RD1 locus was shown to be deleted from all BCG strains but present in all virulent strains of M. bovis and Mycobacterium tuberculosis studied. Subsequent work has shown that this deletion played a major role in the attenuation of BCG (38, 46). However, complementation of BCG with RD1 does not restore virulence to wild-type levels, suggesting that other attenuating mutations exist. Indeed, all BCG strains contain a frameshift mutation in the phoT gene; inactivation of phoT in M. bovis attenuates the strain, and so this phoT mutation may also contribute to the loss of virulence of BCG (15). This observation demonstrates that single nucleotide polymorphisms (SNPs) may play a significant part in the attenuation of BCG.
A major step toward defining the molecular basis of attenuation in BCG was the completion of the genome sequence of M. bovis BCG Pasteur (9). Genomic comparison of BCG Pasteur with M. bovis 2122/97 (22) identified a range of mutational differences, including deletions, duplications, and SNPs. In further work the configuration of two large duplications, DU1 and DU2, was shown to vary across BCG daughter strains, in a manner that was congruent with the previous BCG phylogeny defined by Behr and colleagues using deletions (7, 9, 41).
From the complete chromosome sequences, 736 SNPs were identified between the BCG Pasteur vaccine strain and the virulent M. bovis strain 2122/97 (9). However, only those SNPs that are unique to all BCG strains are good candidates for mutations involved in the attenuation of BCG; such SNPs presumably occurred during the attenuation of BCG after it was derived from wild-type M. bovis. The ideal experiment would be a comparison using the chromosome of the wild-type progenitor from which BCG strains were derived; unfortunately, this strain is unavailable. The M. bovis 2122/97 strain is derived from the clonal complex of M. bovis common in the British Isles (provisionally called Eu1 [53]), while the BCG strain is derived from the French lineage, which is phylogenetically distinct from the Eu1 clonal complex. Many of the 736 SNPs that differ between BCG Pasteur and M. bovis 2122/97 may be lineage specific to either BCG or the Eu1 clonal complex and therefore unlikely to be involved in the attenuation of BCG.
To identify those SNPs that are unique to BCG strains and therefore possible candidates for the attenuation of BCG, we mapped the phylogenetic position of all SNPs identified between M. bovis 2122/97 and BCG Pasteur across a population of United Kingdom and French M. bovis isolates, as well as BCG daughter strains. We used a high-throughput SNP screening methodology to screen hundreds of SNPs across the population and generated a distribution of SNPs across British, French, and BCG M. bovis strains.
It has previously been shown that some SNPs unique to BCG have functional effects: a nonsynonymous SNP (nsSNP) in the mmaA3 gene, which encodes a mycolic acid methyltransferase, results in the loss of methoxymycolic acids from late strains of BCG (5); an SNP in sigK leads to reduced synthesis of MPB83 and MPB70 in late BCG strains (11); and an SNP in the pykA gene reverts a null mutation in M. bovis and allows BCG to grow on glycerol (33). Finally, SNPs in the cyclic AMP receptor protein (CRP) transcriptional regulator in some BCG strains affect the binding of the regulator to DNA (4, 30). Following these examples, we highlight SNPs that may impact on phenotypic differences between virulent M. bovis and BCG.
MATERIALS AND METHODS
SNP identification.
The sequences of M. bovis 2122/97 and M. bovis BCG Pasteur were compared using the DIFFSEQ application from the EMBOSS package (http://emboss.sourceforge.net/). This tool is less complex than many genome comparison tools but can be used here, as the two genomes are entirely colinear. This allowed three classes of mutations to be identified: (i) transitions and transversions; (ii) insertions or deletions (InDels); and (iii) “block” substitutions, where a block of sequence of >1 bp replaces another. The InDels were further subdivided into two groups, the mIns and mDels, which are the insertions or deletions of a single base, respectively. We defined SNPs as the total number of transitions and transversions (736) plus the mIns and mDels (46), which gave a total of 782 positions to be investigated. Each SNP was initially verified by checking the original sequence trace files from the BCG Pasteur and M. bovis 2122/97 sequencing projects.
Bacterial strains.
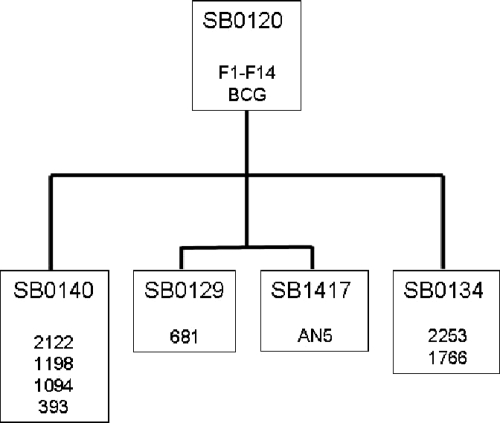
Strains are shown in Table 1, with a spoligotype phylogeny shown in Fig. 1 to illustrate the relationships across the strains. British M. bovis strains were selected from the VLA Weybridge strain collection to represent the British M. bovis population structure. French isolates were selected from the strain collection of the Agence Française de Sécurité Sanitaire des Aliments (AFSSA), such that the spoligotype of the strain was the same as that of BCG (SB0120, as defined by the international M. bovis spoligotype database [www.mbovis.org]). BCG daughter strains were obtained from the VLA Weybridge strain collection, Marcel Behr (McGill University, Montreal, Canada), or the Statens Serum Institut (Copenhagen, Denmark). A total of 34 strains were examined.
TABLE 1.
Strains used in this study
| M. bovis strain | Source | Spoligotype | VNTR | Comment |
|---|---|---|---|---|
| Non-BCG strains | ||||
| 2122/97 | VLA Weybridge, United Kingdom | SB0140 | 8555*33.1 | Genome-sequenced strain |
| F1.2 | AFSSA, Alfort, France | SB0120 | 5654*33.1 | French clinical isolate |
| F3 | AFSSA, Alfort, France | SB0120 | 5553*33.1 | French clinical isolate |
| F4 | AFSSA, Alfort, France | SB0120 | 5554*43.1 | French clinical isolate |
| F5 | AFSSA, Alfort, France | SB0120 | 5554*33.1 | French clinical isolate |
| F6 | AFSSA, Alfort, France | SB0120 | 5554*33.1 | French clinical isolate |
| F7 | AFSSA, Alfort, France | SB0120 | NDa | French clinical isolate |
| F8 | AFSSA, Alfort, France | SB0120 | 4554*33.1 | French clinical isolate |
| F9 | AFSSA, Alfort, France | SB0120 | 5554*33.1 | French clinical isolate |
| F10 | AFSSA, Alfort, France | SB0120 | 5554*33.1 | French clinical isolate |
| F11 | AFSSA, Alfort, France | SB0120 | 5554*33.1 | French clinical isolate |
| F12 | AFSSA, Alfort, France | SB0120 | 5554*33.1 | French clinical isolate |
| F13 | AFSSA, Alfort, France | SB0120 | 5554*33.1 | French clinical isolate |
| F14 | AFSSA, Alfort, France | SB0120 | 5554*33.1 | French clinical isolate |
| 2253 | VLA Weybridge, United Kingdom | SB0134 | 3534*33.1 | United Kingdom clinical isolate |
| 1766 | VLA Weybridge, United Kingdom | SB0134 | 3534*33.1 | United Kingdom clinical isolate |
| 681 | VLA Weybridge, United Kingdom | SB0129 | 6554*23.1 | United Kingdom clinical isolate |
| 1198 | VLA Weybridge, United Kingdom | SB0140 | 6554*33.1 | United Kingdom clinical isolate |
| 1094 | VLA Weybridge, United Kingdom | SB0140 | 7554*33.1 | United Kingdom clinical isolate |
| 393 | VLA Weybridge, United Kingdom | SB0140 | 7554*33.1 | United Kingdom clinical isolate |
| AN5 | VLA Weybridge, United Kingdom | SB1417 | 6554*33.2 | Tuberculin production strain |
| BCG strains | ||||
| Pasteur | VLA Weybridge, United Kingdom | SB0120 | 556233.1 | Genome-sequenced strain |
| Sweden | Marcel Behr, McGill University, Canada | SB0120 | 5553*33.1 | Vaccine strain |
| Tokyo | VLA Weybridge, United Kingdom | SB0120 | 5553*33.1 | Vaccine strain |
| Denmark | Statens Serum Institut, Denmark | SB0120 | 5552/3*33.1 | Vaccine strain |
| Tice | Marcel Behr, McGill University, Canada | SB0120 | 555233.1 | Vaccine strain |
| Frappier | Marcel Behr, McGill University, Canada | SB0120 | 555133.1 | Vaccine strain |
| Glaxo | VLA Weybridge, United Kingdom | SB0120 | 5553*33.1 | Vaccine strain |
| Russia | VLA Weybridge, United Kingdom | SB0120 | ND | Vaccine strain |
| Beijing | China Agricultural University | SB0120 | ND | Vaccine strain |
| Connaught | VLA Weybridge, United Kingdom | SB0120 | ND | Vaccine strain |
| Birkhaug | Marcel Behr, McGill University, Canada | SB0120 | ND | Vaccine strain |
| Prague | Marcel Behr, McGill University, Canada | SB0120 | ND | Vaccine strain |
| Moreau | Marcel Behr, McGill University, Canada | SB0120 | ND | Vaccine strain |
ND, not determined.
FIG. 1.
Spoligotype phylogeny of M. bovis strains. The figure depicts relationships across the M. bovis strains used in this study based solely on spoligotype. Strain numbers are shown clustered into their spoligotype designations (“SB” numbers, based on the international M. bovis spoligotype database at www.mbovis.org). SB0120 is the most replete M. bovis spoligotype pattern, with all other patterns used in this study derivatives of this pattern. Branch lengths are not phylogenetically informative and are shown merely for the purposes of clustering related strains.
Molecular typing.
Strains were typed by both spoligotyping and variable-number tandem repeat (VNTR) (ETR-A to -F) typing. Spoligotyping and VNTR were performed as described previously (21, 32).
Sequenom genotyping.
Genotyping was performed with IPLEX chemistry, on the Sequenom genotyping platform (Sequenom Inc., San Diego, CA). During the IPLEX reaction, oligonucleotide primers anneal directly adjacent to the SNP of interest. Allele-specific extension products are then produced by single base extension of the oligonucleotide with terminator nucleotides, each of unique mass. Multiplexed IPLEX assays of between 1 and 28 assays per plex were designed to detect 701 single nucleotide base changes using the Sequenom Assay Design v3.0.2.0 package. Genomic DNA was extracted from BCG and other M. bovis strains using a standard cetyltrimethylammonium bromide method and diluted in Tris-EDTA to a final concentration of 4 ng/μl. SNP-containing loci were amplified from genomic DNA by PCR. Unincorporated nucleotides were removed by treatment with shrimp alkaline phosphatase, followed by the IPLEX extension reaction, per the manufacturer's instructions. The allele-specific products resulting from the IPLEX reaction were desalted through the addition of an anion-exchange resin and then analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Genotypes were assigned in real time and then evaluated using the SpectroCALLER and SpectroACQUIRE software (Sequenom), respectively. Selected SNPs were confirmed using standard capillary sequencing.
Phylogenetic analysis.
SNP calls were parsed to extract SNPs specific to each strain and concatenated into a single 701-bp sequence. Sequence alignment was carried out using ClustalW, and phylogenetic analyses were performed using the MEGA 4.0 software. Discrepancies in the data (i.e., call rates lower than 100% or ambiguous calls) were replaced by “?” for the purposes of alignment. Distance-based analysis was conducted by applying the neighbor-joining algorithm using the number of nucleotide differences. A consensus parsimony tree was generated using the maximum parsimony method with bootstrapping.
dN/dS estimations.
The sequence of the common ancestor (anc1) of the British lineage and the French lineage was reconstructed at the polymorphic sites using the M. tuberculosis H37Rv strain as an outgroup. The reconstructed ancestral sequence was then compared to the sequence found in strain 2253 (a representative of the British lineage, SB0134 [Table 1]), and the ratio of the number of nonsynonymous to synonymous changes per site (dN/dS ratio) was calculated for 48 synonymous and nonsynonymous mutations that had happened during the descent of this strain from anc1 to its divergence from the lineage that led to the M. bovis sequenced strain 2122. In a similar way the dN/dS ratio was calculated for 87 synonymous and nonsynonymous mutations in strain F1.2 (representing the French lineage, SB0120 [Table 1]) that happened between anc1 and the divergence of F1.2 from the lineage that leads to BCG Pasteur. Finally, the dN/dS ratio was calculated for 161 synonymous and nonsynonymous mutations between the divergence of strain F1.2 and BCG Russia in the lineage leading to BCG Pasteur. dN/dS ratios were calculated with the S.T.A.R.T 2 package (31) using the Nei-Gojobori method and the Jukes-Cantor correction (43).
RESULTS
Sequenom analysis.
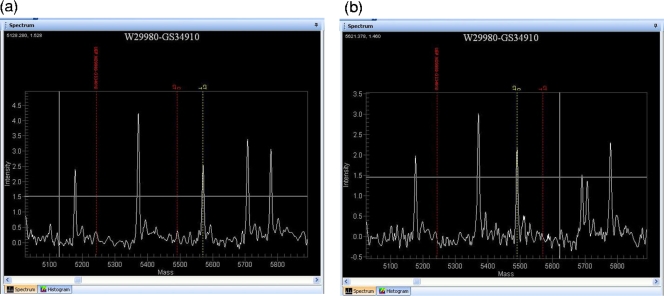
SNPs were queried using oligonucleotides that anneal −1 from the base of interest; allele-specific extension products were then analyzed via matrix-assisted laser desorption ionization mass spectrometry to identify the base at each SNP position across the panel of strains (Fig. 2). From the total of 736 SNPs, 35 SNPs failed and 20 gave ambiguous calls (i.e., many strain-calls missing or both alleles called at the same locus); this gave a total of 681 SNPs that gave usable data. Of these 681 SNPs, 23 were called as invariant across BCG Pasteur and other M. bovis strains, suggesting errors in the original genome sequences. However these 23 SNPs were contained within repeated sequences such as IS1081, REP13E12, and PE-PGRS genes, suggesting that the Sequenom method may also be at fault. To determine whether the sequence or Sequenom calls were correct, we examined the original sequence trace files for the BCG Pasteur and M. bovis 2122/97 genome sequences. This confirmed that 14 of the 23 invariant SNPs were indeed point mutations between BCG Pasteur and M. bovis 2122/97; four SNPs appeared to be errors in M. bovis, while the remaining five SNPs could not be called because of poor sequence reading coverage. As we could not determine the distribution of these 23 SNPs across the remaining strains, they were excluded from the analysis. Hence, we obtained informative data from 658 SNPs. The complete results for all of the 701 SNPs that gave data (including ambiguous calls) are shown in Table S1 in the supplemental material, while the distribution of SNPs across the functional classification of BCG genes is shown in Fig. S1 in the supplemental material.
FIG. 2.
Sequenom output. Images of the MassARRAY TyperAnalyzer v3.3 software (Sequenom) output for two samples. (a) Mass spectrometry output for BCG Pasteur, highlighting a T allele call for SNP GS34910, which is at BCG genome position 565209, located at the start of the sigK gene. (b) Mass spectrometry output for BCG Russia SNP GS34910, showing a C allele call for this SNP. This SNP gives rise to the differential expression of the SigK regulon between early and late BCG strains.
Phylogenetic analysis.
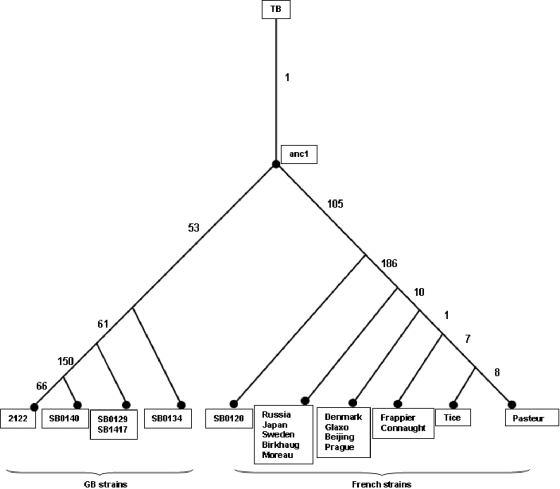
The phylogenetic distribution of SNPs allowed the strains to be clustered into related groupings, showing as expected that M. bovis strains isolated in Britain, those isolated in France, and BCG daughter strains formed three distinct clades (Fig. 3). As the SNPs were selected on comparison of the genomes of BCG Pasteur and M. bovis 2122/97, it should be noted that we generated a “linear” phylogeny, with these two strains being most distant from each other and all other strains falling between them.
FIG. 3.
Distribution of SNPs across British, French, and BCG lineages. A linear phylogeny is shown, where the sequenced strains that were used to derive the SNPs, M. bovis 2122/97 and BCG Pasteur, lie at the extremes. Numbers of SNPs that separate each grouping are shown. Branch lengths are not phylogenetically informative; the figure shows merely the clustering of strains generated by the SNP analysis.
The SNP phylogeny recapitulated BCG genealogies proposed using deletions or duplications (7, 9). Hence, “early” (pre-1927) or “late” (post-1927) BCG strains group together, although the SNPs did not allow the resolution of DU2 groups I and II as defined by Brosch et al. (9). BCG strains Russia, Tokyo, and Moreau (group I) and Sweden and Birkhaug (group II) clustered together. DU2 group III (BCG strains Denmark, Glaxo, Beijing, and Prague) formed a discrete group, with 10 SNPs distinguishing them from groups I and II. One SNP separated BCG Connaught/Frappier from group II, while seven SNPs separated these strains from BCG Tice. Finally, BCG Pasteur had eight strain-specific SNPs. One SNP, a 1-bp insertion at position 842687 (mIns-842687; see Table S1 in the supplemental material), was present in BCG Pasteur and BCG Beijing only. As this SNP is homoplasic, the Sequenom results were verified using standard sequencing and confirmed to be correct. Whether this indicates a selective pressure at this locus is unclear.
BCG Beijing was included in our SNP analysis as this strain had so far, to our knowledge, not been genetically studied. It was known, however, that BCG Beijing was derived from BCG Denmark and shows protective efficacy similar to that of BCG Denmark in animal models (56). The SNP analysis showed, as expected, that it belonged to the DU2-III group with BCG strains Denmark, Prague, and Glaxo. However, comparative genomic hybridization (data not shown) using an M. tuberculosis complex microarray (25) revealed that BCG Beijing had the characteristic RD-Denmark locus intact (41). Hence, it would appear that BCG Beijing was derived from a BCG Danish seed-lot before the RD-Denmark deletion had occurred. Using high-resolution NimbleGen arrays, Leung et al. also recently screened the genomes of BCG Beijing and 12 other BCG strains for InDels and also placed BCG Beijing in the BCG Denmark-derived clade (37).
The M. bovis progenitor attenuated by Calmette and Guérin is not available, having been lost from the Institut Pasteur archives. Therefore, we selected a panel of French M. bovis strains on the basis that they had the same spoligotype as BCG (SB0120) and similar VNTR profiles. Hence, the SNP screen showed minimal variation between the 13 French M. bovis strains. However, they were distinguished from the British strains by 158 SNPs and from BCG by 186 SNPs; we will deal below with the functional implications of these latter 186 SNPs.
The British strains were chosen on the basis of our detailed knowledge of the population structure of M. bovis, with molecular types SB0129, SB0134, and SB0140 representing the major groups of M. bovis in Britain (Fig. 1). In agreement with previous phylogenies of M. bovis based on spoligotypes and VNTR (53, 57), types SB0129 and SB0134 are more closely related to French strains than to the SB0140 clonal complex. The sequenced strain, 2122/97, showed 66 unique SNPs compared to other strains with the same SB0140 pattern.
One hundred fifty-eight SNPs separated all of the French strains from all of the British M. bovis strains. Using BLASTn (3), we determined the status of each of these SNPs in an outgroup, the genome sequence of M. tuberculosis H37Rv, to establish where the SNPs occurred in the evolution of the British and French lineages (Fig. 3). This analysis showed that 53 SNPs had occurred during the evolution of the British M. bovis branch (to SB0134), with 105 SNPs on the French branch. Hence, it appears that the British SB0134 strains are more similar to the common ancestor of the British-French lineage (anc1 in Fig. 3) than to the French strains selected in this study.
Determining the ratio of synonymous and nonsynonymous mutations per site between sequences (dN/dS ratio) can indicate the strength and direction of selection. Purifying selection is normally stronger on nonsynonymous changes, reducing the dN/dS ratio and giving values of <1; in contrast, positive or directional selection would give values for dN/dS of >1. Strains of BCG have been cultured in vitro for long periods, and therefore, dN/dS calculations may reveal differences in selection pressures between in vivo and in vitro growth. Calculations of dN/dS ratios gave values of 0.48 for anc1 (Fig. 3) to strain 2253 (SB0134, 48 SNPs), 0.68 from anc1 to strain F1.2 (SB0120, 87 SNPs), and 0.63 for the branch between the divergence of the F1.2 wild strain and the divergence of BCG Russia (161 SNPs). For such closely related strains, the interpretation of high dN/dS values is complex and the values are open to several interpretations (48). Nevertheless, two conclusions can be reached from our data. First, there appears to be no evidence for increased relaxed selection during the in vitro cultivation of BCG strains compared to in vivo-isolated M. bovis strains (0.63 in vitro versus 0.68 in vivo). Second, the relatively high dN/dS ratios are further evidence for relaxed purifying selection within the M. tuberculosis complex, presumably in response to a low effective population size (53). This latter point has been elegantly demonstrated by Hershberg et al. (29), who performed a multilocus sequencing analysis on a worldwide collection of 108 M. tuberculosis complex strains. The average pairwise dN/dS ratio across these strains for 370 SNPs was 0.57, comparable to the values that we obtained and, they concluded, likely a consequence of reduced selective constraint.
Interestingly, the ugpAEBC region contains SNPs that differentiate the British, French, and BCG lineages. For example, taking the M. bovis 2122/97 genome sequence as the reference, SNP mIns-3095955 is a 1-bp insertion in ugpA that is present in SB0134 and SB0120 M. bovis (i.e., including BCG); this polymorphism is in fact a 1-bp deletion from type SB0129, AN5, and M. bovis 2122/97 that frameshifts ugpA; hence, SNP mIns-3095955 delineates SB0129 and its descendants. Similarly, SNP 3093642 is an nsSNP in ugpB found only in the British SB0140 clonal complex. Finally, SNPs 3092465 and 3093840 are found only in BCG strains (see Table S1 in the supplemental material). Hence, focused sequencing of these SNPs provides a simple way to cluster British, French, and BCG strains. Possible functional implications of these mutations are discussed below.
Functional inferences.
Of the 186 SNPs identified between virulent M. bovis strains and BCG, 115 are nonsynonymous and 55 are synonymous, 13 are intergenic, and three are located in pseudogenes. While synonymous SNPs (sSNPs) may have functional consequences in rare cases (34), nsSNPs, frameshifts, or intergenic SNPs that affect gene expression or protein structure are the most likely source of phenotypic variation.
Virulence.
In a global analysis of genes required for in vivo survival, Sassetti and Rubin identified 194 major candidate genes (50); eight of these genes contain nsSNPs in BCG (Table 2). While many of these nsSNPs encode conservative amino acid substitutions, some may have functional consequences. For example, kdpD encodes the histidine kinase sensor of the two-component system KdpDE that regulates turgor pressure and potassium homeostasis. The BCG kdpD allele contains two nsSNPs, P83S and N776D, the latter of which is a conservative substitution. The N-terminal domain of KdpD contains two Walker nucleotide binding motifs which are important in ATP binding (27). While the P83S substitution does not disrupt these motifs, the P83 residue is conserved across KdpD homologues in many bacterial species (27). Functional analysis of the BCG KdpD protein is therefore warranted.
TABLE 2.
Genes defined as essential in vivo by transposon site hybridization that contain nsSNPs across all BCG strainsc
| Gene | Rv coding sequence | BCG coding sequence | SNP | Function |
|---|---|---|---|---|
| nrp | Rv101 | BCG0134 | L1365M | Nonribosomal peptide synthase |
| senX3 | Rv0490 | BCG0531 | F109S | Two-component sensor |
| kdpD | Rv1028c | BCG1085c | P83S; N776Da | Two-component sensor |
| murI | Rv1338 | BCG1400 | R154L | Glutamate racemase |
| lysX | Rv1640c | BCG1679c | D769Ea | Lysyl-tRNA synthase |
| pks12 | Rv2048c | BCG2067c | S2964R | Mannosyl-β-1-phosphoisoprenoid synthase |
| fadE22 | Rv3061c | BCG3086c | K488Ea,b; S497Cb | Acyl coenzyme A dehydrogenase |
| Rv3335c | Rv3335c | BCG3406c | A86Va | Integral membrane protein |
| Rv3616c | Rv3616c | BCG3680c | A4Va | ESX-1 secreted antigen |
Conservative substitution.
Position with reference to H37Rv protein sequence as M. bovis 2122/97 allele is frameshifted.
Transposon site hybridization was performed as described previously (50).
The SenX3 histidine kinase is also implicated in virulence (44, 50) and contains an F109S mutation; its linked response regulator, the RegX3 protein, also contains an nsSNP (A18T). The RegX3 mutation occurs at an alanine residue which is conserved across many similar two-component regulators. The function of SenX3-RegX3 in M. bovis BCG is unknown, but in Mycobacterium smegmatis SenX-RegX regulates the expression of the high-affinity pstSCAB phosphate uptake system (23, 24). If SenX3-RegX3 also functions in phosphate control in BCG, and as the pst high-affinity system appears to be nonfunctional in BCG, the accumulation of mutations in the senX3-regX3 locus may be indicative of relaxed selection acting at this locus.
It is worth noting that the pks12 gene, which encodes a mannosyl-β-1-phosphoisoprenoid synthase (40), contains 11 nsSNPs between BCG and other M. bovis strains; however, only one of these SNPs, S2964R, is present in all BCG strains but absent from all other M. bovis strains studied. Furthermore, there is no evidence of any structural difference between the mannosyl-β-1-phosphoisoprenoid synthases synthesized by BCG and those synthesized by M. tuberculosis (40), suggesting that this accumulation of nsSNPs in pks12 has no functional effect.
Lesions in metabolism.
The M. bovis genome encodes 17 cytochrome P450 oxidase (CYP) enzymes, with an 18th gene, the CYP142 gene, frameshifted (22). All BCG strains contain a frameshifted CYP123 gene, with an extended C-terminal portion that would be expected to disrupt correct protein folding and function. The role of mycobacterial CYP enzymes is still being elucidated, but they are expected to be integral to lipid metabolism. The function of CYP123 is unknown, but it was found to be upregulated in response to heat stress (55). The genes for three further CYP enzymes, the CYP126, CYP128, and CYP135B1 genes, contain nsSNPs, but their functional consequences are unknown. CYP128 was shown to be essential for in vitro growth of M. tuberculosis (49), so presumably the L203F substitution present in the BCG protein has no major functional effects.
M. bovis and M. bovis BCG strains cannot catabolize alanine due to a frameshift mutation in the aldA gene, which encodes alanine dehydrogenase. Chen and colleagues have also shown that BCG strains exhibit defects in serine metabolism, with BCG Pasteur and Frappier being unable to catabolize serine (12). The genetic basis for this defect in serine catabolism remains unidentified; indeed, sdaA, which encodes serine deaminase, is identical between M. bovis and M. bovis BCG strains, which suggests a regulatory defect (12). Analysis of SNPs that are shared between BCG Frappier and Pasteur did not reveal any obvious candidates that would explain the serine catabolism phenotype.
The M. tuberculosis complex contains three systems for the biosynthesis of trehalose, namely, the OtsAB, TreS, and TreXYZ systems (14, 42). It appears that the OtsAB system is the principal pathway for trehalose biosynthesis in M. tuberculosis; inactivation of the TreXYZ system had no in vitro or in vivo effect on growth (42). The M. bovis 2122/97 strain contains an internal deletion in the treY gene that leads to an inactive protein product (22, 26); however, this mutation is present only in strains that are closely related to 2122/97. It is surprising, therefore, that all BCG strains have an independent mutation in the TreXYZ system, with treZ frameshifted. Hence, the same biosynthetic pathway is mutated in both wild-type virulent M. bovis and in vitro-attenuated BCG. As the TreXYZ pathway is not required for virulence, its loss may simply reflect the removal of biosynthetic redundancy.
Growth on glycerol.
A key metabolic selection placed on the M. bovis progenitor of BCG was the utilization of glycerol as a carbon source. We have previously shown that M. bovis strains contain an nsSNP in the gene encoding pyruvate kinase (pykA) that prevents the conversion of phosphoenolpyruvate to pyruvate and hence blocks glycolysis from feeding into the tricarboxylic acid cycle (22, 33). BCG strains all contain an Asp220Glu mutation that restores activity to pyruvate kinase, a mutation that was selected by the glycerol-based medium used by Calmette and Guérin.
In a chemostat analysis of Escherichia coli strains grown on glycerol, mutations in glpK (encoding glycerol kinase) that increased enzyme efficiency were selected for (28). The glpK gene of M. bovis 2122/97 is frameshifted, while that of BCG is in frame (22, 33). This latter mutation was not, however, selected for during BCG's in vitro growth, as French and related British M. bovis strains have an in-frame glpK; hence, it is merely a mutation in M. bovis 2122/97. The bovine tuberculin production strain, M. bovis AN5, was included in our SNP screen since, like BCG, it is a glycerol-adapted strain of M. bovis (45); SNPs shared between BCG and AN5 may favor growth of M. bovis on glycerol. However, apart from the previously described Asp220Glu mutation in pykA we could identify only one homoplasic SNP shared between BCG and AN5 (SNP 1369616), which was located in the gene for PPE18. However, it appears unlikely that this represents a glycerol-adaptive mutation.
As noted above, the ugpAEBC locus contains a number of SNPs with functional consequences. Hence, ugpA is frameshifted in British type SB0129 and the SB0140 clonal complex because of a 1-bp insertion; the resulting protein will not localize correctly in the membrane and is therefore nonfunctional. All BCG strains have an independent null mutation in the same operon, with ugpB containing an in-frame stop codon, and they also contain an nsSNP in ugpC. Hence, in British and BCG lineages the UgpAEBC transporter is nonfunctional. This is interesting because ugpAEBC encodes a glycerol-3-phosphate transporter that in related actinobacteria is responsive to phosphate starvation (35); hence, BCG contains null mutations in both the high-affinity Pst phosphate uptake system and the Ugp system. This may reflect BCG's in vitro growth conditions but will undoubtedly impair in vivo phosphate acquisition and hence may be implicated in attenuation.
Transcriptional regulators.
Comparison of the transcriptomes of M. bovis 2122/97 and BCG Pasteur revealed that 133 genes showed a minimum twofold difference in expression across the strains (9). It is probable that some of these expression differences reflect differences between the United Kingdom and French clades of M. bovis rather than a difference between virulent and attenuated strains. However, the genes for three transcriptional regulators, BCG3734, BCG3145, and BCG2507c, show nsSNPs across all BCG strains compared to virulent M. bovis strains, mutations which may explain global expression differences between M. bovis BCG and M. bovis.
BCG3734 encodes the CRP, a global gene regulator. Previous work has shown that there are two nsSNPs in BCG3734 compared to the M. bovis and M. tuberculosis genes, which encode E178K and L47P substitutions (54). These mutations have been shown to enhance the binding of BCG CRP to its DNA binding sites (4, 30); however, this enhanced binding does not appear to play a role in attenuation of BCG (30).
The LuxR family regulator BCG2507c contains an N-terminal adenylate/guanylate cyclase catalytic domain, a putative ATPase domain (COG3903 superfamily), and a C-terminal helix-turn-helix domain. The D535E mutation in BCG2507c falls in the ATPase domain; as it is a conservative substitution, its functional consequences are expected to be minimal.
BCG3145 is a member of the AfsR/DnrI/SARP (Streptomyces antibiotic regulatory protein) class of transcriptional regulators. This class also contains EmbR, the regulator of three arabinosyltransferases that are the targets of the front-line tuberculosis drug ethambutol (8). The structure of EmbR has been elucidated, revealing DNA binding, bacterial transcriptional activation (BTA), and forkhead-associated domains (1). While BCG3145 lacks the forkhead-associated domain, the E159G mutation in BCG3145 mutates to glycine a conserved glutamic acid residue located in a tetratricopeptide repeat in the BTA domain (region T3). Tetratricopeptide repeat domains are associated with protein-protein interactions (16), and a conserved core (helices T1 to T7) of the BTA domain seems to be required for proper function of SARP family proteins (1, 51). Hence, the E159G mutation may affect the ability of BCG3145 to regulate transcription.
Cycloserine resistance.
Growth on d-cycloserine can be used to differentiate M. bovis from M. bovis BCG strains, with M. bovis being sensitive to 0.02 mg/ml cycloserine while M. bovis BCG strains are resistant. The molecular basis for this phenotype is unknown. The emergence of cycloserine resistance is usually due to mutations in the alrA gene encoding alanine racemase or the ddlA gene encoding d-alanyl-d-alanine ligase (20); however, the sequences of alrA and ddlA in M. bovis BCG and other M. bovis strains are identical, as are their expression levels (9). The other possibility is that the cycloserine transporter CycA (which also transports d-serine, d-alanine, and glycine) is defective for cycloserine transport in BCG. CycA does in fact contain an nsSNP across all BCG strains, introducing a G122S mutation. It is noteworthy that in all sequenced mycobacterial CycA proteins this position is occupied by glycine or sometimes, in other bacteria, by alanine. David showed that resistance to cycloserine in M. tuberculosis can be due to defective transport of the antibiotic (17), and so it is tempting to speculate that the G122S mutation plays some role in the inherent cycloserine resistance of BCG. This would also suggest that BCG may be defective in d-alanine, glycine, and d-serine transport, but this has not been reported.
DISCUSSION
Phylogenetic relationships based on SNPs identified by comparing two sequenced strains will inevitably place these latter strains as most distant in any phylogeny (2, 58). Phylogenies with these characteristics can therefore be deemed “preselected” or “linear” phylogenies. This is evident in our phylogeny (Fig. 3). However, this does not invalidate the resulting tree; our prior knowledge of the population structure of M. bovis allowed us to select strains which encompass the diversity within the population and to address our primary question of which SNPs were acquired during the initial derivation of BCG. Indeed, the SNP phylogeny is congruent with trees constructed using spoligotyping and deletion typing (52, 53). The one divergence from previous trees is the positioning of BCG Tice and Frappier; BCG Tice and Pasteur shared six SNPs not seen in Frappier, hence placing Tice closer to BCG Pasteur. In a previous schema BCG Frappier had been positioned closer to BCG Pasteur than Tice (9). However, Rosenthal is known to have mixed BCG Pasteur with BCG Tice in the early 1950s to correct for overattenuation (19). From the SNP data presented here, it appears that this mixture of BCG Tice and BCG Pasteur resolved as a variant of BCG Pasteur. In a recent DNA array-based comparative study of BCG vaccine strains, Leung et al. note that BCG Tice has a distinctive duplication (DU-Tice), providing a further marker to differentiate BCG Tice (37).
We describe a range of SNPs with putative functional effects in this study. While it is straightforward to ascribe functional effects to SNPs that, for example, frameshift genes, predicting the impact of intergenic SNPs or nsSNPs is problematic. We have therefore been conservative in our interpretation of the SNP data and instead see them as the basis for focused experimental validation. Furthermore, we are mindful that 35 SNP detection reactions did not generate usable data, as is common in high-throughput projects. Hence, it is possible that informative SNPs are missing from the data set presented here, but future scans will revisit these SNPs.
What are the implications of this research for the BCG vaccine? The fact that BCG daughter strains are genetically distinct is evident from our study and previous work; however, how these genetic differences translate into variation in vaccine efficacy requires considerable functional analyses. Indeed, clarity is first needed on the question of the efficacy of BCG substrains. In a recent review of published data on the efficacy of different BCG strains in human and animal studies, Ritz and colleagues showed that BCG Pasteur, Denmark, and Glaxo were associated with better protection against challenge in the mouse model while BCG Denmark appeared the best in guinea pig models (47). However, standardization of animal protocols for such variables as the dose and route of vaccine administration, interval between immunization and challenge, and dose and route of challenge strain(s) is needed if true head-to-head comparisons of BCG daughter strains are to be made. Similarly, results from human studies suggest that there are distinct differences in immunogenicity across BCG strains, although the lack of standardization across trials again complicates interpretation. For example, in a study of neonatal vaccination in Mexico using BCG strains Brazil, Denmark, and Japan, it appeared that BCG Japan was the least immunogenic (59); however, in a South African study of neonatal vaccination using BCG strains Japan and Denmark, BCG Japan was more immunogenic than BCG Denmark (18).
Against this background it is difficult to connect genetic differences in BCG strains to vaccine efficacy. However, this is not to say that linkages cannot be made, once clear phenotypes are defined. An elegant example of the link between BCG genotype and phenotype was provided by Leung et al. (37), who disclosed a deletion in the BCG Moreau strain that explained the lack of phenolic glycolipid and phthiocerol dimycocerosates from this strain (13). As these latter lipids are potent immunomodulators, their absence from BCG Moreau may explain the lack of BCG-associated complications reported with this strain (13). Hence, the SNP differences defined herein will provide a rich source of genetic variation that can be mapped to functional differences across BCG strains.
Using high-throughput SNP screening, we have therefore identified SNPs that refine the phylogeny of British and French M. bovis strains, confirm previous genealogies of BCG vaccines, and define a minimal set of SNPs between virulent M. bovis strains and the attenuated BCG. These SNPs will therefore facilitate future studies of M. bovis phylogeography and the molecular basis for the attenuation of BCG.
Supplementary Material
Acknowledgments
This work was funded by the Department of Environment, Food and Rural Affairs (project SE3224); the Fondation Raoul Follereau; the Wellcome Trust; and SystemsX.ch. Xiangmei Zhou was funded by BBSRC grant CPA1497, “Genomic and Post-genomics of Salmonella and Mycobacterium as paradigms of intracellular pathogens,” awarded to Paul Barrow, University of Nottingham, United Kingdom.
We also thank one anonymous referee for helpful comments.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 16 March 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alderwick, L. J., V. Molle, L. Kremer, A. J. Cozzone, T. R. Dafforn, G. S. Besra, and K. Futterer. 2006. Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 1032558-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbon, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 1853392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bai, G., M. A. Gazdik, D. D. Schaak, and K. A. McDonough. 2007. The Mycobacterium bovis BCG cyclic AMP receptor-like protein is a functional DNA binding protein in vitro and in vivo, but its activity differs from that of its M. tuberculosis ortholog, Rv3676. Infect. Immun. 755509-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behr, M. A., B. G. Schroeder, J. N. Brinkman, R. A. Slayden, and C. E. Barry III. 2000. A point mutation in the mma3 gene is responsible for impaired methoxymycolic acid production in Mycobacterium bovis BCG strains obtained after 1927. J. Bacteriol. 1823394-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behr, M. A., and P. M. Small. 1997. Has BCG attenuated to impotence? Nature 389133-134. [DOI] [PubMed] [Google Scholar]
- 7.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 2841520-1523. [DOI] [PubMed] [Google Scholar]
- 8.Belanger, A. E., G. S. Besra, M. E. Ford, K. Mikusova, J. T. Belisle, P. J. Brennan, and J. M. Inamine. 1996. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. USA 9311919-11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosch, R., S. V. Gordon, T. Garnier, K. Eiglmeier, W. Frigui, P. Valenti, S. Dos Santos, S. Duthoy, C. Lacroix, C. Garcia-Pelayo, J. K. Inwald, P. Golby, J. N. Garcia, R. G. Hewinson, M. A. Behr, M. A. Quail, C. Churcher, B. G. Barrell, J. Parkhill, and S. T. Cole. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. USA 1045596-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calmette, A., and C. Guérin. 1909. Sur quelques propriétés du bacille tuberculeux d'origine, cultivé sur la bile de boeuf glycérinée. C. R. Acad. Sci. Paris 149716-718. [Google Scholar]
- 11.Charlet, D., S. Mostowy, D. Alexander, L. Sit, H. G. Wiker, and M. A. Behr. 2005. Reduced expression of antigenic proteins MPB70 and MPB83 in Mycobacterium bovis BCG strains due to a start codon mutation in sigK. Mol. Microbiol. 561302-1313. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J. M., D. C. Alexander, M. A. Behr, and J. Liu. 2003. Mycobacterium bovis BCG vaccines exhibit defects in alanine and serine catabolism. Infect. Immun. 71708-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, J. M., S. T. Islam, H. Ren, and J. Liu. 2007. Differential productions of lipid virulence factors among BCG vaccine strains and implications on BCG safety. Vaccine 258114-8122. [DOI] [PubMed] [Google Scholar]
- 14.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 15.Collins, D. M., R. P. Kawakami, B. M. Buddle, B. J. Wards, and G. W. de Lisle. 2003. Different susceptibility of two animal species infected with isogenic mutants of Mycobacterium bovis identifies phoT as having roles in tuberculosis virulence and phosphate transport. Microbiology 1493203-3212. [DOI] [PubMed] [Google Scholar]
- 16.D'Andrea, L. D., and L. Regan. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28655-662. [DOI] [PubMed] [Google Scholar]
- 17.David, H. L. 1971. Resistance to d-cycloserine in the tubercle bacilli: mutation rate and transport of alanine in parental cells and drug-resistant mutants. Appl. Microbiol. 21888-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davids, V., W. A. Hanekom, N. Mansoor, H. Gamieldien, S. J. Gelderbloem, A. Hawkridge, G. D. Hussey, E. J. Hughes, J. Soler, R. A. Murray, S. R. Ress, and G. Kaplan. 2006. The effect of bacille Calmette-Guerin vaccine strain and route of administration on induced immune responses in vaccinated infants. J. Infect. Dis. 193531-536. [DOI] [PubMed] [Google Scholar]
- 19.Dubos, R. J., and C. H. Pierce. 1956. Differential characteristics in vitro and in vivo of several substrains of BCG. I. Multiplication and survival in vitro. Am. Rev. Tuberc. 74655-666. [DOI] [PubMed] [Google Scholar]
- 20.Feng, Z., and R. G. Barletta. 2003. Roles of Mycobacterium smegmatis d-alanine:d-alanine ligase and d-alanine racemase in the mechanisms of action of and resistance to the peptidoglycan inhibitor d-cycloserine. Antimicrob. Agents Chemother. 47283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 1441189-1196. [DOI] [PubMed] [Google Scholar]
- 22.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 1007877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebhard, S., S. L. Tran, and G. M. Cook. 2006. The Phn system of Mycobacterium smegmatis: a second high-affinity ABC-transporter for phosphate. Microbiology 1523453-3465. [DOI] [PubMed] [Google Scholar]
- 24.Glover, R. T., J. Kriakov, S. J. Garforth, A. D. Baughn, and W. R. Jacobs, Jr. 2007. The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J. Bacteriol. 1895495-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golby, P., K. A. Hatch, J. Bacon, R. Cooney, P. Riley, J. Allnutt, J. Hinds, J. Nunez, P. D. Marsh, R. G. Hewinson, and S. V. Gordon. 2007. Comparative transcriptomics reveals key gene expression differences between the human and bovine pathogens of the Mycobacterium tuberculosis complex. Microbiology 1533323-3336. [DOI] [PubMed] [Google Scholar]
- 26.Gordon, S. V., K. Eiglmeier, T. Garnier, R. Brosch, J. Parkhill, B. Barrell, S. T. Cole, and R. G. Hewinson. 2001. Genomics of Mycobacterium bovis. Tuberculosis (Edinburgh) 81157-163. [DOI] [PubMed] [Google Scholar]
- 27.Heermann, R., K. Altendorf, and K. Jung. 2003. The N-terminal input domain of the sensor kinase KdpD of Escherichia coli stabilizes the interaction between the cognate response regulator KdpE and the corresponding DNA-binding site. J. Biol. Chem. 27851277-51284. [DOI] [PubMed] [Google Scholar]
- 28.Herring, C. D., A. Raghunathan, C. Honisch, T. Patel, M. K. Applebee, A. R. Joyce, T. J. Albert, F. R. Blattner, D. van den Boom, C. R. Cantor, and B. O. Palsson. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 381406-1412. [DOI] [PubMed] [Google Scholar]
- 29.Hershberg, R., M. Lipatov, P. M. Small, H. Sheffer, S. Niemann, S. Homolka, J. C. Roach, K. Kremer, D. A. Petrov, M. W. Feldman, and S. Gagneux. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt, D. M., J. W. Saldanha, J. F. Brennan, P. Benjamin, M. Strom, J. A. Cole, C. L. Spreadbury, and R. S. Buxton. 2008. Single nucleotide polymorphisms that cause structural changes in the cyclic AMP receptor protein transcriptional regulator of the tuberculosis vaccine strain Mycobacterium bovis BCG alter global gene expression without attenuating growth. Infect. Immun. 762227-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics (Oxford) 171230-1231. [DOI] [PubMed] [Google Scholar]
- 32.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keating, L. A., P. R. Wheeler, H. Mansoor, J. K. Inwald, J. Dale, R. G. Hewinson, and S. V. Gordon. 2005. The pyruvate requirement of some members of the Mycobacterium tuberculosis complex is due to an inactive pyruvate kinase: implications for in vivo growth. Mol. Microbiol. 56163-174. [DOI] [PubMed] [Google Scholar]
- 34.Kimchi-Sarfaty, C., J. M. Oh, I. W. Kim, Z. E. Sauna, A. M. Calcagno, S. V. Ambudkar, and M. M. Gottesman. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315525-528. [DOI] [PubMed] [Google Scholar]
- 35.Kocan, M., S. Schaffer, T. Ishige, U. Sorger-Herrmann, V. F. Wendisch, and M. Bott. 2006. Two-component systems of Corynebacterium glutamicum: deletion analysis and involvement of the PhoS-PhoR system in the phosphate starvation response. J. Bacteriol. 188724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagranderie, M. R., A. M. Balazuc, E. Deriaud, C. D. Leclerc, and M. Gheorghiu. 1996. Comparison of immune responses of mice immunized with five different Mycobacterium bovis BCG vaccine strains. Infect. Immun. 641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung, A. S., V. Tran, Z. Wu, X. Yu, D. C. Alexander, G. F. Gao, B. Zhu, and J. Liu. 2008. Novel genome polymorphisms in BCG vaccine strains and impact on efficacy. BMC Genomics 9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis, K. N., R. Liao, K. M. Guinn, M. J. Hickey, S. Smith, M. A. Behr, and D. R. Sherman. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J. Infect. Dis. 187117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 1781274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsunaga, I., A. Bhatt, D. C. Young, T. Y. Cheng, S. J. Eyles, G. S. Besra, V. Briken, S. A. Porcelli, C. E. Costello, W. R. Jacobs, Jr., and D. B. Moody. 2004. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J. Exp. Med. 2001559-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mostowy, S., A. G. Tsolaki, P. M. Small, and M. A. Behr. 2003. The in vitro evolution of BCG vaccines. Vaccine 214270-4274. [DOI] [PubMed] [Google Scholar]
- 42.Murphy, H. N., G. R. Stewart, V. V. Mischenko, A. S. Apt, R. Harris, M. S. McAlister, P. C. Driscoll, D. B. Young, and B. D. Robertson. 2005. The OtsAB pathway is essential for trehalose biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 28014524-14529. [DOI] [PubMed] [Google Scholar]
- 43.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3418-426. [DOI] [PubMed] [Google Scholar]
- 44.Parish, T., D. A. Smith, G. Roberts, J. Betts, and N. G. Stoker. 2003. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 1491423-1435. [DOI] [PubMed] [Google Scholar]
- 45.Paterson, A. B. 1948. The production of bovine tuberculoprotein. J. Comp. Pathol. 58302-313. [DOI] [PubMed] [Google Scholar]
- 46.Pym, A. S., P. Brodin, R. Brosch, M. Huerre, and S. T. Cole. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46709-717. [DOI] [PubMed] [Google Scholar]
- 47.Ritz, N., W. A. Hanekom, R. Robins-Browne, W. J. Britton, and N. Curtis. 2008. Influence of BCG vaccine strain on the immune response and protection against tuberculosis. FEMS Microbiol. Rev. 32821-841. [DOI] [PubMed] [Google Scholar]
- 48.Rocha, E. P., J. M. Smith, L. D. Hurst, M. T. Holden, J. E. Cooper, N. H. Smith, and E. J. Feil. 2006. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J. Theor. Biol. 239226-235. [DOI] [PubMed] [Google Scholar]
- 49.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 4877-84. [DOI] [PubMed] [Google Scholar]
- 50.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 10012989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheldon, P. J., S. B. Busarow, and C. R. Hutchinson. 2002. Mapping the DNA-binding domain and target sequences of the Streptomyces peucetius daunorubicin biosynthesis regulatory protein, DnrI. Mol. Microbiol. 44449-460. [DOI] [PubMed] [Google Scholar]
- 52.Smith, N. H., J. Dale, J. Inwald, S. Palmer, S. V. Gordon, R. G. Hewinson, and J. M. Smith. 2003. The population structure of Mycobacterium bovis in Great Britain: clonal expansion. Proc. Natl. Acad. Sci. USA 10015271-15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, N. H., S. V. Gordon, R. de la Rua-Domenech, R. Clifton-Hadley, and R. G. Hewinson. 2006. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat. Rev. Microbiol. 4670-681. [DOI] [PubMed] [Google Scholar]
- 54.Spreadbury, C. L., M. J. Pallen, T. Overton, M. A. Behr, S. Mostowy, S. Spiro, S. J. Busby, and J. A. Cole. 2005. Point mutations in the DNA- and cNMP-binding domains of the homologue of the cAMP receptor protein (CRP) in Mycobacterium bovis BCG: implications for the inactivation of a global regulator and strain attenuation. Microbiology 151547-556. [DOI] [PubMed] [Google Scholar]
- 55.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 1483129-3138. [DOI] [PubMed] [Google Scholar]
- 56.Wang, G. Z., V. Balasubramanian, and D. W. Smith. 1988. The protective and allergenic potency of four BCG substrains in use in China determined in two animal models. Tubercle 69283-291. [DOI] [PubMed] [Google Scholar]
- 57.Winder, C., S. V. Gordon, J. Dale, R. G. Hewinson, and R. Goodacre. 2006. Metabolic fingerprints of Mycobacterium bovis cluster with molecular type: implications for genotype-phenotype links. Microbiology 1522757-2765. [DOI] [PubMed] [Google Scholar]
- 58.Worobey, M. 2005. Anthrax and the art of war (against ascertainment bias). Heredity 94459-460. [DOI] [PubMed] [Google Scholar]
- 59.Wu, B., C. Huang, L. Garcia, A. Ponce de Leon, J. S. Osornio, M. Bobadilla-del-Valle, L. Ferreira, S. Canizales, P. Small, M. Kato-Maeda, A. M. Krensky, and C. Clayberger. 2007. Unique gene expression profiles in infants vaccinated with different strains of Mycobacterium bovis bacille Calmette-Guerin. Infect. Immun. 753658-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.