Abstract
Ehrlichia chaffeensis is an obligately intracellular bacterium that exhibits tropism for mononuclear phagocytes forming cytoplasmic membrane-bound microcolonies called morulae. To survive and replicate within phagocytes, E. chaffeensis exploits the host cell by modulating a number of host cell processes, but the ehrlichial effector proteins involved are unknown. In this study, we determined that p47, a secreted, differentially expressed, tandem repeat (TR) protein, interacts with multiple host proteins associated with cell signaling, transcriptional regulation, and vesicle trafficking. Yeast two-hybrid analysis revealed that p47 interacts with polycomb group ring finger 5 (PCGF5) protein, Src protein tyrosine kinase FYN (FYN), protein tyrosine phosphatase non-receptor type 2 (PTPN2), and adenylate cyclase-associated protein 1 (CAP1). p47 interaction with these proteins was further confirmed by coimmunoprecipitation assays and colocalization in HeLa cells transfected with p47-green fluorescent fusion protein (AcGFP1-p47). Moreover, confocal microscopy demonstrated p47-expressing dense-cored (DC) ehrlichiae colocalized with PCGF5, FYN, PTPN2, and CAP1. An amino-terminally truncated form of p47 containing TRs interacted only with PCGF5 and not with FYN, PTPN2, and CAP1, indicating differences in p47 domains that are involved in these interactions. These results demonstrate that p47 is involved in a complex network of interactions involving numerous host cell proteins. Furthermore, this study provides a new insight into the molecular and functional distinction of DC ehrlichiae, as well as the effector proteins involved in facilitating ehrlichial survival in mononuclear phagocytes.
Human monocytotropic ehrlichiosis is an emerging life-threatening tick-borne zoonosis caused by the obligately intracellular gram-negative bacterium Ehrlichia chaffeensis. E. chaffeensis exhibits tropism for mononuclear phagocytes, replicates within cytoplasmic vacuoles that have early endosomal characteristics, and survives by evading and/or suppressing the activation of innate host defenses (4, 22, 23). Escape of phagocyte killing involves modulation of numerous host cell processes, but the ehrlichial effector proteins involved in the cellular reprogramming strategy to create a permissive host are currently undefined.
E. chaffeensis has two morphologically characterized types: a small dense-cored (DC) form characterized by a dense nucleoid and a large replicating form, the reticulate cell (RC), that has uniformly dispersed nucleoid filaments (33). DC ehrlichiae attach and enter the host cell, undergoing rapid transformation to the RC that replicates and matures to the DC form within 3 days (33, 51). The molecular characteristics that distinguish DC from RC forms are not well defined; however, differential expression of two well-characterized immunoreactive tandem repeat (TR) proteins, p120 and p47, on the surface of the DC cells and extracellularly within the ehrlichial endocytic vacuole has been demonstrated (12, 34).
Some of the molecularly characterized major immunoreactive proteins of E. chaffeensis include p47, p120, p200, and variable-length PCR target protein (12, 26, 31, 49). Three of these proteins (p120, p47, and variable-length PCR target protein) contain TRs, are strongly acidic (pI 4 to 5), exhibit high serine/threonine content, contain predicted sites for posttranslational modifications (glycosylation/phosphorylation), and are secreted, suggesting that they are involved in host interactions. In addition, major B-cell epitopes have been identified within the TRs in these proteins (12, 26, 49). Orthologs of E. chaffeensis p47 have been identified, including immunoreactive TR proteins p36 and mucin-like protein (Erum1110) of Ehrlichia canis and Ehrlichia ruminantium, respectively (12).
Entry of E. chaffeensis involves interaction between the pathogen and host that induces cellular signaling events including protein cross-linking by transglutaminase, tyrosine phosphorylation, and phospholipase C-γ2 (PLC-γ2) activation leading to increased levels of inositol 1,4,5-triphosphate (IP3), and cytosolic free calcium (25). Intracellular survival and proliferation of E. chaffeensis involve modulation of gene transcription, activation, and suppression of tyrosine and mitogen-activated protein kinase (MAPK) activity, downregulation of Toll-like receptors and transcription factors, inhibition of apoptosis, lysosomal fusion, and endosomal maturation, and upregulation of transferrin receptor gene expression in the phagocyte (3, 21-25, 52). Antiehrlichial activity of gamma interferon (IFN-γ) is also inhibited by blocking of tyrosine phosphorylation of Janus kinase (Jak) and signal transducer and activator of transcription (Stat) signaling by E. chaffeensis (22). However, the ehrlichial proteins involved in facilitation of entry, inhibition of apoptosis, and suppression and inhibition of cellular defense mechanisms have not been defined.
To further investigate the role of E. chaffeensis TR proteins in pathobiology, the objective of this study was to identify molecular E. chaffeensis p47-host interactions. We hypothesized that p47 is an ehrlichial effector protein that interacts with multiple host cell proteins essential for cellular entry and survival. In this study, we have identified multiple host proteins with distinct molecular functions that interact with p47, suggesting that it plays an important and complex role in reprogramming host cell processes to create a hospitable environment for ehrlichial survival.
MATERIALS AND METHODS
Cell culture and cultivation of E. chaffeensis.
E. chaffeensis (Arkansas strain) was cultivated in human monocyte leukemia cells (THP-1). THP-1 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 1% HEPES buffer (Sigma Chemical, St. Louis, MO), 1% sodium pyruvate (Sigma) at 37°C in a humidified 5% CO2 atmosphere. E. chaffeensis-infected cells were maintained in 2 to 5% serum-containing medium supplemented with 1% HEPES buffer and 1% sodium pyruvate at 37°C in a humidified 5% CO2 atmosphere. The level of ehrlichial infection was assessed by Diff-Quick staining. For transfection experiments, HeLa cells were propagated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (HyClone) and 1% antibiotic/antimycotic solution (Invitrogen).
Antibodies.
Rabbit anti-p47 antibody was prepared against keyhole limplet hemocyanin-conjugated 19-mer TR peptide (ASVSEGDAVVNAVSQETPA), and rabbit anti-PCGF5 antibody was prepared against a keyhole limpet hemocyanin-conjugated 19-mer peptide (PKVDEEGDENEDDKDYHRS) by commercial vendors (Biosynthesis, Lewisville, TX, and GenScript, Piscataway, NJ, respectively). Other antibodies used in this study were rabbit anti-human protein tyrosine phosphatase, non-receptor type 2 (PTPN2; Proteintech Group, Chicago, IL), rabbit anti-human FYN (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-human FYN (p59 FYN; Sigma Chemical, St. Louis, MO), mouse anti-adenylate cyclase-associated protein 1 (CAP1) (Abnova, Walnut, CA), rabbit anti-green fluorescent protein (GFP), and mouse anti-GAL4 DNA-binding domain (GAL4-BD) tag (Clonetech, Mountain View, CA).
Plasmid construction.
To examine E. chaffeensis p47 interactions, a bait plasmid was constructed by fusing full-length p47 with GAL4-BD in pGBKT7 (Clontech, Mountain View, CA). The coding region of p47 was amplified by PCR from E. chaffeensis genomic DNA using a forward primer that included a 5′ EcoRI site (5′-GGCGAATTCATGCAGGTTGATGTTGG) and reverse primer with a 5′ SalI site (5′-GGCGTCGACCTATTACCAAATTGAGCAGC) (restriction enzyme sites in boldface), and ligated into the complementary sites of pGBKT7 plasmid containing the GAL4-BD. The resulting bait plasmid pGBKT7-p47 was transformed into yeast (Saccharomyces cerevisiae) strain AH109 (Clontech) and selected by growth on minimal synthetically defined medium for yeast (SD) lacking l-tryptophan (SD/−Trp). The expression of bait protein p47 was examined by Western blotting using the lysates from pGBKT7-p47-transformed yeast strain AH109. A strategy similar to that described above for the full-length p47 was used to generate amino (N)-terminal (p471-380) and carboxy (C)-terminal (p47361-842) E. chaffeensis p47 fragments. E. chaffeensis genomic DNA was amplified with a forward primer containing a 5′ EcoRI site (5′-GGCGAATTCATGCAGGTTGATGTTGG) and reverse primer with a 5′ SalI site (5′-GGCGTCGACGCATTTCCTTCAAGAACTG) for the N-terminal region of p47 and a forward primer containing a 5′ EcoRI site (5′-GGCGAATTCCCAGTTCTTGAAGGAAATG) and a reverse primer with a 5′ SalI site (5′-GGCGTCGACCTATTACCAAATTGAGCAGC) for the C-terminal region of p47. Each PCR product was cloned into the EcoRI and SalI sites of pGBKT7 containing the GAL4-BD.
p47 expression in yeast.
To examine p47 expression in yeast, a single isolated pGBKT7-p47-transformed colony was grown overnight in 5 ml SD/−Trp medium until the optical density at 600 nm (OD600) reached 0.6. The cells were centrifuged at 1,000 × g for 5 min at 4°C, and the cell pellet was resuspended in 100 μl prewarmed cracking buffer (8 M urea, 5% sodium dodecyl sulfate [SDS], 40 mM Tris-HCl [pH 6.8], 0.1 mM EDTA, 0.4 mg/ml bromophenol blue, 1% β-mercaptoethanol, 50 mM phenylmethylsulfonyl fluoride, 6.2 μg/ml pepstatin A, 23.1 μg/ml aprotinin, 1.9 μM leupeptin, 9 mM benzamidine) per 7.5 OD600 units of cells. The cell suspension was transferred to a 1.5-ml microcentrifuge tube containing 80 μl of glass beads (425 to 600 μm; Sigma). Samples were heated at 70°C for 10 min, vortexed vigorously for 1 min, and centrifuged at 14,000 rpm for 5 min at 4°C. Supernatant was boiled for 3 to 5 min, and SDS-polyacrylamide gel electrophoresis and Western immunoblotting were performed as previously described (28). Briefly, yeast extracts were resolved on 4 to 12% Bis-Tris gels and transferred to a nitrocellulose membrane. Immunoblot detection of expressed recombinant p47 (pGBKT7-p47; GAL4-BD fused to p47) protein in yeast was performed with mouse anti-GAL4-BD antibody (1:2,000). Bound primary antibodies were detected with alkaline phosphatase-conjugated anti-mouse immunoglobulin G [IgG(H+L)] secondary antibody (Kirkegaard & Perry Laboratories, Gaithersburg, MD) and visualized after incubation with BCIP/NBT (5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium) substrate.
Yeast two-hybrid assays.
Yeast two-hybrid screening was performed using Matchmaker Two-Hybrid System 3 according to the manufacturer's protocol (Clontech). The pretransformed human bone marrow Matchmaker cDNA library was constructed in pGADT7, fused with GAL4-activation domain (GAL4-AD), and then transformed in S. cerevisiae strain Y187 (Clontech) and selected with SD/−Leu. Library clones expressing interacting prey proteins were screened by yeast mating. Positive clones were selected on SD quadruple-dropout (QDO) medium (SD/−Ade/−His/−Leu/−Trp) and confirmed on QDO medium supplemented with 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal) (Clontech). QDO-positive clones were then isolated, and the prey plasmids were sequenced in the UTMB Protein Chemistry Laboratory.
Confirmation of positive interactions and prey plasmid rescue.
To confirm positive interactions, the library plasmids responsible for activation of reporters were rescued and reisolated two or three times on SD double-dropout (DDO) medium (SD/−Leu/−Trp) containing X-α-Gal. The segregated library plasmid was rescued from the positive clones grown on QDO using the Yeastmaker yeast plasmid isolation kit (Clontech). The prey plasmid was isolated from transformed Escherichia coli cells selected on LB plus 100 μg/ml ampicillin. To distinguish positive from false-positive interactions, competent S. cerevisiae strain AH109 cells were cotransformed with the bait plasmid (pGBKT7-p47) and the prey plasmid. In another set of experiments, competent AH109 cells were cotransformed with empty bait plasmid (pGBKT7-empty) and the prey plasmid. The positive interactions were confirmed by selection on DDO and QDO media supplemented with X-α-Gal after 3 to 5 days of incubation. The competent AH109 cells were cotransformed with empty bait (pGBKT7-empty) and the prey plasmid and were selected on DDO and QDO media.
Transfection and immunoprecipitation.
Two mammalian expression vectors, pAcGFP1-C and pProLabel-C, were used for generation of N-terminal-tagged AcGFP1-bait and ProLabel prey fusion proteins. The mammalian expression vector pAcGFP1 encodes GFP, while the pProLabel mammalian expression vector encodes the ProLabel tag (∼6 kDa; β-galactosidase). The bait p47, Nterm p47, or Cterm p47 was cloned in-frame downstream of the AcGFP1 coding sequence (at SalI/HindIII) and expressed as a C-terminal AcGFP1 fusion protein. The prey gene was cloned into SalI/BamHI-linearized pProLabel vector in-frame downstream of the ProLabel tag and expressed as a C-terminal ProLabel tag fusion protein. Top10 E. coli (Invitrogen, Carlsbad, CA) or Fusion blue (Clontech) chemically competent cells were transformed according to the manufacturer's protocol, and positive transformants were screened by PCR, restriction digestion, and DNA sequencing. AcGFP1-p47, AcGFP1-Nterm p47, or AcGFP1-Cterm p47 and ProLabel-tagged prey fusion proteins were coexpressed in mammalian cells (HeLa cells), and interacting proteins were immunoprecipitated with the Matchmaker chemiluminescent coimmunoprecipitation (Co-IP) kit according to the manufacturer's protocol (Clontech). Briefly, HeLa cells were contransfected using Lipofectamine (Invitrogen) with pAcGFP1-p47, pAcGFP1-Nterm p47, or pAcGFP1-Cterm p47 and pProLabel-tagged prey fusion constructs, and posttransfection expression of GFP was confirmed after 48 h. The cells were washed twice with 1× phosphate-buffered saline (PBS), and the cell pellet was collected and resuspended in cell lysis buffer (2 × 106 cells/ml) including phenylmethylsulfonyl fluoride and a cocktail of protease inhibitors (Pierce, Rockford, IL) and incubated in ice for 30 min. The cell lysate was centrifuged at 10,000 × g for 20 min at 4°C, the supernatant was collected, and the protein concentration was determined by the bicinchoninic acid method (Pierce). Anti-AcGFP1 polyclonal antibody was added to 500 μg/ml of lysate and incubated for 2 h at 4°C. After the antibody incubation, lysate was added to washed protein A/G agarose beads at a 1:20 volume of 50% bead slurry and the mixture was incubated for overnight at 4°C. Beads were collected by centrifugation (5,000 × g for 10 s), washed 5× with wash buffer 1, and then 4× with wash buffer 2. For ProLabel detection of protein-protein interaction, each sample of beads was resuspended in 80 μl of lysis/complementation buffer, and the entire content of beads and buffer was transferred to a well in a 96-well assay plate (Costar, Corning, NY). To each well, 30 μl of substrate mix was added and ProLabel activity measured using the Veritas microplate luminometer (Turner Biosystems, Sunnyvale, CA) at 0-, 10-, 15-, 20-, 25-, 30-, 45-, 60-, 90-, and 120-min intervals.
Confocal microscopy.
Untransfected and transfected (pAcGFP1-p47 and pAcGFP1-empty) HeLa cells collected 2 days posttransfection were washed twice with 1× PBS and fixed in 3.7% paraformaldehyde at room temperature for 15 min. Cells were permeabilized and blocked with a mixture of 0.1% Triton X-100 and 1% bovine serum albumin (Sigma) in 1× PBS for 1 h at room temperature. Uninfected and E. chaffeensis-infected THP-1 cells (2 to 3 days postinfection) were cytocentrifuged onto glass slides, fixed in 3.7% paraformaldehyde for 20 min, and permeabilized and blocked with a mixture of 1% Triton X-100 and 5% bovine serum albumin in 1× PBS for 1 h at room temperature. Cells were stained with anti-PCGF5, anti-FYN, anti-PTPN2 (1:50), or anti-CAP1 (1:100) antibodies for 1 h at room temperature. Slides were washed and incubated with Alexa Fluor 568 goat anti-rabbit or goat anti-mouse IgG(H+L) secondary antibodies (1:100) (Molecular Probes, Eugene, OR) for 30 min. Slides were then washed and mounted with ProLong antifade (Invitrogen), and fluorescent images were obtained at the Optical Imaging Laboratory at UTMB using a Zeiss LSM 510 META laser-scanning confocal microscope (Germany). The confocal images were further analyzed by use of fluorescence intensity profiles. A fluorescence intensity profile represents the pixel values in a digitized section along a user-defined area, displayed in a diagram. The electrical signal from the detector was digitized to a numerical value between 0 and 250 arbitrary units.
Statistics.
Statistical difference between control and experimental groups were assessed with the two-tailed Student's t test. Significance is indicated by a P value of <0.05.
RESULTS
The E. chaffeensis TR unit contains a common motif.
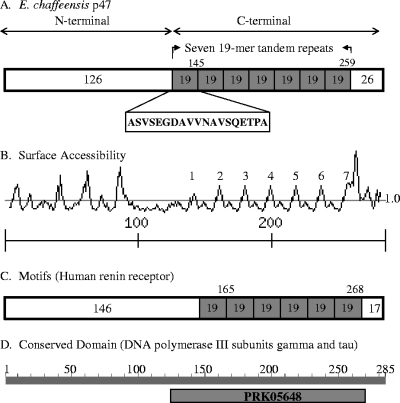
E. chaffeensis p47 is a 285-amino-acid, highly acidic (pI 4.18) protein containing seven 19-mer TRs (ASVSEGDAVVNAVSQETPA) that encompasses the majority of the carboxy-terminal portion (about 47% of full protein) of the protein (Fig. 1A). Each of the seven TRs is predicted to be surface accessible (Fig. 1B). Moreover, the TR unit of p47 has substantial homology with a motif identified in the renin receptor protein (http://pfam.sanger.ac.uk/search/) (Fig. 1C). Renin receptor protein is a multifunctional protein that has been referenced by other names related to its multifunctional role including, ATPase H+ transporting lysosomal accessory protein 2 (ATP6AP2), and endoplasmic reticulum-localized type I transmembrane adaptor protein (CAPER). In addition, a conserved domain search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) identified conservation of multi-domains (i.e., domain models that are computationally detected and likely to contain multiple single domains) in DNA polymerase III subunits gamma and tau (PRK05648: bit score, 37.46; E-value, 0.003) and showed substantial homology to the entire TR-containing C-terminal region (124 to 273 amino acids) of E. chaffeensis p47 (Fig. 1D) (27).
FIG. 1.
Schematic representation of E. chaffeensis p47 and regions of homology with renin receptor and DNA polymerase III subunits. (A) The amino (N)-terminal region is shaded (white bar) and precedes the seven 19-mer (ASVSEGDAVVNAVSQETPA) TRs (47% of the total protein sequence) region (gray bars). (B) The surface accessibility curve (13) predicts that each of the seven 19-mer TRs (numbered 1 to 7 over the curve) is surface exposed. (C) The C-terminal region of p47 contains six renin receptor-like motifs (14). (D) The DNA polymerase III subunit gamma and tau conserved domain (multi-domain PRK05648) was identified from NCBI Conserved Domain Database (CDD) (1). The computationally detected multi-domains are present in the same region of C-terminal region of p47 that contains the seven 19-mer TRs.
Analysis of E. chaffeensis p47 interactions using a yeast two-hybrid system.
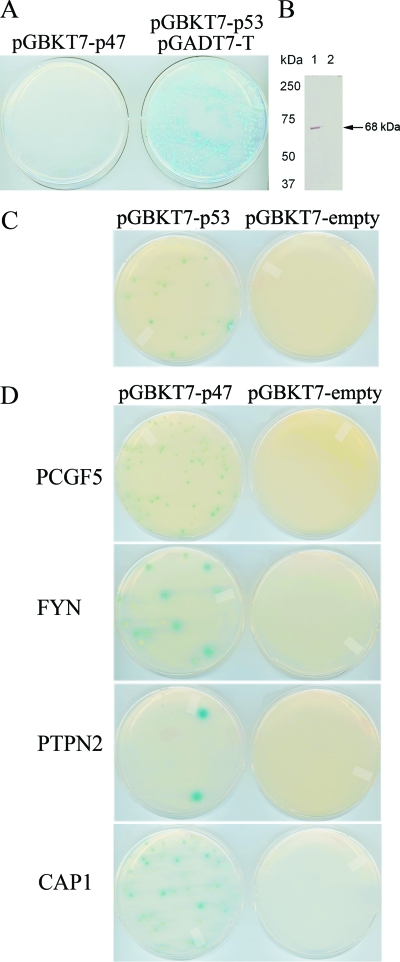
To confirm that the bait (E. chaffeensis p47) did not autonomously activate (autoactivate) the reporter genes in yeast strain AH109 in the absence of a prey protein, AH109 cells were transformed with E. chaffeensis p47/Gal4-BD (plasmid pGBKT7-p47) and plated on triple-dropout (TDO) medium (SD/−Ade/−His/−Trp) supplemented with X-α-Gal. Colonies were not observed after 3 to 5 days of incubation, confirming a lack of autoactivation by p47 (Fig. 2A). To examine toxicity, AH109 cells transformed with pGBKT7 (empty) and pGBKT7-p47 were grown in both solid and liquid media. No significant differences in yeast growth in solid or liquid medium containing bait and empty plasmids was observed, demonstrating that the bait was nontoxic to yeast. E. chaffeensis p47/Gal4-BD fusion protein expression in yeast was confirmed by transforming yeast AH109 with pGBKT7-p47, extracting yeast protein (urea/SDS method), and by Western immunoblotting with an anti-DNA-BD monoclonal antibody (Fig. 2B).
FIG. 2.
Yeast two-hybrid human bone marrow library clones that interacted with E. chaffeensis p47. (A) E. chaffeensis p47 was screened for autoactivation in S. cerevisiae strain AH109 containing pGBKT7-p47 and a positive control (AH109/Y187 diploid with pGBKT7-p53 [murine p53] and pGADT7-T [simian virus 40 large T antigen] generated by mating the S. cerevisiae strains AH109 [pGBKT7-53] and Y187 [pGADT7-T]) and selected with TDO medium. (B) Expression of p47/GAL4-BD (arrow; p47 size = 47 kDa + 21-kDa GAL4-BD tag [68 kDa total]) was determined by immunoblotting of yeast cell extracts transformed with pGBKT7-p47 (lane 1) or pGBKT7-empty (lane 2) using anti-GAL4-BD antibody. (C) To identify possible interacting clones, diploids were selected on QDO medium. A positive interaction was detected by using a diploid containing pGBKT7-p53 and pGADT7-T (plate on the left side of panel C), and the diploid containing pGBKT7-empty and pGADT7-T was used as a negative control (plate on right side of panel C). (D) S. cerevisiae strain AH109 expressing pGBKT7-p47 was mated with S. cerevisiae strain Y187 expressing PCGF5, FYN, PTPN2, and CAP1 (plates on the left side of panel D). S. cerevisiae strain AH109 expressing pGBKT7-empty was mated with S. cerevisiae strain Y187 expressing PCGF5, FYN, PTPN2, and CAP1 (plates on the right side of panel D). Diploids were selected as described for panel C.
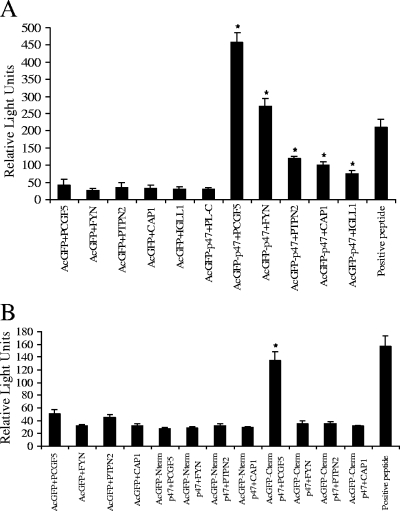
Through yeast two-hybrid screening using yeast mating, 7.68 million diploid clones were screened and about 4% mating efficiency was achieved. In the preliminary screening, 207 yeast colonies were observed on QDO medium supplemented with X-α-Gal identifying potential positive clones that exhibited bait and prey protein-protein interactions. One hundred four colonies were selected for yeast colony PCR followed by DNA sequencing to eliminate the duplicate clones and identify host-interacting partners. The 15 most abundant clones identified in order of frequency were PCGF5, Ig lambda-like polypeptide 1 (IGLL1), Stat6, CYP4F3, IgM light chain, PTPN2, LMAN1, Stat5a, RAI1, UBB, L30, PTPN1, LGALS3, KARS, and ribosomal protein L4. To identify the true interactions and eliminate the false positives, cotransformation assays were performed in yeast with 33 candidate prey plasmids, including the most abundant clones and some clones that were less abundant. Interactions were confirmed with 7 of 33 prey proteins: Polycomb group ring finger 5 (PCGF5), Src protein-tyrosine kinase FYN (FYN), PTPN2, CAP1, IGLL1, NW_001838484.1 and NT_005612.15 (Fig. 2C and D). To confirm the yeast two-hybrid results, we independently examined the interaction of p47 with these prey proteins in mammalian cells by Co-IP and confirmed a direct interaction between p47 and five prey proteins (Table 1). The relative strengths of the interaction between p47 and PCGF5, FYN, PTPN2, CAP1, and IGLL1 were 12, 12, 5, 3, and 3 times higher than those of the respective controls (in the absence of p47; AcGFP1-empty) (Fig. 3A). These results identified p47 as an interacting partner since in the absence of p47 the relative physical interaction values were very low and comparable to each other among the prey proteins (23 to 38 relative light units [RLU]). The full-length p47 interacted with each prey protein specifically and differentially, which is demonstrated by very high relative physical interaction values (468 RLU for PCGF5 to 74 RLU for IGLL1) (Fig. 3A). To confirm the specificity of the protein-protein interaction, the pAcGFP1-p47 and pProLabel-empty vectors were cotransfected to HeLa cells and Co-IP was performed. The relative low RLU (30) value indicated that there was no protein-protein interaction between AcGFP-p47 and pProLabel-empty and demonstrated that the presence of specific prey protein was required for p47-prey protein interaction (Fig. 3A). These data confirmed the yeast two-hybrid results and also indicated that p47 interacted most strongly with PCGF5 and FYN, followed by PTPN2, CAP1, and IGLL1. The top four candidates (PCGF5, FYN, PTPN2, and CAP1) were examined further for interactions with p47.
TABLE 1.
Summary of human proteins that interact with E. chaffeensis p47 as determined by yeast two-hybrid and Co-IP assays
| Interacting protein | GenBank accession no. | Component(s) | Properties/functions |
|---|---|---|---|
| Homo sapiens Polycomb group ring finger 5 (PCGF5) | NM_032373.3 | Centrosome | DNA-dependent regulation of transcription, metal ion binding, protein binding, zinc ion binding, RING (really interesting new gene) finger domain with “cross-brace” motif |
| Homo sapiens FYN oncoprotein related to SRC, FGR, and YES (FYN), transcript variant 1 | NM_002037.3 | Cytosol, endosome, plasma membrane | ATP binding, manganese ion binding, nucleotide binding, protein tyrosine kinase activity, T-cell receptor signaling pathway, calcium ion transport, protein amino acid phosphorylation |
| Homo sapiens protein tyrosine phosphatase, non-receptor type 2 (PTPN2), transcript variant 3 | NM_080423.1 | Cytoplasm | Hydrolase and phosphatase activity, protein tyrosine phosphatase activity, receptor activity, protein amino acid dephosphorylation |
| CAP, adenylate cyclase-associated protein 1 (yeast CAP1), transcript variant 2 | NM_001105530.1 | Cortical actin cytoskeleton, cytoplasm, plasma membrane | Actin binding, activation of adenylate cyclase activity, cytoskeleton organization and biogenesis, signal transduction, establishment and/or maintenance of cell polarity |
| Homo sapiens Ig lambda-like polypeptide 1 (IGLL1), transcript variant 1 | NM_020070.2 | Extracellular region, membrane | Immune response, involved in transduction of signals for cellular proliferation, differentiation from the pro-B-cell to the pre-B-cell stage |
FIG. 3.
E. chaffeensis p47 interaction with multiple human proteins with distinct functional properties. AcGFP (control, without insert) and AcGFP-p47 were coexpressed with ProLabel (control, without insert) and ProLabel-PCGF5, -FYN, -PTPN2, -CAP1, -IGLL1 as fusion proteins in HeLa cells. ProLabel activity was measured at different time intervals after addition of substrate. The ProLabel tag associated with AcGFP-p47 fusion protein and enzyme acceptor combine to form an active enzyme that cleaves the chemiluminescent substrate. The resulting signal is detected with a luminometer as RLU. The ProLabel tag is the limiting constituent and is governed by its interaction with AcGFP-p47 fusion protein. (A) Relative strength and interaction of AcGFP and AcGFP-p47 with ProLabel-PCGF5, -FYN, -PTPN2, -CAP1, and -IGLL1. The results are from three independent experiments. The values are means ± standard deviations shown as RLU. An asterisk indicates that the RLU of AcGFP-p47+PCGF5, AcGFP-p47+FYN, AcGFP-p47+PTPN2, AcGFP-p47+CAP1, AcGFP-p47+IGLL1 were significantly higher than those of AcGFP+PCGF5, AcGFP+FYN, AcGFP+PTPN2, AcGFP+CAP1, and AcGFP+IGLL1 (P < 0.005). (B) Relative strength and interaction of AcGFP, AcGFP-Nterm p47, and AcGFP-Cterm p47 with PL-PCGF5, -FYN, -PTPN2, and -CAP1. The results are from three independent experiments. The values are means ± standard deviations shown as RLU. The RLU of AcGFP-Ctermp47+PCGF5 was significantly higher than those of AcGFP+PCGF5 (*, P < 0.005). AcGFP, pAcGFP1 without insert; AcGFP-p47, pAcGFP1-p47; PL-C, ProLabel without insert; PCGF5, FYN, PTPN2, CAP1, and IGLL1, PL-PCGF5, PL-FYN, PL-PTPN2, PL-CAP1, and PL-IGLL1, respectively; Nterm p47, amino-terminal p47; Cterm p47, carboxy-terminal p47.
C-terminal domain of E. chaffeensis p47-containing TRs interacts with PCGF5.
The majority of the C-terminal domain of p47 is comprised of seven 19-amino-acid (ASVSEGDAVVNAVSQETPA) TRs (47% of the total protein sequence). To define the p47 domain that interacts with the identified host proteins, the N-terminal (p471-380) and C-terminal (p47361-842) regions of p47 were cloned in-frame downstream of the pAcGFP1 coding sequence (at SalI/HindIII) and expressed as a C-terminal AcGFP1 fusion protein. pAcGFP1-p471-380 and pAcGFP1-p47361-842 were cotransfected with library plasmid into HeLa cells. Forty-eight hours posttransfection, cells expressing GFP were lysed and Co-IP was performed on the cell lysates. The Co-IP results demonstrated that only p47361-842 (C-terminal region) containing the seven 19-mer TRs interacted with PCGF5, but no interaction was observed with FYN, PTPN2, and CAP1 (Fig. 3B). However, the relative interaction with the p47 C-terminal construct with PCGF5 was weaker (more than 2 times compared to the control) than the relative interaction of full-length p47 with PCGF5 (12 times compared to control). N-terminal p47 did not exhibit any substantial interaction with PCGF5, FYN, PTPN2, and CAP1 (Fig. 3B), although the full-length p47 strongly interacted with PCGF5, FYN, PTPN2, and CAP1 (Fig. 3A).
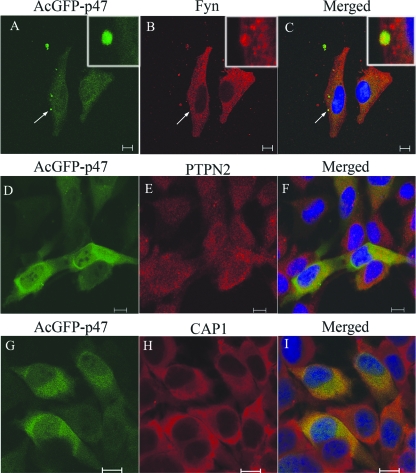
Recombinant E. chaffeensis p47 (pAcGFP1-p47) colocalizes with FYN, PTPN2, and CAP1.
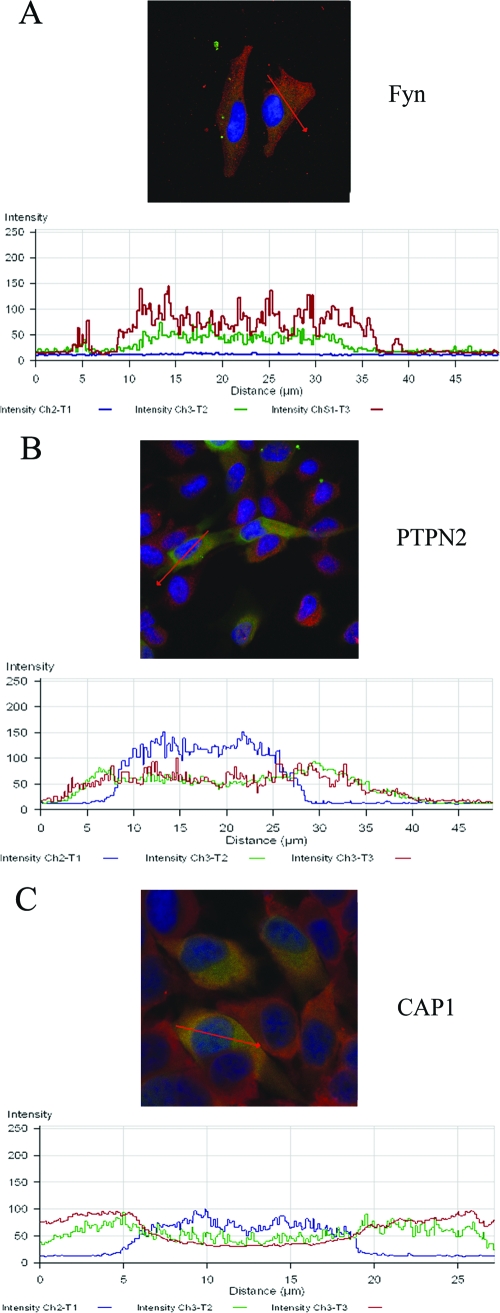
Interactions between p47 and three interacting proteins (FYN, PTPN2, and CAP1) were examined in transfected mammalian cells (HeLa). Immunofluorescent confocal microscopy revealed that recombinant p47 colocalized with FYN, PTPN2, and CAP1 (Fig. 4A to I). Recombinant E. chaffeensis p47 and host cell protein-tyrosine kinase FYN exhibited primarily diffuse cytoplasmic and perinuclear colocalization in the cell (Fig. 4A to C), although a few punctate colocalized spots were observed associated with plasma membrane and cytoplasm (Fig. 4A to C, also see insets for details). In the case of PTPN2, both cytoplasmic and nuclear colocalization of recombinant p47 and PTPN2 was detected in a small punctulated pattern (Fig. 4D to F). Although most of PTPN2-p47 interaction was associated with cytoplasm (Fig. 4E), some was localized in the nucleus of the cell (Fig. 4F). A diffused colocalization of p47 and CAP1 was observed exclusively in the cytoplasm (Fig. 4G to I). The confocal intensity profile (a profile shown as a red line in the confocal image in Fig. 5) collected across the transfected HeLa cells stained with FYN, PTPN2, and CAP1 indicated a similar distribution pattern of recombinant p47 (green) and FYN, PTPN2, and CAP1 (red) (Fig. 5A to C).
FIG. 4.
Colocalization of FYN, PTPN2, and CAP1 with AcGFP-p47 in HeLa cells. pAcGFP-p47-transfected HeLa cells (2 days posttransfection) were labeled and observed by confocal laser-scanning microscopy. The AcGFP-p47 (green) (A, D, and G) and anti-FYN (red; B), anti-PTPN2 (red; E), and anti-CAP1 (red; H) signals were merged, respectively (C, F, and I). FYN, PTPN2, and CAP1 colocalize with p47 (C, F, and I). The arrow indicates the punctulated colocalization of p47 and FYN. In the insets (A, B, and C), the arrow indicates an area highlighted to show the membrane colocalization of p47 and FYN. Small punctulated colocalization of p47 and PTPN2 is observed mostly in the cytoplasm of HeLa cells (F). Bar, 10 μm.
FIG. 5.
Fluorescent confocal microscopy and intensity profiles of HeLa cells expressing AcGFP-p47 (p47-green) fixed and stained with DAPI (blue) and anti-FYN, -PTPN2, or -CAP1 antibodies (red) showing colocalization of p47 with FYN (A), PTPN2 (B), and CAP1 (C). The red line in the confocal image indicates the area selected for fluorescence intensity profile analysis and is displayed in graph form (below). The x axis shows distance (μm), and the y axis shows intensity (0 to 250 arbitrary units).
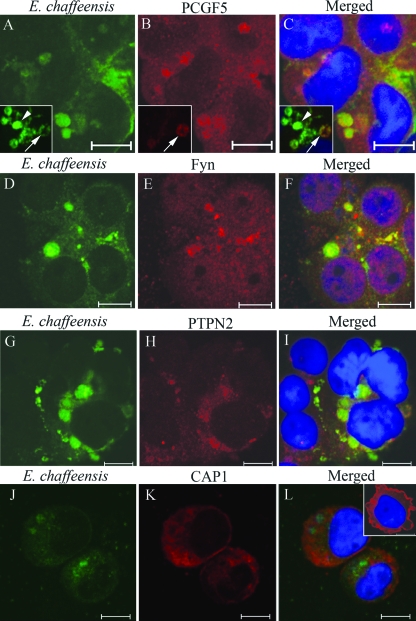
Differentially expressed E. chaffeensis p47 colocalizes with PCGF5, FYN, PTPN2, and CAP1 in E. chaffeensis-infected THP-1 cells.
E. chaffeensis exists in two distinct morphological forms known as RC and DC cells (33). Consistent with our previous report (12), we observed that only DC E. chaffeensis cells were stained by anti-p47 serum compared to anti-Dsb (pan-Ehrlichia marker) that stained all of the ehrlichiae (see insets in Fig. 6A to C). Double-immunofluorescence labeling of E. chaffeensis-infected THP-1 cells revealed that PCGF5, FYN, PTPN2, and CAP1 colocalized only with morulae that stained with anti-p47 antibody (DC ehrlichiae; Fig. 6A to L). The strongest colocalization was observed with PCGF5 and FYN, which exhibited similar distribution and fluorescent intensity (Fig. 6A to F). PTPN2 also exhibited similar distribution, but the fluorescent intensity was weaker (Fig. 6H). Conversely, CAP1 was redistributed in E. chaffeensis cells compared to uninfected cells (Fig. 6L and inset). CAP1 also exhibited increased intensity adjacent to the morulae membrane, but was not observed within the morula space (Fig. 6K and L).
FIG. 6.
Colocalization of E. chaffeensis p47 with PCGF5, FYN, PTPN2, and CAP1 in E. chaffeensis-infected THP-1 cells. THP-1 cells were infected with E. chaffeensis and 3 days postinfection were dually labeled and examined by confocal microscopy. The panels on the right are merged images. PCGF5, FYN, PTPN2, and CAP1 colocalize with E. chaffeensis p47-labeled morulae (yellow, right panels). Differential expression of E. chaffeensis p47 by a subset of ehrlichiae compared to Dsb (constitutively expressed by E. chaffeensis) is revealed by double-immunofluorescent labeling of E. chaffeensis-infected THP-1 cells with anti-E. chaffeensis Dsb (inset in Fig. 6A, green) and anti-p47 (inset in Fig. 6B, red) sera; the merged image (inset in Fig. 6C) demonstrates the singly (green, marked with arrowhead) and dually labeled (yellow, marked with arrow) E. chaffeensis. In the inset (bottom, right panel), a normal uninfected THP-1 cell reveals that CAP1 is mainly associated with plasma membrane, while in the E. chaffeensis-infected THP-1 cell, CAP1 is distributed in cytoplasm and associated with E. chaffeensis-containing morulae. Bar, 10 μm.
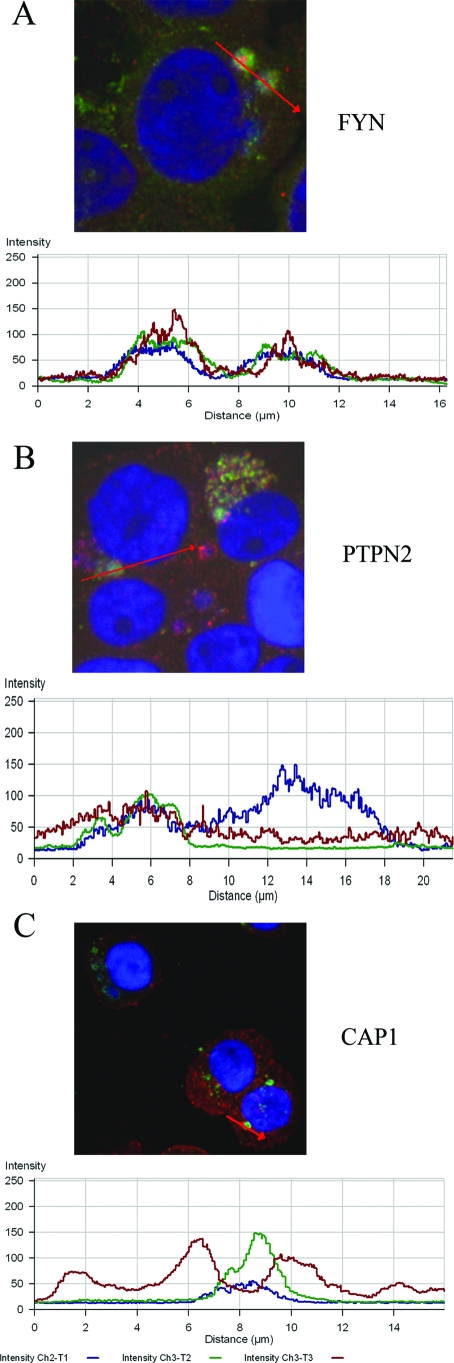
Distribution and colocalization of FYN, PTPN2, and CAP1 with p47-expressing ehrlichiae.
To further examine the distribution and colocalization of proteins FYN, PTPN2, and CAP1 with p47-expressing E. chaffeensis morula in detail, a confocal intensity profile across the colocalized spot in the confocal image was analyzed (Fig. 7A to C). The results demonstrated that p47 (green; Alexa Fluor 488), E. chaffeensis (small blue patch; DAPI [4,6′-diamidino-2-phenylindole] staining of bacterial DNA), and FYN or PTPN2 (red; Alexa Fluor 568) had consistently strong colocalization throughout the morulae and revealed a similar pattern of elevated peaks (green, blue, and red) across the morula profile (Fig. 7A and B). Elevated peaks were detected adjacent to the morula, suggesting increased localization at the morula membrane for CAP1 (Fig. 7C). CAP1 (red) encircled the morula forming a boundary outside the DC ehrlichiae resembling a cup-like structure across the colocalized spot and was not strongly detected in the morula space (Fig. 7C). Of note, CAP1 was mostly associated with cell membrane in normal THP-1 cells (Fig. 6L, inset), while its distribution pattern was diffusely distributed in the cytoplasm in E. chaffeensis-infected THP-1 cells.
FIG. 7.
Fluorescent confocal microscopy and intensity profiles of E. chaffensis-infected THP-1 cells fixed and stained with DAPI (blue), anti-p47 (green) and anti-FYN, -PTPN2, or -CAP1 antibodies (red) showing colocalization of p47 with FYN (A), PTPN2 (B), and CAP1 (C). The red line in the confocal image indicates the area selected for fluorescence intensity profile analysis and is displayed in graph form (below). The x axis shows distance (μm), and the y axis shows the intensity (0 to 250 arbitrary units). FYN (red) and PTPN2 (red) colocalize and associate with p47 (green), as revealed in diagrams A and B, respectively. In panel C, the profile along the E. chaffeensis-infected THP-1 cells displays that CAP1 (red) surrounds the E. chaffeensis-containing morulae stained with p47 (green) and also with DAPI (blue).
DISCUSSION
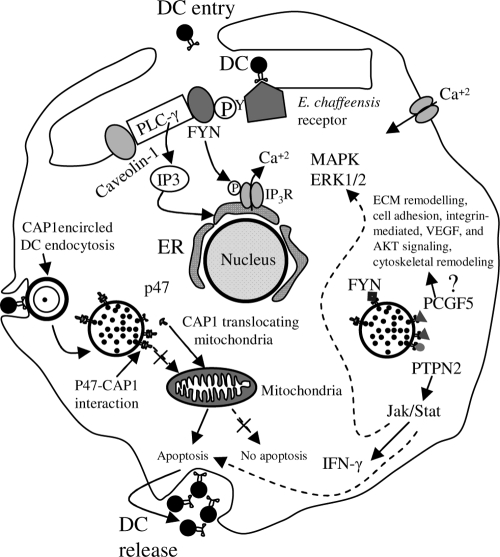
In order to survive as an obligately intracellular bacterium inside mononuclear phagocytes, E. chaffeensis manipulates host cellular processes to create a hospitable environment, averting the activation of the bactericidal machinery, most likely through specific interactions of its surface-expressed and/or secreted effector proteins. In this study, we reveal a novel interaction between the E. chaffeensis TR-containing p47 and multiple host proteins associated with signaling, transcriptional regulation, and vesicle trafficking, including PCGF5, FYN, PTPN2, and CAP1. A proposed model of this interaction is presented in Fig 8. The importance of p47 in pathobiology and immunity is suggested by its differential expression on the surface of DC ehrlichiae, characteristics that include mucin-like features (high S/T content; predicted glycosylation sites), TRs, and strong immunoreactivity. p120 and p47 have been identified on the surface of DC ehrlichiae, in the morula space, and have been associated with the morula membrane (12, 34). The mechanism of secretion appears to be Sec independent, as a signal peptide is not present on p47 and secretion is predicted to occur by a leaderless (Sec independent) secretion system (SecretomeP 2.0). Our results demonstrate that p47 directly interacts with these proteins and that this interaction is specific for the p47-expressing DC form of E. chaffeensis. This is the first study to identify a biologically relevant interaction between an ehrlichial protein and multiple host cell proteins with distinct cellular functions.
FIG. 8.
Proposed model of the role of p47 in E. chaffeensis infection and survival in monocytes based on this study and previous reports (22-24, 51, 52). Binding of E. chaffeensis to its receptor directly or indirectly activates FYN. Activated FYN tyrosine kinase phosphorylates and activates PLC-γ, which hydrolyzes membrane phospholipid PIP2 (phosphatidylinositol biphosphate), resulting in increased level of IP3 and release of Ca2+ from intracellular stores, and Ca2+ influx. FYN may regulate the function of IP3 receptor by phosphorylation and promoting release of Ca2+ from the endoplasmic reticulum. FYN also phosphorylates caveolin-1 involved in ehrlichial entry. p47-PTPN2 interaction modulates cytokine signaling events by exerting negative feedback on the Jak/Stat pathway by dephosphorylation of Jaks and Stats involved in intravacuolar maintenance and survival of E. chaffeensis. p47-PCGF5 interaction modulates gene transcription associated with cell signaling and remodeling of cytoskeleton, facilitating and supporting intracellular survival of E. chaffeensis. CAP1 promotes rapid actin dynamics in conjunction with ADF/cofilin and is required for cell morphology, migration, and endocytosis. Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis, which may play important role in release of DC ehrlichiae from the monocyte.
E. chaffeensis p47 can be divided into two major regions: an N-terminal region consisting of 126 amino acids and a C-terminal TR-containing region (133 amino acids) (12). The entire TR-containing C-terminal region has substantial homology to DNA polymerase III subunits gamma and tau. In addition, the TR sequence (19 amino acids) has homology to a C-terminal motif within the renin receptor/ATP6AP2/CAPER protein, a protein with multiple identities, domains, and functions, including angiotensin generation; activation of signal transduction pathways by inducing phosphorylation of ERK 1, ERK 2, and MAPK; association with vacuolar ATPases; and involvement in cellular proliferation and transformation (9, 32, 38). The significance of this homology is not clear, but the homology of the C-terminal TR region of E. chaffeensis p47 with proteins involved in important cellular protein-protein interactions is consistent with our findings with regard to p47.
In the yeast two-hybrid experiments, PCGF5 protein was the most frequently identified interacting partner and exhibited the strongest protein-protein interaction with p47. This was consistent with the immunofluorescence microscopy results that indicated a strong PCGF5 immunofluorescence and colocalization with p47 expressed by DC ehlichiae in dually stained infected cells. PCGF5 has not been extensively studied, but it appears to be involved in DNA-dependent regulation of transcription, metal ion binding, and protein-protein interactions. PCGF5 has a specialized Zn finger domain consisting of 40 to 60 residues that binds two atoms of zinc and is defined by the “cross-brace” motif involved in protein-protein interactions (6, 17, 37). PCGF5 is related to the Polycomb group proteins (transcriptional repressors) Bmi-1/PCGF4 and Mel-18/PCGF2 (36, 39), which play an important role in the regulation of Hox gene expression, X-chromosome inactivation, tumorigenesis, and self-renewal; maintenance of pleuripotency of stem cells; and stimulation of E3 ubiquitin ligase activity (10, 30, 45). Bmi-1 and Mel-18 regulate gene expression associated with extracellular matrix remodeling, cell adhesion, and integrin-mediated signaling and affect the expression of the constituent members of vascular endothelial growth factor (VEGF), and AKT signaling pathway (46). Thus, p47-expressing DC ehrlichiae may recruit PCGF5 in an effort to modulate host cell gene expression to favor survival. Altered gene expression in E. chaffeensis-infected cells has been reported, but the mechanisms involved are unknown (52). A Co-IP experiment using C- and N-terminal deletion mutants of p47 identified the C-terminal, TR-containing region of E. chaffeensis p47 as the primary interaction domain with PCGF5. However, the interaction between PCGF5 and C-terminal region of p47 was weaker than the interaction between PCGF5 and full-length p47, demonstrating the contribution of the p47 N-terminal region in the interaction. Furthermore, none of the full-p47-interacting partners, including PCGF5, FYN, PTPN2, and CAP1, interacted with the N-terminal region of p47 devoid of TR, further implicating the importance of the both C- and N-terminal domains in these protein-protein interactions.
One of the first events for entry and infection is the recruitment of tyrosine-phosphorylated protein and PLC-γ2 in the early nascent morulae containing the DC ehrlichiae (24, 51). Tyrosine-phosphorylated protein and PLC-γ2 colocalize with caveolin-1 in caveolae containing E. chaffeensis (2, 24). Tyrosine kinases are known to be involved in ehrlichial entry; however, the specific kinases involved have not been determined. A strong p47-FYN interaction, similar to p47-PCGF5, was discovered by a yeast two-hybrid assay and was reflected by a robust immunofluorescence and colocalization of p47 and FYN in dually stained infected cells. FYN is a cytosol, endosome, and membrane-associated, non-receptor protein tyrosine kinase and a member of the Src family that is enriched in caveoalae (2). The association of p47 with FYN and the intracellular DC form of E. chaffeensis suggests that it may be recruited by p47 to facilitate the entry process. FYN specifically phosphorylates caveolin-1 and is required for coxsackievirus internalization and infection via caveolin-associated vesicles to polarized epithelial cells (11). FYN was not observed associated with E. chaffeensis RCs, demonstrating a selective association with DC ehrlichiae that express p47.
Another p47-interacting protein, PTPN2, is a member of the protein tyrosine phosphatase (PTP) family also known as T-cell PTP (TC-PTP) that catalyzes the dephosphorylation of phosphotyrosine peptides and regulates phosphotyrosine levels in signal transduction pathways. PTPs are known to regulate a variety of cellular processes, including cell growth, differentiation, mitotic cycle, and oncogenic transformation (42). Multiple substrates of PTPN2 included colony-stimulating factor CSF-1R, epidermal growth factor receptor, platelet-derived growth factor receptor, IR, p52Shc, Jak1, Jak3, Stat1, Stat3, Stat5a/b, and Stat6 (42). The in vivo and in vitro analyses indicate that PTPN2 could control cytokine signaling events by their negative action on the Jak/Stat pathway (41, 42). The loss of PTPN2 results in Stat5 hyperactivation, increased production of IFN-γ, tumor necrosis factor alpha (7), interleukin-12, and inducible nitric oxide synthase; increased tyrosine phosphorylation and recruitment of a Grb2/Gab2/Shp2 complex to the CSF-1 receptor; and enhanced activation of ERK and may affect transcription factor PU.1 signaling (8, 19, 40, 41, 48). The Jak/Stat pathway is inhibited by E. chaffeensis (22), and our results support the possibility that p47 may be involved in the inhibition of IFN-γ-induced tyrosine phosphorylation of Stat1, Jak1, and Jak2 by interacting with PTPN2.
The localization of CAP1 with the DC morulae was observed adjacent to the morula boundaries (membrane), and CAP1 was not observed inside the morula space as was observed with PCGF5, FYN, and PTPN2. p47 is secreted and has been demonstrated on the morula membrane containing the DC ehrlichiae (12); thus, the interaction between p47 and CAP1 appears to occur at the morula membrane interface. The distribution pattern of CAP1 was remarkably different in E. chaffeensis-infected cells from that in normal cells, where it is primarily associated with the plasma membrane, indicating that the distribution of this protein is altered as a result of E. chaffeensis infection. CAPs were originally identified in yeast as a component of the adenylyl cyclase complex, and yeast cells deficient in CAPs are defective in cytoskeleton organization. Although CAPs do not regulate cyclic AMP in animal cells, their role in regulation of actin remodeling in response to cellular signals is widely conserved. CAP1 is highly conserved monomeric actin binding protein that contains actin (C terminal), adenylyl cyclase, and cofilin (N terminal), and Src homology 3 (SH3) and profilin (central region) binding domains (5, 16, 29). Genetic studies in yeast have implicated CAP1 in vesicle trafficking and endocytosis (18, 35). In mammalian cells, CAP1 is associated with an SH3 domain-dependent mAbp1/dynamin complex involved in receptor-mediated endocytosis (20).
Ehrlichia and Anaplasma spp. are known to inhibit apoptosis, but the molecular mechanisms are not well understood. Ehrlichiae, like chlamydiae, inhibit apoptosis early in infection, but while chlamydiae induce cell death at the end of the infection cycle, ehrlichial exit mechanisms remain undefined (15, 43, 47). Interestingly, CAP1 has also been implicated in promoting apoptosis by functioning as an actin shuttle to mitochondria. Similar to cofilin, BAD, and BAX, CAP1 rapidly translocates to mitochondria independent of caspase activation, where it promotes apoptosis (44, 50). Associations between ehrlichial morula and mitochondria have been consistently observed (33, 51). Thus, p47 and CAP1 interaction may serve a dual function by facilitating endocytosis and vesicle trafficking and promoting apoptosis in the late stages of infection.
Ehrlichiae must interact with the host cell to reprogram host cell defense mechanisms. E. chaffeensis p47 interacts with and appears to facilitate the recruitment of PCGF5, FYN, PTPN2, and CAP1 to morula containing DC E. chaffeensis. This newly identified interaction between a DC ehrlichia-expressed protein, p47, and the multiple host proteins involved in regulation of gene transcription, vesicle trafficking, and cell signaling pathways suggests that identification of additional interacting host cell ligands of other DC cell-expressed proteins will facilitate studies to elucidate the function of this unique class of ehrlichial proteins. Further research is needed to understand the mechanisms which underlie these host-pathogen molecular interactions and the importance of these interactions in E. chaffeensis pathobiology.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (AI 071145).
We are grateful to Rhykka L. Connelly and Thomas Albrecht in the Infectious Disease and Toxicology Optical Imaging Core at UTMB for assistance with confocal microscopy. We also thank David H. Walker and Xue-jie Yu for reviewing the manuscript and providing helpful suggestions.
Editor: A. Camilli
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R. G. 1998. The caveolae membrane system. Annu. Rev. Biochem. 67199-225. [DOI] [PubMed] [Google Scholar]
- 3.Barnewall, R. E., N. Ohashi, and Y. Rikihisa. 1999. Ehrlichia chaffeensis and E. sennetsu, but not the human granulocytic ehrlichiosis agent, colocalize with transferrin receptor and up-regulate transferrin receptor mRNA by activating iron-responsive protein 1. Infect. Immun. 672258-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnewall, R. E., Y. Rikihisa, and E. H. Lee. 1997. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect. Immun. 651455-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertling, E., O. Quintero-Monzon, P. K. Mattila, B. L. Goode, and P. Lappalainen. 2007. Mechanism and biological role of profilin-Srv2/CAP interaction. J. Cell Sci. 1201225-1234. [DOI] [PubMed] [Google Scholar]
- 6.Borden, K. L., and P. S. Freemont. 1996. The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 6395-401. [DOI] [PubMed] [Google Scholar]
- 7.Bourdeau, A., N. Dube, K. M. Heinonen, J. F. Theberge, K. M. Doody, and M. L. Tremblay. 2007. TC-PTP-deficient bone marrow stromal cells fail to support normal B lymphopoiesis due to abnormal secretion of interferon-γ. Blood 1094220-4228. [DOI] [PubMed] [Google Scholar]
- 8.Bourdeau, A., N. Dube, and M. L. Tremblay. 2005. Cytoplasmic protein tyrosine phosphatases, regulation and function: the roles of PTP1B and TC-PTP. Curr. Opin. Cell Biol. 17203-209. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, D. J. 2008. Critical review of prorenin and (pro)renin receptor research. Hypertension 511259-1264. [DOI] [PubMed] [Google Scholar]
- 10.Cao, R., Y. Tsukada, and Y. Zhang. 2005. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20845-854. [DOI] [PubMed] [Google Scholar]
- 11.Coyne, C. B., and J. M. Bergelson. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124119-131. [DOI] [PubMed] [Google Scholar]
- 12.Doyle, C. K., K. A. Nethery, V. L. Popov, and J. W. McBride. 2006. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emini, E. A., J. V. Hughes, D. S. Perlow, and J. Boger. 1985. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 55836-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, S. F., C. Schwarz, J. Vier, and G. Häcker. 2001. Characterization of antiapoptotic activities of Chlamydia pneumoniae in human cells. Infect. Immun. 697121-7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman, N. L., Z. Chen, J. Horenstein, A. Weber, and J. Field. 1995. An actin monomer binding activity localizes to the carboxyl-terminal half of the Saccharomyces cerevisiae cyclase-associated protein. J. Biol. Chem. 2705680-5685. [DOI] [PubMed] [Google Scholar]
- 17.Freemont, P. S. 1993. The RING finger. A novel protein sequence motif related to the zinc finger. Ann. N. Y. Acad. Sci. 684174-192. [DOI] [PubMed] [Google Scholar]
- 18.Geli, M. I., and H. Riezman. 1998. Endocytic internalization in yeast and animal cells: similar and different. J. Cell Sci. 1111031-1037. [DOI] [PubMed] [Google Scholar]
- 19.Heinonen, K. M., F. P. Nestel, E. W. Newell, G. Charette, T. A. Seemayer, M. L. Tremblay, and W. S. Lapp. 2004. T-cell protein tyrosine phosphatase deletion results in progressive systemic inflammatory disease. Blood 1033457-3464. [DOI] [PubMed] [Google Scholar]
- 20.Kessels, M. M., A. E. Engqvist-Goldstein, D. G. Drubin, and B. Qualmann. 2001. Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J. Cell Biol. 153351-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumagai, Y., Z. Cheng, M. Lin, and Y. Rikihisa. 2006. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect. Immun. 745014-5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, E. H., and Y. Rikihisa. 1998. Protein kinase A-mediated inhibition of gamma interferon-induced tyrosine phosphorylation of Janus kinases and latent cytoplasmic transcription factors in human monocytes by Ehrlichia chaffeensis. Infect. Immun. 662514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, M., and Y. Rikihisa. 2004. Ehrlichia chaffeensis downregulates surface Toll-like receptors 2/4, CD14 and transcription factors PU.1 and inhibits lipopolysaccharide activation of NF-kappa B, ERK 1/2 and p38 MAPK in host monocytes. Cell. Microbiol. 6175-186. [DOI] [PubMed] [Google Scholar]
- 24.Lin, M., and Y. Rikihisa. 2003. Obligatory intracellular parasitism by Ehrlichia chaffeensis and Anaplasma phagocytophilum involves caveolae and glycosylphosphatidylinositol-anchored proteins. Cell. Microbiol. 5809-820. [DOI] [PubMed] [Google Scholar]
- 25.Lin, M., M. X. Zhu, and Y. Rikihisa. 2002. Rapid activation of protein tyrosine kinase and phospholipase C-γ2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infect. Immun. 70889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, T., X. Zhang, A. Wakeel, V. L. Popov, and J. W. McBride. 2008. A variable-length PCR target protein of Ehrlichia chaffeensis contains major species-specific antibody epitopes in acidic serine-rich tandem repeats. Infect. Immun. 761572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer, A., J. B. Anderson, M. K. Derbyshire, C. Weese-Scott, N. R. Gonzales, M. Gwadz, L. Hao, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, D. Krylov, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, N. Thanki, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2007. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35D237-D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride, J. W., R. E. Corstvet, S. D. Gaunt, C. Boudreaux, T. Guedry, and D. H. Walker. 2003. Kinetics of antibody response to Ehrlichia canis immunoreactive proteins. Infect. Immun. 712516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriyama, K., and I. Yahara. 2002. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J. Cell Sci. 1151591-1601. [DOI] [PubMed] [Google Scholar]
- 30.Muller, J., and J. A. Kassis. 2006. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16476-484. [DOI] [PubMed] [Google Scholar]
- 31.Nethery, K. A., C. K. Doyle, X. Zhang, and J. W. McBride. 2007. Ehrlichia canis gp200 contains dominant species-specific antibody epitopes in terminal acidic domains. Infect. Immun. 754900-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen, G., F. Delarue, C. Burckle, L. Bouzhir, T. Giller, and J. D. Sraer. 2002. Pivotal role of the renin/prorenin receptor in angiotensin II production and cellular responses to renin. J. Clin. Investig. 1091417-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popov, V. L., S. M. Chen, H. M. Feng, and D. H. Walker. 1995. Ultrastructural variation of cultured Ehrlichia chaffeensis. J. Med. Microbiol. 43411-421. [DOI] [PubMed] [Google Scholar]
- 34.Popov, V. L., X. Yu, and D. H. Walker. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb. Pathog. 2871-80. [DOI] [PubMed] [Google Scholar]
- 35.Qualmann, B., M. M. Kessels, and R. B. Kelly. 2000. Molecular links between endocytosis and the actin cytoskeleton. J. Cell Biol. 150F111-F116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez, C., I. Sanchez, J. A. Demmers, P. Rodriguez, J. Strouboulis, and M. Vidal. 2007. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Cell. Proteomics 6820-834. [DOI] [PubMed] [Google Scholar]
- 37.Saurin, A. J., K. L. Borden, M. N. Boddy, and P. S. Freemont. 1996. Does this have a familiar RING? Trends Biochem. Sci. 21208-214. [PubMed] [Google Scholar]
- 38.Schefe, J. H., M. Menk, J. Reinemund, K. Effertz, R. M. Hobbs, P. P. Pandolfi, P. Ruiz, T. Unger, and H. Funke-Kaiser. 2006. A novel signal transduction cascade involving direct physical interaction of the renin/prorenin receptor with the transcription factor promyelocytic zinc finger protein. Circ. Res. 991355-1366. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz, Y. B., and V. Pirrotta. 2008. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20266-273. [DOI] [PubMed] [Google Scholar]
- 40.Simoncic, P. D., A. Bourdeau, A. Lee-Loy, L. R. Rohrschneider, M. L. Tremblay, E. R. Stanley, and C. J. McGlade. 2006. T-cell protein tyrosine phosphatase (Tcptp) is a negative regulator of colony-stimulating factor 1 signaling and macrophage differentiation. Mol. Cell. Biol. 264149-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simoncic, P. D., A. Lee-Loy, D. L. Barber, M. L. Tremblay, and C. J. McGlade. 2002. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr. Biol. 12446-453. [DOI] [PubMed] [Google Scholar]
- 42.Stuible, M., K. M. Doody, and M. L. Tremblay. 2008. PTP1B and TC-PTP: regulators of transformation and tumorigenesis. Cancer Metastasis Rev. 27215-230. [DOI] [PubMed] [Google Scholar]
- 43.Verbeke, P., L. Welter-Stahl, S. Ying, J. Hansen, G. Hacker, T. Darville, and D. M. Ojcius. 2006. Recruitment of BAD by the Chlamydia trachomatis vacuole correlates with host-cell survival. PLoS Pathog. 2e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, C., G. L. Zhou, S. Vedantam, P. Li, and J. Field. 2008. Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis. J. Cell Sci. 1212913-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, H., L. Wang, H. Erdjument-Bromage, M. Vidal, P. Tempst, R. S. Jones, and Y. Zhang. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431873-878. [DOI] [PubMed] [Google Scholar]
- 46.Wiederschain, D., L. Chen, B. Johnson, K. Bettano, D. Jackson, J. Taraszka, Y. K. Wang, M. D. Jones, M. Morrissey, J. Deeds, R. Mosher, P. Fordjour, C. Lengauer, and J. D. Benson. 2007. Contribution of Polycomb homologues Bmi-1 and Mel-18 to medulloblastoma pathogenesis. Mol. Cell. Biol. 274968-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiong, Q., W. Bao, Y. Ge, and Y. Rikihisa. 2008. Ehrlichia ewingii infection delays spontaneous neutrophil apoptosis through stabilization of mitochondria. J. Infect. Dis. 1971110-1118. [DOI] [PubMed] [Google Scholar]
- 48.You-Ten, K. E., E. S. Muise, A. Itie, E. Michaliszyn, J. Wagner, S. Jothy, W. S. Lapp, and M. L. Tremblay. 1997. Impaired bone marrow microenvironment and immune function in T cell protein tyrosine phosphatase-deficient mice. J. Exp. Med. 186683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, X.-J., P. Crocquet-Valdes, L. C. Cullman, and D. H. Walker. 1996. The recombinant 120-kilodalton protein of Ehrlichia chaffeensis, a potential diagnostic tool. J. Clin. Microbiol. 342853-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zha, J., H. Harada, E. Yang, J. Jockel, and S. J. Korsmeyer. 1996. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L). Cell 87619-628. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, J. Z., V. L. Popov, S. Gao, D. H. Walker, and X. J. Yu. 2007. The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell. Microbiol. 9610-618. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, J.-Z., M. Sinha, B. A. Luxon, and X.-J. Yu. 2004. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect. Immun. 72498-507. [DOI] [PMC free article] [PubMed] [Google Scholar]