Abstract
While in transit within and between hosts, uropathogenic Escherichia coli (UPEC) encounters multiple stresses, including substantial levels of nitric oxide and reactive nitrogen intermediates. Here we show that UPEC, the primary cause of urinary tract infections, can be conditioned to grow at higher rates in the presence of acidified sodium nitrite (ASN), a model system used to generate nitrosative stress. When inoculated into the bladder of a mouse, ASN-conditioned UPEC bacteria are far more likely to establish an infection than nonconditioned bacteria. Microarray analysis of ASN-conditioned bacteria suggests that several NsrR-regulated genes and other stress- and polyamine-responsive factors may be partially responsible for this effect. Compared to K-12 reference strains, most UPEC isolates have increased resistance to ASN, and this resistance can be substantially enhanced by addition of the polyamine cadaverine. Nitrosative stress, as generated by ASN, can stimulate cadaverine synthesis by UPEC, and growth of UPEC in cadaverine-supplemented broth in the absence of ASN can also promote UPEC colonization of the bladder. These results suggest that UPEC interactions with polyamines or stresses such as reactive nitrogen intermediates can in effect reprogram the bacteria, enabling them to better colonize the host.
The urinary tract is normally a sterile environment, and it is both hostile and poorly accessible to most microbes. However, roughly one-half of women in the United States experience a urinary tract infection (UTI) at least once in their lifetime, and one-quarter of affected women endure recurrence (22, 25). More than 80% of UTIs are due to strains of uropathogenic Escherichia coli (UPEC), which are usually presumed to be part-time gut flora that have reached the urinary tract by ascension via the periurethral area (53). Transmission of UPEC among individuals occurs primarily by way of fecal-oral routes and, in some cases, may involve the ingestion of contaminated food products or sexual contact (15, 23, 33, 40, 41, 57). In order to survive and disseminate, UPEC must be able to adapt to multiple environments and stresses both within and outside the host.
When a UPEC infection occurs, recruitment of nitric oxide (NO)-producing neutrophils to the bladder is an important line of defense (26, 48). Within hours of infection, the nitrite levels in the urine increase up to threefold, and eventually the levels of NO within the bladder are 30- to 50-fold higher than those in uninfected controls (39, 48). The high levels of NO are due in part to inducible NO synthase activity, which is upregulated within 6 h after infection (45). A role may also be played by endothelial NO synthase, which is upregulated and activated in the bladder mucosa by E. coli lipopolysaccharide (36) and by the bacteria themselves, which can produce NO with nitrite reductases under low-oxygen-tension conditions (12). NO is a precursor of a variety of reactive nitrogen intermediates (RNIs), such as peroxynitrite and nitrosothiols, which can inflict extensive damage on nucleic acids, lipids, and proteins. Thiols, amines, aromatic residues, heme groups, and iron-sulfur clusters are particularly susceptible to attack by RNIs, making many key metabolic enzymes targets (17, 18). UPEC may also encounter RNIs outside the urinary tract, possibly during passage through the upper gastrointestinal tract, where nitrate (NO3−) and nitrite (NO2−) levels can be very high, or on the surface of meat products, which are often treated with nitrite as a coloring agent and preservative (15, 16, 27, 29, 64).
Adaptive responses that allow a bacterial population to survive one stressful condition can, in some instances, enhance its ability to handle other environmental stresses (2, 5, 31, 32, 42). This cross-protective effect may also potentiate bacterial virulence within a host. Recently, UPEC was found to have the capacity to withstand RNI levels that prevent growth of nonpathogenic E. coli K-12 strains (7, 60). RNI resistance in UPEC is controlled in part by the envelope stress response sigma factor RpoE (σE), the RNA chaperone Hfq, the NO-detoxifying enzyme HmpA, and polyamines (7, 38, 60). Expanding on these findings, we show here that UPEC can transiently adapt to high levels of nitrosative stress via a polyamine-linked mechanism, enabling this pathogen to grow more rapidly after subsequent exposure to RNIs and to better colonize the urinary tract in a mouse UTI model system.
MATERIALS AND METHODS
Bacterial strains and growth curves.
The K-12 reference strain MG1655 and UTI89, a human cystitis isolate, have been described previously (4, 10, 44). Other UPEC isolates were kindly provided by D. A. Low (University of California, Santa Barbara) and W. E. Hooton (University of Washington School of Medicine). All growth experiments were performed at 37°C in 100 mM morpholineethanesulfonic acid (MES)-buffered Luria-Bertani broth (MES-LB broth) (pH 5) with or without added sodium nitrite or cadaverine (Sigma-Aldrich) as indicated below. The cultures used to obtain the data shown in Fig. 1B were grown in 5 ml MES-LB broth in loosely capped borosilicate glass tubes (20 by 150 mm) with shaking (225 rpm, tilted at a 30° angle), and growth was monitored by determining the optical density at 600 nm (OD600) using a Spectronic 20D+ (Thermo). All other growth curves were obtained using 200-μl cultures in 100-well honeycomb plates and a Bioscreen C instrument (Growth Curves USA).
FIG. 1.
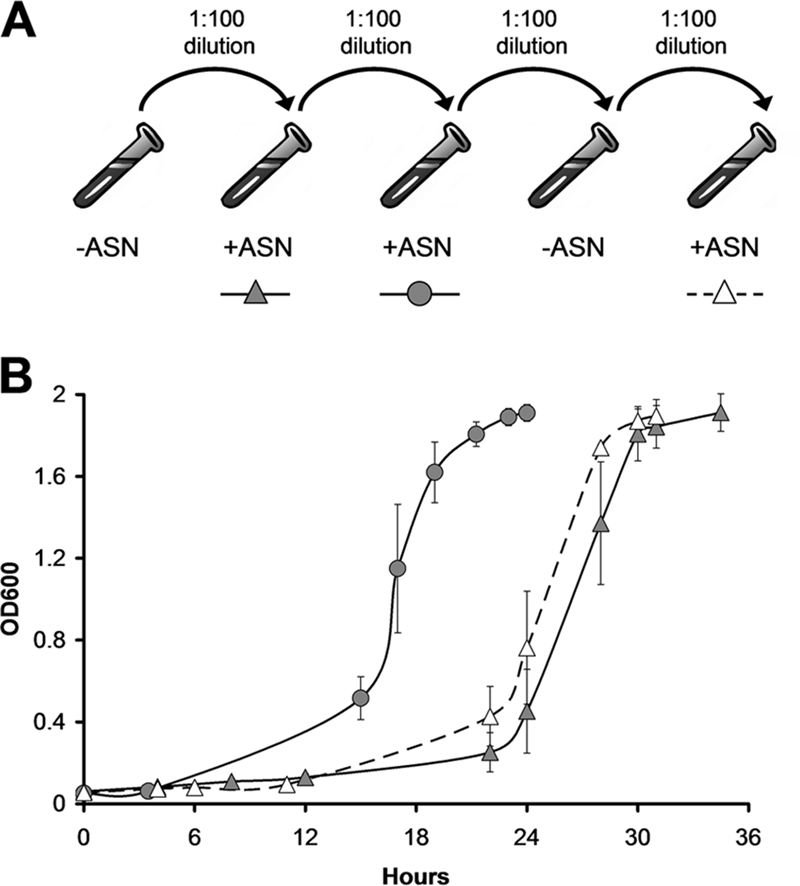
UTI89 adapts to growth in the presence of 3 mM ASN. (A) Diagram of the experimental protocol used. Colonies of UTI89 grown from a freezer stock were used to start overnight cultures in MES-LB broth without ASN. For each round, cultures were grown with shaking in tubes at 37°C and allowed to reach stationary phase before they were subcultured 1:100 into new media with or without ASN, as indicated. Culture growth was monitored by determining the OD600. (B) Growth curves of cultures with ASN indicated in the diagram in panel A, demonstrating the difference in lag time between cultures conditioned with 3 mM ASN and cultures not conditioned with 3 mM ASN. The error bars indicate standard deviations from the means of five independent cultures.
Microarray sample preparation.
Separate colonies of UTI89 grown on LB agar plates from a −80°C freezer stock were used to start overnight shaking cultures in MES-LB broth. Each culture was then diluted 1:100 into 7 ml MES-LB broth with or without 3 mM acidified sodium nitrite (ASN) in loosely capped borosilicate glass tubes (20 by 150 mm) and grown with shaking at 37°C. Growth was monitored until the OD600 was 1.5, at which point cultures were spun down, and the pellets were frozen at −80°C for at least 12 h. RNA was extracted with hot phenol-chloroform and purified by CsCl centrifugation. cDNA was synthesized, fragmented, and labeled using the protocol recommended by Affymetrix.
Affymetrix microarray gene expression analysis.
Fragmented and labeled cDNA (15 μg) was added to 270 μl of hybridization buffer and hybridized to an Affymetrix GeneChip E. coli genome 2.0 array. After 20 h of hybridization at 45°C, the GeneChips were washed, stained, and scanned according to the standard Affymetrix protocol. The arrays were scanned using an Affymetrix GeneChip 3000 scanner enabled for high-resolution scanning, and the raw images were converted to CEL files using Affymetrix GCOS software. Image processing using the GCRMA method for probe-level data (30) was performed using the Bioconductor Package in the R statistical environment (24). The CEL files were analyzed as a group, background corrected using GCRMA (63), and normalized using quantile normalization, and summary measures for probe sets were obtained by median polish. Transcripts were categorized (Fig. 2) based on literature searches and gene information drawn from EcoCyc (37; http://ecocyc.org/) and Affymetrix NetAffx (http://www.affymetrix.com/analysis/index.affx).
FIG. 2.
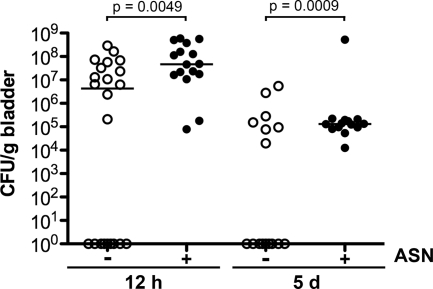
Enhanced colonization of the bladder by ASN-conditioned UPEC. Adult female CBA/J mice were inoculated with 107 CFU of UTI89 that had been grown to an OD600 of 1.5 in MES-LB broth with or without 3 mM ASN. Bacterial titers in bladder homogenates were determined at 12 h or 5 days postinoculation. The horizontal bars indicate median values for the groups. The P values were determined using Fisher's exact test (n = 13 to 22 mice).
Quantitative RT-PCR.
Primers for real-time PCR (RT-PCR), which were designed using the D-LUX Designer application available on the Invitrogen website, were made so that their G+C content was approximately 50% and they amplified regions 56 to 65 bp long. The primers used include norV_289FL (5′-CGGGAGAACTGATGGCACAAATTCCCG-3′), norV_289FL/307RU (5′-CGAGTCGATAGCGTTGGCAGT-3′), nrdH_153FL (5′-CAACTGCTCAGGGCTTTCGTCAGTTG-3′), nrdH_153FL/164Rub (5′-CCAGCTAAGATCGCCTGCAA-3′), hmpA_743RL (5′-CGGCCCATAAAGAAATCACCTGCCG-3′), and hmpA_743RL/732FU (5′-AATGTTGGCGATGTCGTGAAA-3′). RNA collected for microarray analysis and stored at −80°C was used to make cDNA according to the protocol recommended by Affymetrix. Control reactions with no reverse transcriptase were also performed. cDNA was used as a template for RT-PCR with a Roche LightCycler 480, using a Roche LightCycler 480 DNA SYBR green I master kit according to the manufacturer's protocol. In addition to triplicate reactions for experimental cDNA samples, controls without reverse transcriptase, and controls without cDNA, dilutions of genomic DNA were used to generate a standard curve for each primer set. Quantitation was carried out by performing LightCycler 480 Abs Quant/2nd Derivative Max analysis.
Mouse infections.
Female CBA/J mice (Jackson Labs) that were approximately 7 to 9 weeks old were used in infection studies in accordance with IACUC-approved protocols. UTI89 from frozen stock was grown with shaking overnight at 37°C in MES-LB broth and then subcultured 1:100 in 7 ml MES-LB broth with or without 3 mM ASN or 3 mM cadaverine as indicated below. The cultures were incubated at 37°C with shaking until the OD600 was 1.5. Bacteria were then pelleted by centrifugation for 8 min at 8,000 × g, washed three times, and resuspended in phosphate-buffered saline. Each mouse was briefly anesthetized using isofluorane and inoculated via a transurethral catheter with 50 μl of a bacterial suspension containing approximately 1 × 107 CFU of control, ASN-conditioned, or cadaverine-treated UTI89. Twelve hours or 5 days later, bladders were collected, weighed, and homogenized, and the homogenates were serially diluted and plated on LB agar plates to determine the number of bacteria per gram of tissue. Mouse experiments were repeated at least twice with similar results (total combined data are presented below).
Analysis of type 1 pilus expression.
Expression of type 1 pili by UTI89 was assessed in two ways. First, the ability of UTI89 to agglutinate Saccharomyces cerevisiae was qualitatively determined by mixing 20 μl of each bacterial strain (OD600, 1.5) with 200 μl of a 1% suspension of baker's yeast in phosphate-buffered saline on glass slides. In addition, the orientation of the fim switch was determined using a previously described inverse PCR protocol (8). Chromosomal DNA was prepared from control, ASN-conditioned, and cadaverine-treated cultures (OD600, ∼1.5) using a Promega Wizard genomic DNA kit. Five hundred nanograms of each genomic DNA sample was used as a template in a separate reaction designed to detect the fim switch in its on and off orientations. The reactions used the same outside primer (5′-CGACAGCAGAGCTGGTCGCTC-3′) with one of two inside primers. The primer 5′-GTAAATTATTTCTCTTGTAAATTAATTTCACATCACCTCCGC-3′ was used to detect fim in the off orientation, while the reverse complement was used to detect fim in the on orientation. At least three separate cultures were tested for each group, and the ratios of the intensities of bands resulting from the PCRs were compared to determine relative on and off levels of the fim switch.
Statistics.
Results from the mouse experiments and in vitro growth assays were analyzed by Fisher's exact and Mann-Whitney two-tailed t tests, respectively, using Prism 5.01 software (GraphPad Software). P values less than 0.05 are considered significant.
Microarray data accession number.
Complete microarray data have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE15319.
RESULTS
Adaptation of UPEC to growth in the presence of ASN.
The ASN system is widely used to generate RNIs in vitro (20, 43). When added to MES-LB broth (pH 5), sodium nitrite is converted to nitrous acid, which spontaneously forms NO and other RNIs (3, 62). In the initial subculture from MES-LB broth to MES-LB broth with 3 mM ASN, UPEC cystitis isolate UTI89 has a long lag phase before the start of exponential growth, as shown in Fig. 1. When the bacteria are allowed to reach stationary phase and are then subcultured again into new broth containing ASN, the lag phase is considerably shorter, indicating that the bacteria are now better suited to grow in the presence of ASN, either due to conditioning or due to acquisition of one or more mutations (Fig. 1). To distinguish between these possibilities, we subcultured the short-lag-phase bacteria in broth without ASN, allowed the culture to reach stationary phase, and again challenged the bacteria by subculture with 3 mM ASN. After one round of growth in the absence of ASN, the lag phase returned to the original, longer period (Fig. 1). Thus, a reduction in the length of the lag phase is dependent on previous growth under stress conditions, indicating that adaptation via a conditioning effect, and not via mutation, occurs.
Enhanced colonization of the bladder by ASN-conditioned UPEC.
For some pathogens, increased resistance to certain environmental stresses correlates with enhanced virulence potential (2, 5). Our results indicating that UTI89 can be conditioned to better withstand the stresses generated by in vitro growth in the presence of ASN raised the possibility that ASN-conditioned UPEC may also have an increased capacity to colonize the urinary tract. To address this possibility, adult female CBA/J mice were inoculated via a transurethral catheter with UTI89 that had been grown with shaking to an OD600 of 1.5 in MES-LB broth with or without 3 mM ASN. Bladders were collected at 12 h and 5 days postinoculation, and bacterial titers were determined by serial dilution of tissue homogenates. Bacteria grown in the absence of ASN failed to colonize about 50% of the mice, while the ASN-conditioned bacteria effectively colonized all of the animals at both time points examined (Fig. 2).
A key virulence factor that enables UPEC to effectively colonize the urinary tract is type 1 pili (also called type 1 fimbriae [for a review, see reference 14]). These peritrichous, phase-variable, multisubunit filaments promote UPEC adherence to and invasion of bladder epithelial cells by binding host mannose-containing glycoprotein receptors. Differential expression of type 1 pili by ASN-conditioned UPEC could conceivably account for the enhanced capacity of these microbes to colonize the bladder. However, ASN-conditioned and control bacteria performed equally well in mannose-sensitive, type 1 pilus-dependent yeast agglutination assays. In addition, semiquantitative PCR analysis of the reversible orientation-dependent genetic switch that controls the phase-variable transcription of the fim genes involved in the biogenesis of type 1 pili also demonstrated that ASN conditioning had no significant effect on type 1 pilus expression (data not shown). These results indicate that growth in ASN primes UPEC to better colonize the bladder independent of any overt effects on type 1 pilus expression.
Transcriptional profile of ASN-conditioned UPEC.
To better understand how UTI89 is able to adapt to growth in the presence of ASN and how this facilitates bacterial colonization of the bladder, gene expression profiles of ASN-conditioned UTI89 were analyzed using microarrays. Bacteria from two independent overnight starter cultures were diluted 1:100 into MES-LB broth with or without 3 mM ASN and grown to an OD600 of 1.5, at which point RNA was isolated from each culture and processed separately for hybridization to Affymetrix GeneChip E. coli genome 2.0 arrays. These arrays contain probe sets to detect transcripts from the E. coli K-12 lab strain MG1655, UPEC isolate CFT073, and two enteropathogenic E. coli isolates, O157:H7 strain EDL933 and O157:H7 strain Sakai. Although UTI89 is not specifically represented on the arrays (or any currently available commercial array), our unpublished genomic analysis indicates that only about 271 of the predicted genes in UTI89 are not represented on the chip, making the arrays a valuable tool for study of this pathogenic isolate.
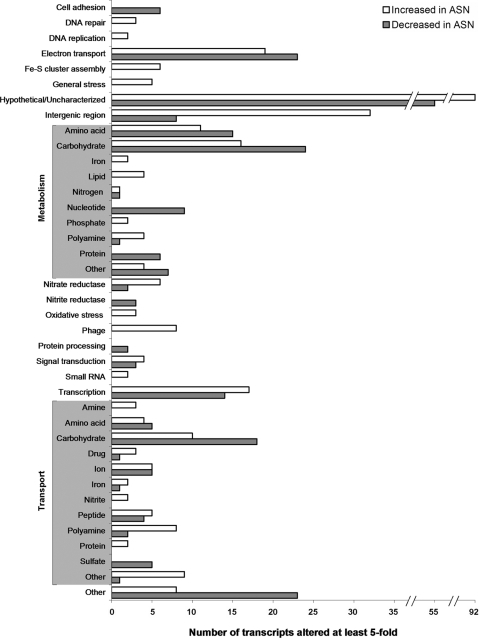
Analysis of biological duplicates using the microarrays showed that there are extensive differences in the transcriptomes of ASN-conditioned UTI89 and untreated controls. A total of 304 genes were upregulated at least fivefold in cultures containing ASN, while 244 genes were downregulated ≥5-fold. A summary of these genes according to functional category is shown in Fig. 3, and the genes included in each category are listed in Table S1 in the supplemental material. Hypothetical and uncharacterized genes constitute the most numerous class affected, with 55 genes downregulated and 92 genes upregulated at least fivefold. The 20 most highly upregulated and downregulated transcripts detected after growth of UTI89 in the presence of ASN are shown in Tables 1 and 2, respectively. Notably, no change in expression of the major type 1 pilus subunit FimA gene was detected in ASN-conditioned UTI89, although transcription of some of the genes encoding the minor type 1 pilus subunits was somewhat elevated. Enhanced expression of other major pilus-related genes, including those encoding P pili and curli, was not detected. In contrast, the expression of numerous genes involved in the transport and metabolism of carbohydrates was highly altered (Fig. 3), perhaps indicating that there was a shift in energy utilization strategies in response to RNI stress-induced respiratory chain damage. These changes may also reflect a shift toward metabolic and catabolic pathways (e.g., the acquisition or generation of substrates for the citric acid cycle) that enhance production of reducing equivalents like NADH and NADPH used by many of the proteins implicated in the detection and detoxification of RNIs. The ASN-induced alterations in carbohydrate transport and metabolism did not correlate with any overt changes in either bacterial colony morphology or encapsulation.
FIG. 3.
Functional classification of transcripts affected ≥5-fold by 3 mM ASN in the growth medium, as determined by microarray analysis. Some transcripts were identified by multiple probe sets listed in Table S1 in the supplemental material, but for clarity each transcript is shown only once.
TABLE 1.
Twenty most upregulated transcripts in the presence of ASN
| Gene | UTI89 locus tag | Function | Ratioa | Probe setb |
|---|---|---|---|---|
| norV | C3072 | Flavorubredoxin | 6,198 | 1768449_s_at |
| 472 | 1768764_s_at | |||
| 284 | 1767683_at | |||
| norW | C3073 | Flavorubredoxin oxidoreductase | 5,572 | 1766754_s_at |
| yeaR | C1995 | Putative tellurite resistance protein | 1,552 | 1768944_s_at |
| ytfE | C4818 | Iron-sulfur cluster repair | 624 | 1761805_s_at |
| hcr | C0876 | Hybrid cluster protein oxidoreductase | 761 | 1761136_s_at |
| yoaG | C1994 | Hypothetical protein | 394 | 1762744_s_at |
| None | C1993 | Hypothetical protein | 308 | 1762869_s_at |
| gabP | C3018 | 4-Aminobutyrate permease | 258 | 1764229_s_at |
| yciF | C1527 | Putative structural protein | 267 | 1762401_s_at |
| None | None | Intergenic region | 178 | 1764414_s_at |
| yibH | C4135 | Putative inner membrane protein | 166 | 1764883_s_at |
| yciE | C1526 | Hypothetical protein | 148 | 1768587_s_at |
| narV | C1683 | Respiratory nitrate reductase 2 gamma chain | 122 | 1763823_at |
| yciG | C1528 | Hypothetical protein | 110 | 1761873_s_at |
| glcA | C3391 | Glycolate transporter | 104 | 1759891_s_at |
| nark | C1420 | Nitrate/nitrite antiporter | 70 | 1764323_s_at |
| None | C4930 | Hypothetical protein | 73 | 1760306_at |
| None | C1068 | Hypothetical protein | 67 | 1761754_s_at |
| None | C1702 | Putative sensor kinase | 59 | 1768460_s_at |
| hcp | C0876 | Hybrid cluster protein | 57 | 1766925_s_at |
Ratio of the expression in cultures containing ASN to the expression in cultures not containing ASN.
When different probe sets for a single gene yielded different results, all probe sets are listed.
TABLE 2.
Twenty most downregulated transcripts in the presence of ASN
| Gene | UTI89 locus tag | Function | Ratioa | Probe setb |
|---|---|---|---|---|
| ynfE | C1774 | Putative dimethyl sulfoxide reductase chain | 0.002 | 1760079_s_at |
| ompW | C1456 | Outer membrane protein W precursor | 0.005 | 1763696_s_at |
| dmsA | C0967 | Anaerobic dimethyl sulfoxide reductase subunit | 0.008 | 1765899_s_at |
| dmsB | C0968 | Anaerobic dimethyl sulfoxide reductase subunit | 0.008 | 1766933_s_at |
| ynfG | C1776 | Probable anaerobic dimethyl sulfoxide reductase chain | 0.008 | 1760438_s_at |
| tdcG | C3547 | l-Serine dehydratase 1 | 0.008 | 1761379_s_at |
| 0.019 | 1767162_s_at | |||
| hybB | C3417 | Probable Ni/Fe hydrogenase cytochrome subunit | 0.009 | 1766320_s_at |
| ompF | C1001 | Outer membrane protein F precursor | 0.010 | 1767981_s_at |
| nrfC | C4662 | Formate-dependent nitrite reductase subunit | 0.010 | 1765634_s_at |
| tnaB | C4262 | Low-affinity tryptophan permease | 0.011 | 1760880_at |
| dmsC | C0969 | Anaerobic dimethyl sulfoxide reductase chain C | 0.012 | 1763364_s_at |
| tdcF | C3548 | Hypothetical protein | 0.012 | 1760738_s_at |
| ydhY | C1866 | Putative ferredoxin-like protein | 0.013 | 1760450_s_at |
| alsA | C4682 | d-Allose transport ATP-binding protein | 0.015 | 1762416_at |
| 0.019 | 1761939_s_at | |||
| ynfH | C1777 | Putative dimethyl sulfoxide reductase subunit | 0.015 | 1762268_s_at |
| ynfF | C1775 | Putative dimethyl sulfoxide reductase subunit | 0.015 | 1764834_s_at |
| yedF | C2131 | Hypothetical protein | 0.016 | 1765812_s_at |
| None | None | Intergenic region | 0.017 | 1768754_s_at |
| yeiC | C2440 | Hypothetical kinase | 0.019 | 1759557_at |
| nrfA | C4660 | Formate-dependent nitrite reductase subunit | 0.019 | 1762253_s_at |
Ratio of the expression in cultures containing ASN to the expression in cultures not containing ASN.
When different probe sets for a single gene yielded different results, all probe sets are listed.
Not surprisingly, the expression of many of the genes shown previously to be rapidly upregulated in E. coli K-12 strains upon exposure to various inducers of nitrosative stress is also significantly elevated in ASN-conditioned UTI89 (21, 28, 35, 43, 49). These genes include norVW (encoding flavorubredoxin and its cognate oxidoreductase; upregulated 6,198- and 5,572-fold, respectively), ytfE (encoding a protein involved in repair of iron-sulfur clusters; upregulated 624-fold), nrdH (encoding a ribonucleotide reductase; upregulated 24.2-fold), hmpA (encoding an NO-detoxifying flavohemoglobin; upregulated 20.9-fold), sufA (encoding an iron-sulfur cluster assembly protein; upregulated 6.4-fold), soxS (encoding a regulator of oxygen stress responses; upregulated 4.2-fold), and other genes involved in iron utilization, detoxification reactions, and nitrate-nitrite metabolism. Increased expression was verified by quantitative RT-PCR for three of these genes, norV, hmpA, and nrdH, which showed 12,717-, 27-, and 8-fold increases, respectively, compared to the 6,198-, 20-, and 24-fold increases determined with the microarrays. It should be noted that our array experiments were not designed to directly probe the immediate response of UTI89 to RNIs. Instead, we collected RNA once the OD600 of the bacteria reached 1.5 to globally assess the status of ASN-conditioned UTI89. In addition to RNIs, these late-log-phase bacteria also likely encounter many other stresses, including nutrient and oxygen limitations. Of note, growth of UTI89 in the presence of ASN to mid-log phase (OD600, ∼0.5) rather than to late log phase (OD600, 1.5) did not significantly enhance bacterial colonization of the bladder (see Fig. S1 in the supplemental material). These data suggest that growth to late log phase helps condition UTI89 for survival within the host in addition to, and perhaps independent of, ASN effects.
Several genes whose expression is altered during the transition of E. coli from an aerobic environment to a microaerobic environment (47) are differentially expressed in ASN-conditioned UTI89, as are many genes that are regulated by the oxygen- and NO-sensitive transcriptional regulator FNR (11). Several FNR-regulated genes, including hmpA and ytfE, are also controlled in part by NsrR, an NO-sensitive member of the Rrf2 family of transcription factors (6, 19, 52, 59). Of the top 20 most upregulated genes in ASN-conditioned UTI89 (Table 1), 5 are repressed by active NsrR. In addition to ytfE, these genes include yeaR (encoding a putative tellurite resistance protein; upregulated 1,552-fold), yoaG (encoding a protein with an unknown function; upregulated 394-fold), and hcp-hcr (encoding a hydroxylamine reductase and its associated oxidoreductase; upregulated 57.2- and 761-fold, respectively). Transcription of the NsrR-repressed gene ygbA, which encodes another hypothetical protein, was also substantially induced (22-fold). A total of 6 of the 12 genes or operons shown in previous studies to be repressed by NsrR in E. coli were upregulated in our assays (6, 19, 50). Interestingly, expression of these six NsrR regulon members (yeaR, hcp-hcr, ytfE, yeaR-yoaG, ygbA, and hmpA) is also elevated in UPEC isolate CFT073 and/or the asymptomatic bacteriuria E. coli strain 83972 either in the urinary tract of mice or human volunteers or when the bacteria are grown in urine (54, 58). These findings suggest that many, but not necessarily all, members of the NsrR regulon have key roles in the adaptation of UPEC to both RNIs in vitro and stresses encountered in vivo within the urinary tract. At least one NsrR-repressed gene, hmpA, has a clear role in the resistance of UPEC to RNIs in vitro (data not shown) (60), but deciphering the specific contributions of NsrR and members of its regulon to UPEC colonization and survival within the host requires further investigation.
Cadaverine enhances UPEC colonization of the bladder.
By microarray analysis, we found that the expression of multiple genes involved in either the transport or metabolism of polyamines was altered fivefold or more in ASN-conditioned UTI89 (Fig. 3; see Table S2 in the supplemental material). Polyamines are ubiquitous polycationic molecules that can modulate myriad cellular functions, including bacterial stress response and virulence cascades (51, 56). Major polyamines produced by bacteria, as well as most other forms of life, include putrescine, spermidine, and cadaverine. In E. coli, putrescine has been shown to significantly enhance the expression of 309 genes collectively referred to as the polyamine modulon (65). We found that 111 (36%) of these genes are upregulated in ASN-conditioned UTI89 twofold or more (see Table S2 in the supplemental material).
Previously, we reported that exposure of UTI89 to ASN stimulates cadaverine production, causing levels of this polyamine to increase within 3 h nearly fivefold compared to controls (7). Cadaverine synthesis is controlled by CadC, an acid-inducible transcriptional regulator of the cad operon, which consists of cadB (encoding a lysine-cadverine antiporter) and cadA (encoding a lysine decarboxylase) (61). Disruption of any of the cad genes abrogates cadaverine synthesis by UTI89 and severely attenuates bacterial growth in the presence of 3 mM ASN (7). In contrast, addition of exogenous cadaverine or other polyamines enhances growth of both wild-type UTI89 and the cad mutants in the presence of ASN. This effect was not due to polyamine-mediated quenching of NO radicals or reduction of the mutation frequency. As determined by microarray analysis, transcription of the cad genes was not elevated in ASN-conditioned UTI89, suggesting that the cad gene products may be transiently induced early during growth in the presence of ASN but not later, as the bacteria approach stationary phase. Interestingly, expression of a CadA homologue, the so-called “constitutive” lysine decarboxylase encoded by ldcC as part of the polyamine modulon, was increased 6.2-fold in ASN-conditioned UTI89 (see Table S2 in the supplemental material).
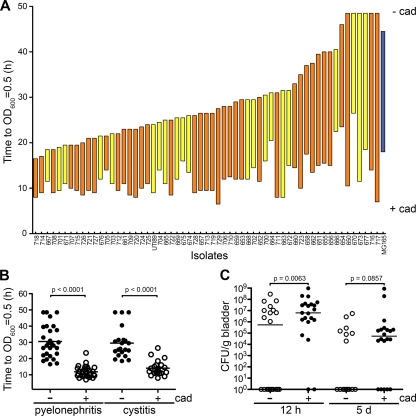
These results suggest that polyamines and members of the polyamine modulon, many of which are stress response genes, are important regulators of UPEC RNI resistance and possibly host colonization. To examine this possibility, we first determined if the ability of cadaverine to enhance the growth of UTI89 in the presence of 3 mM ASN was unique to this UPEC isolate. Twenty-eight pyelonephritis and 21 cystitis UPEC isolates were subcultured into MES-LB broth containing 3 mM ASN with or without 3 mM cadaverine, and the time that it took each culture to reach an OD600 of 0.5 was determined. The concentration of cadaverine used in these assays is less than the levels that are excreted by UTI89 grown in MES-LB broth (7). As shown in Fig. 4A, all but seven of the UPEC isolates grew better in the presence of 3 mM ASN than the K-12 reference strain MG1655. The addition of exogenous cadaverine significantly accelerated the growth of all strains, including MG1655, reducing the time required to reach an OD600 of 0.5 in ASN-containing broth by 7 to >41 h (Fig. 4A and B). Overall, cadaverine had an equalizing effect on the growth of all strains tested, such that the cultures all reached an OD600 of 0.5 in the presence of ASN at more similar rates. No significant differences were observed between the cystitis and pyelonephritis isolates in these assays. Notably, 3 mM cadaverine had no effect on UPEC growth rates in MES-LB broth in the absence of ASN (data not shown) (7).
FIG. 4.
Cadaverine stimulates UPEC growth in the presence of ASN and colonization of the bladder. (A) MG1655 (blue), 28 pyelonephritis isolates (orange), and 21 cystitis isolates (yellow) were diluted from overnight MES-LB broth cultures into fresh medium containing 3 mM ASN with or without 3 mM cadaverine. The top of each bar (− cad) indicates the time required for the individual isolate to reach an OD600 of 0.5 in ASN-containing broth, while the bottom of each bar (+ cad) indicates the time required to reach the same OD600 with both ASN and cadaverine present. (B) Data in panel A grouped to more clearly show the overall effect of cadaverine (cad) on growth of the UPEC isolates in the presence of 3 mM ASN. The P values were calculated using the Mann-Whitney U test. (C) Adult female CBA/J mice were inoculated with 107 CFU of UTI89 that had been grown to an OD600 of 1.5 in MES-LB broth with or without 3 mM cadaverine. Bacterial titers in bladder homogenates were determined at 12 h and 5 days postinoculation. The P values were determined using Fisher's exact test (n = 14 to 22 mice). The horizontal bars in panels B and C indicate median values for the groups.
Considering the effects of cadaverine on UPEC growth in ASN-containing broth and the ability of ASN-conditioned bacteria to better colonize the host, we next asked if growth of UPEC in the presence of 3 mM cadaverine (without ASN) was sufficient to enhance UPEC colonization of the bladder. Adult female CBA/J mice were inoculated via a transurethral catheter with UTI89 that had been grown with shaking to an OD600 of 1.5 in MES-LB broth with or without 3 mM cadaverine. Bladders were collected at 12 h and 5 days postinoculation, and bacterial titers were calculated by serial dilution of tissue homogenates. As seen with ASN-conditioned UTI89, bacteria grown in cadaverine-supplemented broth were much more effective at colonizing the bladder than the bacteria grown in MES-LB broth alone (Fig. 4C). More than 90% of the mice inoculated with cadaverine-treated UTI89 were colonized, while only ∼50% of the mice inoculated with control untreated UTI89 harbored the pathogen at the 12-h time point. A similar, although less substantial, difference was observed at 5 days postinoculation.
DISCUSSION
UPEC likely comes across numerous stresses both within the urinary tract and elsewhere while it is in transit through and between hosts. Transmission via a fecal-oral route alone involves bacterial passage through several diverse and potentially hostile environments, ranging from the oral pharyngeal cavity, stomach, and intestines to the vaginal and/or periurethral regions prior to ascension into the urinary tract. In addition to RNIs, potential stresses generated at these sites include oxygen radicals, antibacterial peptides, shearing forces, variations in osmolarity and pH, and changes in oxygen and nutrient availability. Results presented here indicate that RNIs, and probably other stresses, encountered by UPEC prior to entry into the urinary tract can significantly impact the outcome of an infection. Our data show that UPEC can transiently adapt to high concentrations of ASN and that ASN-conditioned bacteria can better colonize the bladder, possibly via effects on multiple NsrR-regulated genes and other stress- and polyamine-responsive factors.
Compared to a K-12 reference strain, most UPEC isolates have increased resistance to ASN, and this can be substantially augmented by addition of cadaverine (Fig. 4). RNIs, as generated by ASN, can induce cadaverine production by UPEC (7), and cadaverine on its own in the absence of ASN can also promote UPEC colonization of the bladder. These results suggest that ASN may prime UPEC for increased survival within the urinary tract in part by stimulating polyamine production. Microarray-based experiments described here indicate that a significant fraction of the polyamine modulon is upregulated in ASN-conditioned strain UTI89. Many of these genes encode stress response factors that may enable UPEC to better handle the multitude of host defenses in play during the course of a UTI. Polyamines may also increase the virulence potential of UPEC by modifying biofilm formation, affecting the conductivity of outer membrane porins, and/or altering the expression of secreted toxins and iron-chelating agents (51, 56). Notably, polyamines are abundant within the intestinal tract (13, 46) and can also be found in substantial quantities within vaginal secretions (9, 34), where they may interact with and phenotypically modulate UPEC prior to entry into the urinary tract. UPEC may also encounter high levels of polyamines within the urinary tract, where urine polyamine concentrations can be elevated as a consequence of pregnancy or UTI (1, 55).
Our data indicate that UTI89 grown to late log phase with shaking in MES-LB broth cultures have only about a 50% chance of effectively colonizing the mouse urinary tract, suggesting that successful infections in this experimental setup are stochastic events, possibly dependent upon random on and off switching of one or more genes within individual bacteria. By maintaining a phenotypically diverse population under relatively hospitable conditions (i.e., aerated MES-LB broth cultures), UPEC may enhance its chance of survival should its environment take a turn for the worse. We suggest that ASN and cadaverine can act as signals, causing a phenotypic shift of the culture population as a whole so that it possibly becomes less diverse but is overall better suited to handle the rigors of the urinary tract. During a natural infection, UPEC interactions with polyamines or stresses such as RNIs may similarly reprogram the microbes, enhancing their ability to colonize the host. Our microarray analysis indicates that this reprogramming, or conditioning phenomenon, likely involves alteration of multiple regulatory and metabolic pathways, including those involved in stress responses, carbohydrate transport and catabolism, and membrane permeability.
While preexposure of UPEC to RNIs and polyamines can facilitate UPEC colonization of the bladder, it is likely that bacterial adaptation to other environmental factors and stressors can have similar or even opposing effects. For example, we found that acclimatization of UTI89 to osmotic stress during growth in high-salt medium (LB broth with 5% NaCl) renders the pathogen unable to effectively colonize the bladder, in sharp contrast to results obtained with ASN-conditioned UPEC (data not shown). Thus, different stresses encountered by UPEC both within and outside the urinary tract can affect the establishment and progression of a UTI in profoundly different ways. Ultimately, greater understanding of how UPEC assimilates and responds to assorted environmental cues and stresses may bring to light novel approaches to prevent and combat UTIs.
Supplementary Material
Acknowledgments
This study was funded by NIH grants DK068585 and DK069526. J.M.B. was supported in part by NIH genetics training grant T32-GM07464.
We are grateful to R. B. Weiss and D. M. Dunn (University of Utah) for their expert help in acquiring, processing, and analyzing the microarray data. We also thank R. R. Kulesus for assistance with processing genomic data and D. A. Low (University of California, Santa Barbara) and W. E. Hooton (University of Washington School of Medicine) for providing many of the UPEC isolates.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 2 March 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Andersson, A. C., S. Henningsson, and E. Rosengren. 1978. Increased formation of diamines and polyamines in the pregnant rat. J. Physiol. 285311-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, D. L. 1996. Preservation microbiology and safety: evidence that stress enhances virulence and triggers adaptive mutations. Trends Food Sci. Technol. 791-95. [Google Scholar]
- 3.Benjamin, N., and R. Dykhuizen. 1999. Nitric oxide and epitheilial host defense, p. 215-230. In F. C. Fang (ed.), Nitric oxide and infection. Kluwer Academic/Plenum Publishers, New York, NY.
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Bo Andersen, J., B. B. Roldgaard, B. B. Christensen, and T. R. Licht. 2007. Oxygen restriction increases the infective potential of Listeria monocytogenes in vitro in Caco-2 cells and in vivo in guinea pigs. BMC Microbiol. 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bower, J. M., and M. A. Mulvey. 2006. Polyamine-mediated resistance of uropathogenic Escherichia coli to nitrosative stress. J. Bacteriol. 188928-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan, A., P. Roesch, L. Davis, R. Moritz, S. Pellett, and R. A. Welch. 2006. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect. Immun. 741072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, K. C., P. S. Forsyth, T. M. Buchanan, and K. K. Holmes. 1979. Amine content of vaginal fluid from untreated and treated patients with nonspecific vaginitis. J. Clin. Investig. 63828-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, S. L., C. S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooton, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 1035977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Constantinidou, C., J. L. Hobman, L. Griffiths, M. D. Patel, C. W. Penn, J. A. Cole, and T. W. Overton. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 2814802-4815. [DOI] [PubMed] [Google Scholar]
- 12.Corker, H., and R. K. Poole. 2003. Nitric oxide formation by Escherichia coli. Dependence on nitrite reductase, the NO-sensing regulator Fnr, and flavohemoglobin Hmp. J. Biol. Chem. 27831584-31592. [DOI] [PubMed] [Google Scholar]
- 13.Deloyer, P., G. Dandrifosse, C. Bartholomeus, N. Romain, M. Klimek, J. Salmon, P. Gerard, and G. Goessens. 1996. Polyamine and intestinal properties in adult rats. Br. J. Nutr. 76627-637. [DOI] [PubMed] [Google Scholar]
- 14.Dhakal, B. K., R. R. Kulesus, and M. A. Mulvey. 2008. Mechanisms and consequences of bladder cell invasion by uropathogenic Escherichia coli. Eur. J. Clin. Investig. 38(Suppl. 2)2-11. [DOI] [PubMed] [Google Scholar]
- 15.Duncan, C., H. Li, R. Dykhuizen, R. Frazer, P. Johnston, G. MacKnight, L. Smith, K. Lamza, H. McKenzie, L. Batt, D. Kelly, M. Golden, N. Benjamin, and C. Leifert. 1997. Protection against oral and gastrointestinal diseases: importance of dietary nitrate intake, oral nitrate reduction and enterosalivary nitrate circulation. Comp. Biochem. Physiol. Part A 118939-948. [DOI] [PubMed] [Google Scholar]
- 16.Dykhuizen, R. S., R. Frazer, C. Duncan, C. C. Smith, M. Golden, N. Benjamin, and C. Leifert. 1996. Antimicrobial effect of acidified nitrite on gut pathogens: importance of dietary nitrate in host defense. Antimicrob. Agents Chemother. 401422-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, F. C. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2820-832. [DOI] [PubMed] [Google Scholar]
- 18.Fang, F. C. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Investig. 992818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filenko, N., S. Spiro, D. F. Browning, D. Squire, T. W. Overton, J. Cole, and C. Constantinidou. 2007. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J. Bacteriol. 1894410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firmani, M. A., and L. W. Riley. 2002. Reactive nitrogen intermediates have a bacteriostatic effect on Mycobacterium tuberculosis in vitro. J. Clin. Microbiol. 403162-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flatley, J., J. Barrett, S. T. Pullan, M. N. Hughes, J. Green, and R. K. Poole. 2005. Transcriptional responses of Escherichia coli to S-nitrosoglutathione under defined chemostat conditions reveal major changes in methionine biosynthesis. J. Biol. Chem. 28010065-10072. [DOI] [PubMed] [Google Scholar]
- 22.Foxman, B. 1990. Recurring urinary tract infection: incidence and risk factors. Am. J. Public Health 80331-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foxman, B., S. D. Manning, P. Tallman, R. Bauer, L. Zhang, J. S. Koopman, B. Gillespie, J. D. Sobel, and C. F. Marrs. 2002. Uropathogenic Escherichia coli are more likely than commensal E. coli to be shared between heterosexual sex partners. Am. J. Epidemiol. 1561133-1140. [DOI] [PubMed] [Google Scholar]
- 24.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griebling, T. L. 2005. Urologic diseases in America project: trends in resource use for urinary tract infections in women. J. Urol. 1731281-1287. [DOI] [PubMed] [Google Scholar]
- 26.Haraoka, M., L. Hang, B. Frendeus, G. Godaly, M. Burdick, R. Strieter, and C. Svanborg. 1999. Neutrophil recruitment and resistance to urinary tract infection. J. Infect. Dis. 1801220-1229. [DOI] [PubMed] [Google Scholar]
- 27.Hill, M. J. 1991. Nitrates and nitrites in food and water. Woodhead Publishing Limited, Cambridge, United Kingdom.
- 28.Hyduke, D. R., L. R. Jarboe, L. M. Tran, K. J. Chou, and J. C. Liao. 2007. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. USA 1048484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iijima, K., J. Grant, K. McElroy, V. Fyfe, T. Preston, and K. E. McColl. 2003. Novel mechanism of nitrosative stress from dietary nitrate with relevance to gastro-oesophageal junction cancers. Carcinogenesis 241951-1960. [DOI] [PubMed] [Google Scholar]
- 30.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4249-264. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins, D. E., S. A. Chaisson, and A. Matin. 1990. Starvation-induced cross protection against osmotic challenge in Escherichia coli. J. Bacteriol. 1722779-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins, D. E., J. E. Schultz, and A. Matin. 1988. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J. Bacteriol. 1703910-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, J. R., and P. Delavari. 2002. Concurrent fecal colonization with extraintestinal pathogenic Escherichia coli in a homosexual man with recurrent urinary tract infection and in his male sex partner. Clin. Infect. Dis. 35E65-E68. [DOI] [PubMed] [Google Scholar]
- 34.Jones, B. M., M. al-Fattani, and H. Gooch. 1994. The determination of amines in the vaginal secretions of women in health and disease. Int. J. STD AIDS 552-55. [DOI] [PubMed] [Google Scholar]
- 35.Justino, M. C., J. B. Vicente, M. Teixeira, and L. M. Saraiva. 2005. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 2802636-2643. [DOI] [PubMed] [Google Scholar]
- 36.Kang, W. S., F. J. Tamarkin, M. A. Wheeler, and R. M. Weiss. 2004. Rapid up-regulation of endothelial nitric-oxide synthase in a mouse model of Escherichia coli lipopolysaccharide-induced bladder inflammation. J. Pharmacol. Exp. Ther. 310452-458. [DOI] [PubMed] [Google Scholar]
- 37.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulesus, R. R., K. Diaz-Perez, E. S. Slechta, D. S. Eto, and M. A. Mulvey. 2008. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 763019-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lundberg, J. O., I. Ehren, O. Jansson, J. Adolfsson, J. M. Lundberg, E. Weitzberg, K. Alving, and N. P. Wiklund. 1996. Elevated nitric oxide in the urinary bladder in infectious and noninfectious cystitis. Urology 48700-702. [DOI] [PubMed] [Google Scholar]
- 40.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 3451007-1013. [DOI] [PubMed] [Google Scholar]
- 41.Manges, A. R., S. P. Smith, B. J. Lau, C. J. Nuval, J. N. Eisenberg, P. S. Dietrich, and L. W. Riley. 2007. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case-control study. Foodborne Pathog. Dis. 4419-431. [DOI] [PubMed] [Google Scholar]
- 42.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 1741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukhopadhyay, P., M. Zheng, L. A. Bedzyk, R. A. LaRossa, and G. Storz. 2004. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. USA 101745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulvey, M. A., J. D. Schilling, and S. J. Hultgren. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 694572-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mysorekar, I. U., M. A. Mulvey, S. J. Hultgren, and J. I. Gordon. 2002. Molecular regulation of urothelial renewal and host defenses during infection with uropathogenic Escherichia coli. J. Biol. Chem. 2777412-7419. [DOI] [PubMed] [Google Scholar]
- 46.Osborne, D. L., and E. R. Seidel. 1990. Gastrointestinal luminal polyamines: cellular accumulation and enterohepatic circulation. Am. J. Physiol. 258G576-G584. [DOI] [PubMed] [Google Scholar]
- 47.Partridge, J. D., G. Sanguinetti, D. P. Dibden, R. E. Roberts, R. K. Poole, and J. Green. 2007. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J. Biol. Chem. 28211230-11237. [DOI] [PubMed] [Google Scholar]
- 48.Poljakovic, M., M. L. Svensson, C. Svanborg, K. Johansson, B. Larsson, and K. Persson. 2001. Escherichia coli-induced inducible nitric oxide synthase and cyclooxygenase expression in the mouse bladder and kidney. Kidney Int. 59893-904. [DOI] [PubMed] [Google Scholar]
- 49.Pullan, S. T., M. D. Gidley, R. A. Jones, J. Barrett, T. M. Stevanin, R. C. Read, J. Green, and R. K. Poole. 2007. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: unaltered methionine biosynthesis indicates lack of S nitrosation. J. Bacteriol. 1891845-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rankin, L. D., D. M. Bodenmiller, J. D. Partridge, S. F. Nishino, J. C. Spain, and S. Spiro. 2008. Escherichia coli NsrR regulates a pathway for the oxidation of 3-nitrotyramine to 4-hydroxy-3-nitrophenylacetate. J. Bacteriol. 1906170-6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhee, H. J., E. J. Kim, and J. K. Lee. 2007. Physiological polyamines: simple primordial stress molecules. J. Cell. Mol. Med. 11685-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodionov, D. A., I. L. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 1e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronald, A. 2002. The etiology of urinary tract infection: traditional and emerging pathogens. Am. J. Med. 113(Suppl. 1A)14S-19S. [DOI] [PubMed] [Google Scholar]
- 54.Roos, V., and P. Klemm. 2006. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect. Immun. 743565-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Satink, H. P., J. Hessels, A. W. Kingma, G. A. van den Berg, F. A. Muskiet, and M. R. Halie. 1989. Microbial influences on urinary polyamine excretion. Clin. Chim. Acta 179305-314. [DOI] [PubMed] [Google Scholar]
- 56.Shah, P., and E. Swiatlo. 2008. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 684-16. [DOI] [PubMed] [Google Scholar]
- 57.Smith, J. L., P. M. Fratamico, and N. W. Gunther. 2007. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis. 4134-163. [DOI] [PubMed] [Google Scholar]
- 58.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 726373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spiro, S. 2007. Regulators of bacterial responses to nitric oxide. FEMS Microbiol. Rev. 31193-211. [DOI] [PubMed] [Google Scholar]
- 60.Svensson, L., B. I. Marklund, M. Poljakovic, and K. Persson. 2006. Uropathogenic Escherichia coli and tolerance to nitric oxide: the role of flavohemoglobin. J. Urol. 175749-753. [DOI] [PubMed] [Google Scholar]
- 61.Watson, N., D. S. Dunyak, E. L. Rosey, J. L. Slonczewski, and E. R. Olson. 1992. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J. Bacteriol. 174530-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woolford, G., R. J. Casselden, and C. L. Walters. 1972. Gaseous products of the interaction of sodium nitrite with porcine skeletal muscle. Biochem. J. 13082P-83P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, Z., and R. A. Irizarry. 2005. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J. Comput. Biol. 12882-893. [DOI] [PubMed] [Google Scholar]
- 64.Xu, J., X. Xu, and W. Verstraete. 2001. The bactericidal effect and chemical reactions of acidified nitrite under conditions simulating the stomach. J. Appl. Microbiol. 90523-529. [DOI] [PubMed] [Google Scholar]
- 65.Yoshida, M., K. Kashiwagi, A. Shigemasa, S. Taniguchi, K. Yamamoto, H. Makinoshima, A. Ishihama, and K. Igarashi. 2004. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J. Biol. Chem. 27946008-46013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.