Abstract
The virulence of bacterial pathogens is a complex process that requires the dynamic expression of many genes for the pathogens to invade and circumvent host defenses, as well as to proliferate in vivo. In this study, we employed a large-scale screen, signature-tagged mutagenesis (STM), to identify Streptococcus pyogenes virulence genes important for pathogenesis within the host. Approximately 1,200 STM mutants were created and screened using the zebrafish infectious disease model. The transposon insertion site was identified for 29 of the 150 mutants that were considered attenuated for virulence. Previously reported streptococcal virulence genes, such as mga, hasA, amrA, smeZ, and two genes in the sil locus, were identified, confirming the utility of the model for revealing genes important for virulence. Multiple genes not previously implicated in virulence were also identified, including genes encoding putative transporters, hypothetical cytosolic proteins, and macrolide efflux pumps. The STM mutant strains display various levels of attenuation, and multiple separate insertions were identified in either the same gene or the same locus, suggesting that these factors are important for this type of acute, invasive infection. We further examined two such genes, silB and silC of a putative quorum-sensing regulon, and determined that they are significant virulence factors in our model of necrotizing fasciitis. sil locus promoter expression was examined under various in vitro conditions, as well as in zebrafish tissues, and was found to be differentially induced. This study was a unique investigation of S. pyogenes factors required for successful invasive infection.
Severe Streptococcus pyogenes infections, such as streptococcal toxic shock syndrome and necrotizing fasciitis, result in high mortality rates that range from 25% to 50% (7, 43). The severity and incidence of these invasive S. pyogenes diseases have increased since the 1980s despite what had been considered near eradication of S. pyogenes infections in the previous decades of the 20th century due to the development of antibiotics (13, 43). The reasons for this resurgence remain vague, despite numerous reports indicating the prevalence of particular strains associated with severe S. pyogenes infections (9, 13). Recent studies assessed factors that are present in various serotypes, but they failed to determine specific pathogen and host factors that are common denominators of invasive S. pyogenes disease (34, 55). Moreover, from the host perspective, Kotb et al. eloquently illustrated that certain human leukocyte class II alleles confer a predisposition to severe S. pyogenes disease (29). Undoubtedly, the complexity and versatility of this organism, particularly its remarkable ability to alter its broad spectrum of virulence factors in order to survive in different environments, are the foundation for its success.
Although in vitro studies have permitted analysis of virulence determinants, the conditions are selective, as the systems cannot completely mimic host-pathogen interactions. In recent years, new methods have been developed to examine the expression and regulation of bacterial virulence factors in vivo. One of these genetic strategies, signature-tagged mutagenesis (STM), was generated to identify proteins that are required for successful infection or in vivo survival (19). STM requires the creation of several libraries of mutants via random transposon mutagenesis. The mutant strains in each library possess a unique molecular signature or tag, supplied by the transposon, which is easily identifiable either by PCR using primers specific to the tag or by hybridization. Therefore, the STM scheme allows multiple clones to be screened simultaneously with an animal model, resulting in a high-throughput and relatively cost-effective search for important pathogenic determinants. The mutant clones that are not recovered after passage through the host are defective in the ability to survive in vivo and thus are attenuated for virulence.
Since the first STM screen, applied to Salmonella enterica serovar Typhimurium (19), over 40 STM studies have been reported, encompassing an array of bacterial pathogens, diseases, and host model systems (for reviews, see references 2 and 54). STM studies have identified predicted factors directly related to virulence mechanisms, such as adhesins, invasins, and transcriptional regulators. Factors have also been identified that were not previously implicated in virulence or that lack homology to any genes with known or predicted functions. Furthermore, considering the numerous STM screens that have been conducted, there is little overlap of results, reflecting not only the importance of large-scale searches but also the variability of virulence determinants required for different stages and types of infection.
We were able to optimize the conditions necessary to successfully conduct this first reported study of STM applied to S. pyogenes. Of utmost importance to this genetic approach is a suitable model of infection. This study employed a vertebrate animal model, the zebrafish (Danio rerio), which, when infected intramuscularly with S. pyogenes, develops a disease imitating a necrotizing fasciitis disease state (40). The scope of this STM study mirrors that of other studies of bacterial pathogens in that we identified previously known virulence factors, as well as unsuspected determinants. Some of the novel factors have high homology to known genes, although their functions or roles in virulence have not been characterized. Such factors include those of the sil locus, which were originally identified in a mutagenesis study of S. pyogenes employing a murine model of necrotizing fasciitis (23). The importance of silC and silB in virulence was clearly demonstrated, as an organism with targeted mutations in the sil locus proved to be highly attenuated (23). Although the precise function of silC has not been defined, it is hypothesized to be involved in transcriptional regulation of other virulence factors involved in invasive S. pyogenes disease (14). We sought to further characterize the sil locus by examining the infection profile using our streptococcus-zebrafish model of severe disease and to better understand the environmental conditions under which factors of the sil locus might be induced.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Plasmids were maintained in Escherichia coli TOP10 cells (Invitrogen) and cultured aerobically in Luria-Bertani medium at 37°C. When necessary, Luria-Bertani medium was supplemented with 25 μg/ml kanamycin, with 750 μg/ml erythromycin (Erm), or with 1.4% Bacto agar (BBL) to create solid medium. The S. pyogenes M14 HSC5 (18, 35) strain used in this study was cultured anaerobically in Todd-Hewitt medium (BBL) supplemented with 0.2% yeast extract or in this medium supplemented with 2% proteose peptone (BBL) (TP) and incubated in air-tight 15-ml conical tubes at 37°C without shaking. The minimal media used included M9 (M9 salts, 0.2% glucose, 1 mM MgSO4, 100 μM CaCl2, 1% Casamino Acids, 0.3% yeast extract, 0.0001% vitamin B) and C media (32). The antibiotic concentrations used for S. pyogenes cultures were as follows: 500 μg/ml kanamycin and 1 μg/ml Erm. S. pyogenes cultured on solid medium (TP supplemented with 1.4% agar) was incubated in an anaerobic gas chamber with GasPak envelopes (BBL) at 37°C.
Generation of the S. pyogenes transposon mutant library.
Twelve uniquely tagged pJDM-STM vectors were created previously (38) and used to transform the HSC5 strain of S. pyogenes. These vectors contain a transposon, Tn4001 (32), that inserts randomly and in single copy into the S. pyogenes chromosome. Each of the vectors, pJDM-STM-1 to pJDM-STM-12, was isolated from E. coli TOP10 (Invitrogen) cells using a plasmid purification kit (Qiagen), and transformation of S. pyogenes was accomplished using a previously established protocol (6). Thus, 12 separate libraries of transformants were created, resulting in approximately 1,200 STM mutants. The mutants were stored at −80°C in 96-well plates with 30% glycerol.
Preparation of streptococci.
One mutant strain from each of the 12 libraries was cultured individually overnight and diluted 1:50 into fresh TP until the optical density at 600 nm (OD600) was 0.265, which corresponds to mid-log phase or 108 CFU/ml. The cultures were washed once, resuspended in fresh TP to a concentration of 108 CFU/ml, combined to form a pool of 12 mutant strains, and diluted to obtain a concentration of 107 CFU/ml. The concentration of the bacterial cells was confirmed by plating serial dilutions onto solid media.
Zebrafish infection.
Prior to infection, zebrafish were maintained as previously reported (40). Zebrafish were anesthetized in Tris-buffered Tricaine (pH 7.0) (168 μg/ml; 3-aminobenzoic acid ethyl ester; Sigma), and using a 0.3-ml U-100 ultrafine syringe, 10 μl of the pooled bacterial culture was injected into the dorsal muscle of four fish, as described previously (40), resulting in a total infectious dose of 105 CFU. The infected fish, as well as fish mock inoculated with sterile medium as a negative control and fish infected with the wild-type strain as a positive control, were placed separately into 400-ml glass beakers containing 225 ml of double-distilled water supplemented with 60 mg/liter of aquatic salts (Instant Ocean) and placed in a glass front incubator at 28°C.
At 24 h postinfection, three of the four fish infected with the pool of mutant bacteria (input pool) were euthanized in 100 ml of a 320-μg/ml Tricaine (Sigma) solution. The spleens and muscle (site of injection) of each fish were aseptically removed, homogenized in 200 μl of phosphate-buffered saline (PBS), and cultured overnight in 10 ml of TP containing Erm at 37°C. Genomic DNA was isolated from bacterial cells from both the input pool and the output pool (homogenate) (Wizard genomic DNA purification kit; Promega). The genomic DNA of both pools was subjected to PCR analysis using primers specific for each of the 12 tags and for the Erm resistance gene, as described previously (38).
The protocols for the zebrafish infections were approved by the Wayne State University Institutional Animal Care and Use Committee and followed all federal regulations regarding the care and use of laboratory animals.
DNA sequencing of tagged mutants.
Each of the potentially attenuated strains was scored by using a general categorization scheme based on the specific organ (muscle and/or spleen) and the number of organs, as well as the number of fish from which the mutant was missing. For example, four mutant strains were identified as missing from the output pools of both organs (spleen and muscle) of all three fish examined, and seven STM mutants were absent from one organ (the spleen) of all three fish. The 29 mutants in the categories with the greatest number of factors, indicating significant attenuation of virulence, were selected for further sequence analysis. Selected mutant strains that were absent from the recovered pool but were present in the inoculum pool were prepared for sequencing using a genomic DNA isolation kit (Qiagen). The specific sites of transposon insertion were determined by purifying chromosomal DNA from an identified mutant and sequencing off of the end of the transposon into the adjacent chromosomal DNA using primer 5′ Tn4001 Rev (5′-CTTGGGTCATGTAAAAGTCCTCTGGGTATG-3′). Sequencing of chromosomal DNA was performed by Fidelity Systems (Gaithersburg, MD). Direct chromosomal sequencing of the transposon insertion, as opposed to subcloning and sequencing, allowed confirmation of single transposon insertions, whereas if multiple insertions had occurred in the chromosome, getting a single pure sequence off of the transposon would not have been possible. Previous reports have demonstrated the ability of a single copy of the Tn4001 transposon to insert into the HSC5 S. pyogenes genome (15, 38). Each sequence was subsequently analyzed by comparison to known sequences in the databases of the National Center for Biotechnology Information.
Competitive assays.
S. pyogenes strains were prepared as explained above, resulting in cultures containing 108 CFU/ml for each STM mutant strain and the wild-type strain. The two strains were mixed at a 1:1 ratio and diluted to obtain a culture containing 107 CFU/ml. Ten microliters of the 107-CFU/ml culture (105 CFU injection) was used to infect zebrafish intramuscularly, and the culture was serially diluted and plated to confirm the input ratio of the mutant to the wild-type strain. At 24 h postinfection, the spleens and muscles of eight euthanized infected zebrafish were aseptically dissected and homogenized. Dilutions of the homogenates were plated onto solid media with and without Erm to determine the output ratio of the mutant to the wild-type strain. The competitive index (CI) was calculated by dividing the output ratio by the input ratio. A CI of <1 indicates that there was attenuation compared to the wild-type strain, which had a CI of 1.
Determination of the LD50.
The 50% lethal dose (LD50) was determined by infecting six zebrafish intramuscularly per inoculum using doses ranging from 1 × 103 to 1 × 106 CFU for each strain analyzed. At least three separate experiments (n = 18) were conducted, and the LD50 was calculated using the equation of Reed and Muench (50).
Construction of an in-frame mutant of the hypothetical cytosolic mutant.
Using the sequence of the MGAS315 strain (accession number NC_004070), we designed primers to amplify a region ∼500 bp upstream of the SPyM3_0029 gene and another ∼460-bp region downstream of this gene. The upstream fragment included the first three codons of the coding region. Similarly, the downstream fragment contained the last three codons of the gene. SalI restriction sites were included in the 3′ primer for the upstream sequence and the 5′ primer for the downstream sequence. The amplified fragments were then digested with SalI, ligated together into the PCR 8/GW/TOPO vector (Invitrogen), and transformed into E. coli TOP10 cells (Invitrogen). The resulting construct was digested with SalI, ligated with a fragment containing a kanamycin resistance cassette cut from vector pABG-5 (17) by SalI restriction digestion, and cloned into E. coli TOP10 cells (Invitrogen). The ∼2,500-bp fragment containing the first and last three codons of hyp-0029 interrupted with the kanamycin resistance cassette was then ligated into the E. coli-streptococcus shuttle vector pJRS233 (44). This vector was used to replace the wild-type allele in S. pyogenes HSC5 using a previously described method (53). The chromosomal structure of the mutant allele was confirmed by PCR.
sil locus promoter-reporter fusion construct.
The sil locus promoter construct was created by PCR amplification of the ∼300-bp region of the sil locus using the HSC5 chromosome as the template and primers 5′silpro BamHI (CGC GGA TCC CTG AAG CCA CCC GTT TTC) and 3′silpro EcoRI (CCG GAA TTC CCT CTA AGA CAA AAA TAT TC). The fragment was cloned into pMNN1, a derivative of pABG5 that contains the gene encoding the PhoZ alkaline phosphatase of Enterococcus faecalis under the control of the rofA promoter but maintains PhoZ on the cell membrane (17, 41). The PCR fragment containing the sil locus promoter was cloned into the BamHI and EcoRI sites, replacing the rofA promoter on pMNN1. The plasmid was propagated in E. coli TOP10 cells (Invitrogen), and the purified plasmid was then used to transform the HSC5 strains as described previously (6). The negative control included a vector that contained the phoZ reporter gene without a promoter.
Alkaline phosphatase liquid assay.
Ten-milliliter portions of the cultures were grown in TP overnight in 15-ml screw-cap conical tubes at 37°C. The growth conditions varied according to the assay; for example, a culture was grown at 30°C or in a 50-ml flask in a 37°C shaking incubator, as detailed in Results. Log-phase cultures were treated alongside untreated cultures with 5 mM and 7 mM H2O2 or were centrifuged and resuspended in wild-type supernatant and cultured for an additional 2 h at 37°C. Culture concentrations were normalized to an OD600 of 0.750 with TP, and 50 μl of each normalized sample was placed into the wells of a 96-well plate in triplicate along with 200 μl of 1-mg/ml p-nitrophenyl phosphate (Sigma) suspended in 1 M Tris (pH 8). Following incubation in the dark for 1 h at room temperature, three optical densities were determined: OD405, OD550, and OD600. Activity was determined by using the following formula: [OD405 − (1.75 × OD550)]/(volume × time × OD600) × 1,000. Each assay was repeated a minimum of three times.
Histology of S. pyogenes-infected zebrafish.
Zebrafish were inoculated intramuscularly as described above, and histology sections were prepared as reported previously (45). Tissue sections from the zebrafish infected with the reporter construct were stained with an antiserum to detect S. pyogenes (Lee Laboratories, Grayson, GA) at a dilution of 1:500 and incubated for 2 h in a humidified atmosphere at room temperature, after which they were washed with PBS and stained with goat anti-rabbit antibody conjugated to Alexa Fluor 488 (Molecular Probes) at a dilution of 1:2,500 for 30 min. Following extensive washing in PBS, the sections were stained with antibody against alkaline phosphatase (a gift from Michael Caparon) at a dilution of 1:10 and incubated as described above for 2 h. Staining with a secondary goat anti-rabbit antibody conjugated to Alexa Fluor 568 (Molecular Probes) at a dilution of 1:2,500 for 30 min followed further washing in PBS.
Statistical analysis.
Statistical significance was determined using a two-tailed paired t test with the StatView statistical analysis software (SAS Institute Inc., Cary, NC).
RESULTS
Generation of S. pyogenes signature-tagged transposon mutants.
Plasmids pJDM-STM-1 through pJDM-STM-12 (38), each carrying a modified Tn4001 transposon (32) and 1 of 12 unique 34-bp oligonucleotides inserted between two unique restrictions sites, were used for STM of the S. pyogenes M14 HSC5 strain. Each vector was used to transform S. pyogenes, resulting in 12 separate libraries and approximately 1,200 mutant strains. Previous work with pJDM-STM-1 to pJDM-STM-12 (38), as well as previous reports of Tn4001 in HSC5 (15, 38), demonstrated that transposition with Tn4001 results in single random integration of the transposon into the chromosome.
Screening the STM library.
The STM library was screened in a zebrafish necrotizing fasciitis model system. S. pyogenes injected into the dorsal muscle of the zebrafish produces a largely localized infection (40). Therefore, it was necessary to optimize the technique in order to obtain appropriate inoculum levels to establish an infection in the host but not overwhelm the immune system, as well as to determine from which organs and at what time postinfection a significant bacterial load may be recovered. We determined that 105 CFU was the optimal combined total dose for infection, as it proved to be difficult to recover all 12 mutants from the infection pool with a lower dose and a higher dose overwhelmed the immune system of the host, causing an increased mortality rate. With this infectious dose, a significant bacterial load consisting of a representative group of all 12 injected strains was recovered from the spleen, as well as the muscle, of the fish by 24 h postinjection.
The wild-type strain and one strain from each of the 12 libraries were cultured individually (to screen for growth deficiencies) and allowed to grow to mid-log phase (see Materials and Methods), at which time the concentrations of the mutant strains were normalized to 108 CFU/ml, the mutant strains were pooled (producing the input pool), and a total dose of 105 CFU was used to infect four zebrafish intramuscularly. Three of the four fish used were euthanized at 24 h postinfection. Whole spleens and muscle tissue from the site of injection (approximately 0.5 cm3) were aseptically dissected from the euthanized fish. DNA, isolated from the bacteria recovered from the muscles and spleens of the infected fish (output pool), as well as from the input (in vitro) pool, was then subjected to PCR analysis using primers specific for the 12 individual tags and the Erm resistance gene in the transposon. Strains with PCR fragments not present in the output pool but present in the input pool were identified as mutants that were cleared from the tissue during infection, indicating that there was attenuation of virulence (Fig. 1). The STM screen revealed approximately 150 mutants that were putatively attenuated for virulence.
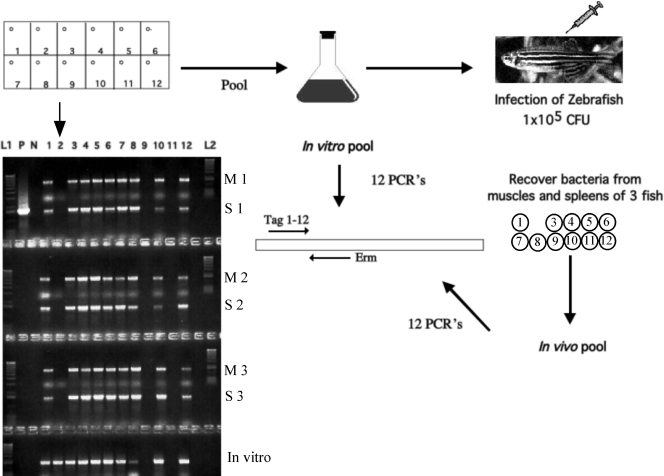
FIG. 1.
Diagram of STM. Mutant strains were pooled, and a mixed inoculum containing 105 CFU was injected into the dorsal muscle of four zebrafish. At 24 h, zebrafish were euthanized, and bacterial DNA was isolated from the homogenized spleens and muscles of three fish. Both input and output pools were subjected to PCR analysis with a specific primer for each of the 12 tags (lanes 1 to 12). The gel shows that the STM-2 clone was missing from the output pools of both organs of all three fish but was present in the input pool. Also missing were STM-9 and STM-11, but since they were also missing from the input (in vitro) pool, they were not considered further. Lanes L1 and L2, DNA ladder; lane C, positive control amplified from an STM plasmid. M, muscle; S, spleen.
Each of the potentially attenuated strains was scored by using a general categorization scheme based on the specific organ(s) and the number of organs, as well as the number of fish from which the mutant was missing. The 29 mutants in the categories with the greatest number of factors, indicating significant attenuation of virulence, were selected for further analysis (Table 1).
TABLE 1.
S. pyogenes sequences identified by STMa
| Category | Strain | Gene(s) (% identity)b | Putative function | Tnp locationc | CId | LD50 (CFU) |
|---|---|---|---|---|---|---|
| Transport or binding | A4-12 | SPyM3_1649 (95) | ABC transporter (salT) | 76/494 | 0.328 | 1 × 105 |
| D10-12 | SPyM3_0233, SPyM1_0271 (96) | ABC transporter substrate-binding protein | 232/281 | 0.0011 | NDg | |
| D11-5 | SPyM3_0013, SPyM1_0014 (97) | Putative amino acid permease | I | 0.0011 | ND | |
| F4-11 | SPyM1_0275 (94) | Serine/threonine sodium transporter | I | 0.004 | ND | |
| C7-12 | SPyM3_0386 (32)e | No homology to S. pyogenes in nucleotide sequence; macrolide efflux protein (mpf) | 12/404 | 0.0174 | 1 × 106 | |
| C1-7 | 45/404 | 0.0747 | ND | |||
| E1-12 | 92/404 | 0.0693 | ND | |||
| A10-12 | 94/404 | 0.0566 | ND | |||
| H8-12 | 95/404 | 0.005 | ND | |||
| E12-8 | 113/404 | 0.0246 | ND | |||
| H11-8 | 136/404 | 0.0564 | ND | |||
| F7-8 | 203/404 | 0.0207 | ND | |||
| F2-9 | 232/404 | 0.1749 | 2 × 103 | |||
| G10-12 | SPyM3_0386 (99) | Macrolide efflux protein (mefE) | 385/391 | 0.0135 | 5 × 105 | |
| G12-1 | 386/391 | 0.1666 | ND | |||
| Cellular processes | A11-8 | SPyM3_1851, SPyM1_1851 (96) | Hyaluronan synthase (hasA) | I | 0.0071 | ND |
| B10-1 | SPyM3_0385, SPyM1_0449 (96) | Nucleotide sugar dehydrogenase | I | 0.076 | ND | |
| B12-7 | AF493605.1 (95)f | Putative sensor histidine kinase (silB) | 199/437 | 0.101 | ND | |
| Secreted proteins | F2-1 | SPyM3_1716 (92) | Mitogenic exotoxin Z (smeZ) | 103/131 | 0.269 | 7.4 × 104 |
| G8-6 | SPyM1_1702 (92) | I | 0.153 | 4.6 × 105 | ||
| Regulators | A12-7 | SPyM3_1728, SPyM1_1720 (91) | trans-acting positive regulator (mga) | I | 0.0434 | ND |
| C1-1 | SPyM1_0612 (94) | Transcriptional activator (amrA) | 193/428 | 0.182 | ND | |
| G5-8 | AF493605.1 (95)f | Putative transcriptional regulator (silC) | 35/39 | 0.307 | ND | |
| Conserved hypothetical proteins | C3-2 | SPyM3_029 (97) | Hypothetical cytosolic protein | 346/446 | 0.0005 | ND |
| C7-7 | 217/446 | 0.0011 | ND | |||
| G4-12 | 346/446 | 0.0074 | ND | |||
| G8-8 | 361/446 | 0.0137 | ND | |||
| F2-5 | SPyM1_0462 (98) | Hypothetical cytosolic protein | 100/274 | 0.0854 | ND | |
| F10-6 | SPyM3_1795, SPyM1_1794 (95) | Conserved hypothetical protein | I | 0.0677 | ND |
For the wild type the CI is 1 and the LD50 is 3×103 CFU.
The locus tag from S. pyogenes serotype M1 strain MGAS5005 (accession number NC_007297) and/or M3 serotype strain MGAS315 (accession number NC_004070) strain is indicated. Unless indicated otherwise, the value in parentheses is the level of identity at the nucleotide level. When two genes are listed for one strain, the percent identity applies to both genes.
Amino acid location of the transposon insertion/total number of amino acids in the protein. “I” indicates that the transposon is inserted in an intergenic region upstream of the protein indicated.
Wild-type and mutant strains were injected at a 1:1 ratio; a CI of <1 indicates attenuation.
Level of identity at the amino acid level.
The accession number rather than the gene is indicated.
ND, not determined.
Analysis of STM attenuated virulence determinants.
The site of the transposon insertion and the genes potentially responsible for the attenuated phenotype were identified via sequence analysis. For the 29 strains sequenced, transposon insertions were identified in six previously determined S. pyogenes virulence factors: two regulators (mga and amrA), a capsule biosynthesis gene (hasA), a superantigen (smeZ), and genes of a putative quorum-sensing regulon (silC, encoding a putative transcriptional regulator, and silB, encoding a histidine kinase) (Table 1). The identification of known virulence factors validated the efficacy of our virulence screen with zebrafish for revealing genes that are important for S. pyogenes pathogenesis. Other categories, listed in Table 1, comprise genes involved in cellular processes and transport and three genes that encode conserved hypothetical proteins.
Multiple single-insertion mutants were identified with transposon insertions in separate regions of the same gene. The majority of the previously published STM studies have reported multiple single-insertion mutants for STM candidates, which may be indicative of transposons that do not insert in an absolutely random manner or may indicate that the isolated genes are critical for in vivo survival in the infectious model and disease state. Since the insertions in most of these genes were found to be in distinct positions, we hypothesize that they do not define hot spots but rather factors that have key functions in this type of S. pyogenes invasive disease.
Two mutants had single insertions in the gene encoding the superantigen SmeZ and in mefE, which encodes a protein with homology to a macrolide efflux pump. Surprisingly, another eight mutants were identified with single insertions in different regions of a gene that we have designated mfp (Table 1). Although mfp has no DNA sequence homology to known genes, at the protein level its product is 55% homologous to a putative macrolide efflux pump. Four independent insertions in a hypothetical cytosolic protein were revealed, and these mutants are among the most highly attenuated STM mutants evaluated (Table 1). The gene encoding this hypothetical protein is upstream of the purB gene, which encodes adenylosuccinate lyase, a key enzyme in the purine biosynthetic pathway. Upon closer examination of the transposon insertions upstream of purB, we questioned whether these insertions actually created polar mutations, disrupting transcription of the downstream purB gene. The hypothetical protein gene is transcribed in the same direction as purB, and the majority of the inserts are toward the 3′ region of the gene. Consequently, a mutant with a nonpolar, in-frame deletion of the gene encoding the hypothetical cytosolic protein was constructed and tested in the zebrafish model. This mutant strain displayed the same degree of virulence in the zebrafish model as the wild-type strain, in contrast to the high level of attenuation of the mutants with transposon insertions in the same gene (data not shown). Furthermore, reverse transcription-PCR analysis revealed that the transposon inserts upstream of purB create a polar mutation, indicating that the attenuated phenotype of these STM mutants is due to a disruption of purB (data not shown).
Two mutants with insertions in a gene that encodes a known secreted protein, SmeZ, were identified in the STM screen. Theoretically, an STM screen would not permit identification of this class of genes, as the interrupted genes encoding the secreted proteins would be transcomplemented by the expression of the genes in the mixed input pool. Therefore, identification of smeZ suggests that the gene product does not diffuse out or implement its mode of action at a considerable distance from the bacterial cell; rather, it remains in close proximity, whereby the inactivation cannot be compensated. These findings emphasize the sensitivity of the screen and suggest that these determinants are critical for the disease state.
Competition assays and determination of the LD50 of STM mutants.
To rule out the possibility that attenuation of the mutant strains was due to a general growth defect, an in vitro growth curve was generated for each of the mutants listed in Table 1, as well as the wild-type strain. The growth assays were performed in rich medium (TP) as well as two types of minimal media, M9 and C media (see Materials and Methods). None of the mutants demonstrated a lower growth rate at any growth phase than the wild type in any of the three media (data not shown).
Virulence attenuation of the STM mutants was confirmed and quantified by CI assays and LD50 experiments, as described in Materials and Methods. Using both of these methods also allowed comparative analysis of the ability of a mutant strain to survive in the presence and in the absence of the wild-type strain.
When the mutants were analyzed for attenuation in the presence of the wild-type strain in the competition assays, a wide range of CIs was observed, from 0.5025 for the smeZ mutant (∼0.5 log) to 0.0005 for the hypothetical cytosolic protein mutant, corresponding to >1,000-fold attenuation (Table 1). Additionally, a range of LD50s were observed with the mfp multiple-insertion mutant, and one of the strains had the lowest LD50, which was approximately the same as that of the wild-type strain. However, we observed much higher LD50s with two other mfp mutant strains, suggesting that certain insertions in this gene may disrupt some portion of its product (e.g., the transmembrane domain) that plays a vital role in its function. Furthermore, we observed a trend in comparisons of the CIs and LD50s of the mutant strains that produce putative secreted proteins. The smeZ mutants had CIs that were significantly lower than the level of attenuation as determined by the LD50 (Table 1). This phenotype was consistent with what would be expected for a strain that may be complemented to some degree by the wild-type strain in the competition assays but displayed a higher level of attenuation when the zebrafish were infected with each strain individually. The same phenotype was observed with the mutant with a mutation in salT, which encodes a putative ABC transporter. The CI computed for the salT mutant revealed a fairly subtle level of attenuation (0.442), whereas the LD50 was 2 logs higher than that of the wild-type strain. In this instance, the presence of the wild-type strain in the CI assay may complement the SalT mutant by providing the missing transported product.
Characterization of the sil locus.
The sil locus gene was identified previously in an S. pyogenes screen utilizing a murine model of necrotizing fasciitis with an M14 strain of S. pyogenes that was isolated from a human patient with the disease (23). The sil locus, comprised of five open reading frames (ORFs), is highly homologous to a quorum-sensing regulon and peptide-sensing system of Streptococcus pneumoniae (12, 23, 25). The first two ORFs, silA and silB, encode a two-component regulatory system, while the last two ORFs, silD and silE, produce ABC transporters (14, 23). Although the silC sequence exhibits no homology to a known sequence, the small peptide encoded by silC is postulated to be a transcriptional regulator of S. pyogenes virulence factors, the transcription of which is suppressed by a signaling molecule transcribed complementary to and overlapping silC, referred to as SilCR, which is found only in some S. pyogenes strains (14). In turn, the small peptide encoded by silC was found to suppress the transcription of silCR, thus forming a transcriptional circuit via a sensing system and coordinately regulating the expression of S. pyogenes virulence factors (14). It should be noted that the HSC5 M14 strain used in this study has a mutation in the start codon of silCR, as described previously for another M14 S. pyogenes strain isolated from a patient with necrotizing fasciitis (23). Consequently, SilCR is not produced in HSC5, and as a result, our transcriptional analysis of the sil locus would not have been influenced by the divergently transcribed peptide. As mentioned above, two of the STM transposon insertions in our study mapped to the sil locus, with an insert in each of the silC and silB genes, suggesting that this locus plays a vital role in this specific type of disease state.
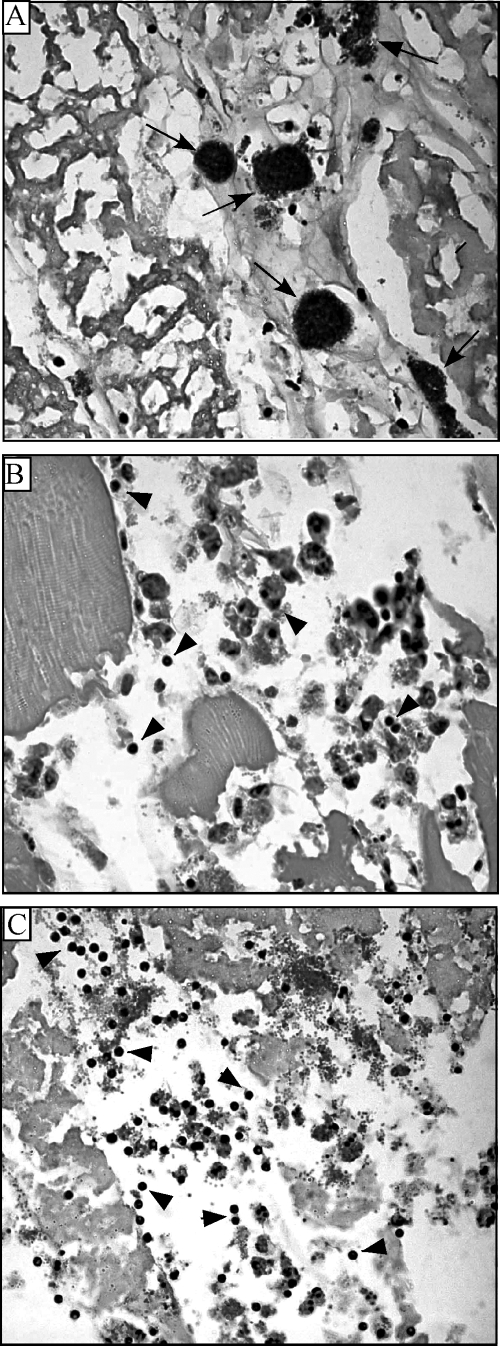
Correlations have been observed between histopathologic characteristics of necrotizing fasciitis infections in animal models and humans. Specifically, there is a lack of neutrophil infiltrate at the site of infection, where a heavy bacterial load has been found (3, 10, 21, 58). The same feature has been observed in our zebrafish necrotizing fasciitis model (40). The wild-type strain, injected intramuscularly, produces significant necrosis of the muscle tissue and few inflammatory cells at the site of infection, where the bacterial cells are in large aggregates (Fig. 2A). In contrast, inflammatory cells are present and closely associated with the bacterial cells in an infection with either a silC or silB mutant (Fig. 2B and C).
FIG. 2.
Zebrafish histology after S. pyogenes infection. Tissue sections were prepared from the dorsal muscle 24 h postinfection with 105 CFU wild-type bacteria (A), a silB mutant (B), and a silC mutant (C) and stained with hematoxylin and eosin. The arrows in panel A point to aggregates of bacteria, and the arrowheads in panels B and C point to inflammatory cells at the site of infection. Magnification, ×1,000.
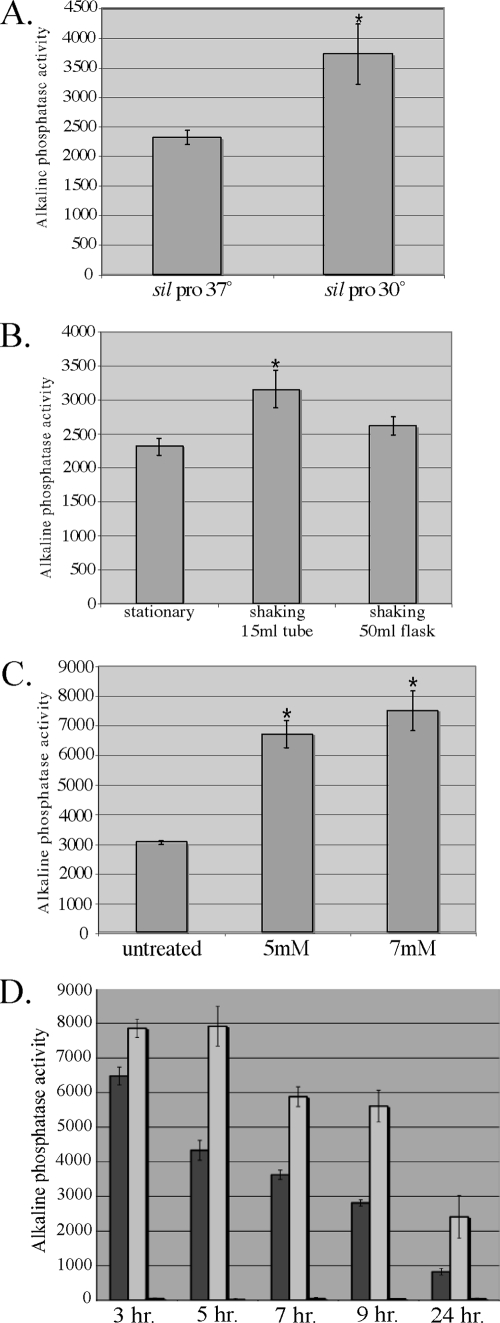
In an effort to further characterize the sil locus, we created a reporter construct by cloning the promoter sequence preceding the sil locus upstream of the phoZ gene, which encodes alkaline phosphatase (17), the expression of which is detected by antibody staining or is measured by a colorimetric assay. Because the sil locus is highly homologous to a quorum-sensing regulon and silC encodes a putative virulence regulator, the expression of the sil locus promoter was examined in a variety of environmental conditions by examining the strain carrying the reporter construct with several in vitro assays. In each assay a negative control vector that lacks a promoter upstream of the phoZ gene was also assayed. We initially examined the effects of temperature variation. Both strains were cultured overnight in normal laboratory medium (TP) at 30°C and 37°C. The cultures were then treated with a phosphatase substrate to gauge alkaline phosphatase activity (as described in Materials and Methods), providing a direct measure of promoter expression. The sil locus promoter was expressed at significantly higher levels (P < 0.0001) at the lower temperature, 30°C (Fig. 3A), indicating that the locus is differentially regulated under certain environmental conditions.
FIG. 3.
Analysis of sil promoter expression. (A) Response to temperature. Overnight cultures of the wild-type strain carrying the sil promoter construct were grown at 37°C and 30°C (*, P < 0.0001). (B) Response to oxygen tension and agitation. The HSC5 wild-type strain carrying the sil promoter construct was cultured in 10 ml of TP under the following conditions: in a 15-ml conical tube without shaking, in a 15-ml conical tube with shaking, or in a 50-ml flask with shaking overnight (*, P < 0.0001). (C) Response to H2O2 treatment. The wild-type strain carrying the sil promoter construct was grown to mid-log phase and then treated with 5 mM or 7 mM H2O2 for 2 h at 37°C (*, P < 0.001). (D) Response to growth phase. The wild-type strain (dark gray bars), the silC mutant strain (light gray bars), and the silB mutant strain (black bars) carrying the sil promoter construct were assayed for activity at different times during growth.
As the in vivo environment is considered to be a high-stress environment, we wanted to determine whether certain stress factors have an effect on sil promoter expression and therefore investigated conditions in which there was increased oxygen tension and there were increasing concentrations of H2O2. Since S. pyogenes growth is optimal under oxygen-limiting conditions, we compared the promoter expression of a 10-ml stationary culture in a 15-ml conical tube at 37°C to that of a 10-ml culture in a 50-ml flask or a 15-ml tube shaking in a water bath at the same temperature. Treatment with the substrate indicated that the level of promoter expression was slightly but significantly (P < 0.0001) higher(1.36-fold) in the culture shaking in the 15-ml tube in the water bath, suggesting that the agitation rather than the increased exposure to oxygen in the 50-ml flask induced the promoter (Fig. 3B). Expression of the promoter was also significantly induced (P < 0.001) upon exposure to two different concentrations of H2O2 compared to the expression in an untreated culture (Fig. 3C). These results demonstrate that sil promoter expression is upregulated when a culture is subjected to specific stressors.
Quorum-sensing molecules are often secreted and autoregulated or are regulated by alternative secreted factors through sensing by an associated sensor kinase-response regulator two-component system. If factors secreted into the supernatant interact with the SilB histidine kinase to cause a change in the sil locus expression, then treatment of a culture carrying the sil promoter construct with the supernatant of a culture in stationary phase may result in a change in sil promoter expression. Therefore, a strain containing the promoter construct grown to early log phase was resuspended with the filtered supernatant of an overnight culture of the parent strain and incubated for 2 h under normal conditions. However, the supernatant treatment resulted in no difference in promoter expression compared to the medium-treated control (data not shown). These results suggest that if silC encodes a secreted molecule that interacts with SilB, the molecule remains cell associated rather than diffusing out to a significant distance from the cell or that no other secreted factors present in the supernatant at this time point regulate the sil locus.
To determine the growth phase in which the sil locus is most highly expressed, we examined the promoter expression at various time points after dilution from an overnight culture into fresh media, including 3, 5, 7, 9, and 24 h (Fig. 3D). Promoter expression was examined over time for the wild-type strain, as well as both the silB and the silC insertion mutant strains. In the wild-type strain, the greatest level of expression was observed at 3 h, and expression decreased in a stepwise fashion at the subsequent time points, indicating that high cell density is not the major factor influencing the upregulation of the sil locus, as observed with most quorum-sensing systems. In fact, the data are consistent with high cell density acting as a negative regulatory factor of sil locus expression. This regulation is lost when sil promoter expression is measured in the silC mutant strain. In addition, for each time point, the expression is higher than that in the wild-type strain (Fig. 3D). These data suggest that SilC plays a role, either directly or indirectly, in regulation of the promoter upstream of the locus. Not surprisingly, expression of the promoter is completely lost when it is measured in the silB mutant strain, as silB encodes the histidine kinase sensor of the two-component system that putatively controls expression of genes in the sil locus.
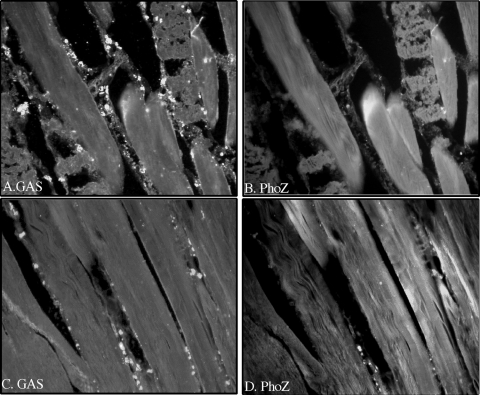
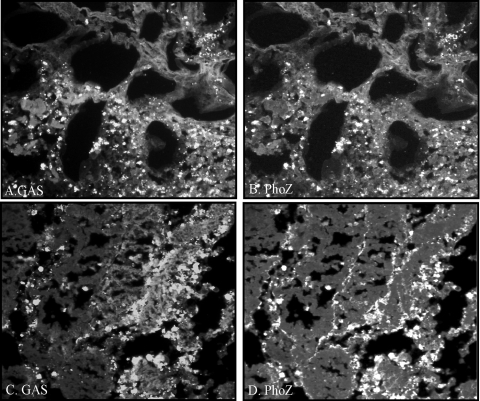
Lastly, to determine if the sil locus promoter was regulated in response to signals in vivo, a strain carrying the promoter construct was injected into the dorsal muscle of the zebrafish, and at 5 and 24 h postinfection the fish were euthanized and preparations were fixed for histological analysis. Major differences were observed between the two time points. Individual cocci stained with an antibody against S. pyogenes were detected at 5 h postinfection in the muscle tissue (Fig. 4A and C). However, many of the bacterial cells were not counterstained with the alkaline phosphatase antibody (Fig. 4B and D), suggesting that the sil promoter is selectively or rather weakly upregulated in the earlier stages of infection. In contrast, a much denser population of S. pyogenes cells was observed at 24 h in the necrotic tissue (Fig. 5A and C), which also stained brightly for alkaline phosphatase (Fig. 5B and D), indicating a high level of expression of the sil promoter at this stage of localized infection in the muscle. These results confirm that sil locus factors not only are expressed in vivo but also are differentially regulated over time.
FIG. 4.
Analysis of sil promoter expression in vivo. Following 5 h of intramuscular infection with 105 CFU of a strain carrying the sil promoter construct, zebrafish sections were prepared and stained with antibodies against S. pyogenes (A and C) and alkaline phosphatase (B and D), as described in Materials and Methods. Magnification, ×400.
FIG. 5.
Analysis of sil promoter expression in vivo. Following 24 h of intramuscular infection with 105 CFU of a strain carrying the sil promoter construct, zebrafish sections were prepared and stained with antibodies against S. pyogenes (A and C) and alkaline phosphatase (B and D), as described in Materials and Methods. Magnification, ×400.
DISCUSSION
The identification of virulence factors in an in vivo study is paramount to gaining a better understanding of pathogenic molecular mechanisms. One of the ways in which this has been achieved in the last decade is by applying a large-scale genetic screen, STM, to an appropriate model of infection. In this study, the zebrafish served as a model of severe S. pyogenes disease, mirroring the type of invasive, necrotic infection observed both macroscopically and microscopically in mammalian models, as well as in humans (21, 40, 58). Our results illustrate the successful application of STM to S. pyogenes and to the identification of expected and novel determinants that are required for the establishment and progression of disease. However, any random transposon mutagenesis study may result in polar mutations, and the virulence phenotypes observed for some of the attenuated mutants may be due to polar mutations. Although many of the insertions identified occur at the end of operons, for the insertions that are not at the end of operons, we cannot rule out the possibility of polarity.
For the 29 STM mutants sequenced, five genes have previously been implicated in virulence: hasA, mga, smeZ, silC, and silB. hasA is the first of three genes that comprise the hyaluronic acid capsule operon and encodes hyaluronan synthase, which adds alternating N-acetyl-d-glucosamine and d-glucoronic acid residues to form the hyaluronic acid polymer of the S. pyogenes capsule. The crucial role of capsule in S. pyogenes pathogenesis has been well established. For example, studies have shown that acapsular mutant strains are unable to defend against phagocytic engulfment (11, 60), and using a murine model of necrotizing fasciitis, Ashbaugh et al. concluded that capsule is critical for the progression of tissue necrosis (1).
S. pyogenes is not unique among pathogens in possessing the ability to coordinately regulate virulence gene expression while it is exposed to different environmental stimuli. Among the global regulatory networks in S. pyogenes are two-component signal transduction systems, quorum-sensing systems, and multigene regulons that are controlled by individual transcriptional regulators. One of the most significant of these “stand-alone” regulators is Mga, an STM mutant identified in this screen. Mga autoregulates and controls the transcription of virulence genes essential for immune evasion, adhesion, and internalization during the exponential phase of growth (24, 30). Moreover, a recent transcriptome analysis revealed that Mga controls the transcription of more than 10% of the S. pyogenes genome, including not only the core regulon virulence genes but also genes putatively involved in sugar metabolism (52). Maximal expression of mga occurs in exponential phase, as well as upon exposure to elevated CO2 levels, increased temperature, and iron-limiting conditions (5, 33, 46). Additionally, AmrA, a putative membrane protein involved in capsular polysaccharide biosynthesis, was discovered to be an important factor for the optimal expression of mga (51). This STM study is the first study to implicate amrA in virulence, as a transposon insertion in this gene resulted in an attenuated phenotype in our model of invasive disease.
One of the hallmarks of S. pyogenes invasive disease involves the toxigenic effects that continue to manifest following intense antibiotic therapy and surgical debridement. The numerous superantigens or exotoxins that the bacterium secretes have been implicated in these clinical outcomes. Superantigens have been proven to elicit a proinflammatory cytokine response and to induce symptoms of toxic shock in different animal models (4, 20, 37). Furthermore, superantigens have been detected in the circulation of patients with invasive S. pyogenes disease (56), as well as at the local site of tissue infection, with a regional cytokine profile that mirrors a typical superantigen response (42). Eleven exotoxins have been described to date, including streptococcal pyrogenic exotoxins A, B, C, F, G, H, and J, as well as various allelic forms of the streptococcal mitogenic exotoxin Z gene (smeZ) (27, 28, 48). smeZ is a chromosomal gene and is present in all S. pyogenes strains investigated. Studies have demonstrated that it is poorly expressed in vitro, with positive transcripts from certain strains observed only at late log phase, yet the superantigen is readily detected in the sera of patients with severe disease (49, 59). Although most superantigens examined trigger inflammatory responses and many are detected in clinical specimens, the smeZ product appears to be the predominant and most potent immunoactive agent (48, 49, 59, 61). Hence, the virulence attenuation of the smeZ toxin mutant, as well as the hasA capsule mutant, the mga global regulator mutant, and the amrA transcriptional activator mutant, was not a surprising finding in this study of S. pyogenes invasive disease and, consequently, served to validate the genetic screen.
We identified another class of genes common to most STM screens and involved in transport, including genes that encode an ABC transporter substrate-binding protein, an amino acid permease, a serine/threonine sodium transporter, and two macrolide efflux pumps, as well as salT, encoding a transporter of the salivaricin locus, a putative lantibiotic-producing operon. Among the most intriguing of the transporters identified that are involved in virulence are the macrolide efflux pumps. One of these, the mefE product, is a known efflux pump belonging to the major facilitator superfamily, and the gene encoding the other, which we have designated mfp, lacks homology to any known nucleotide sequence but has 33% identity and 55% similarity to mefE of S. pyogenes at the amino acid level. We were able to determine via arbitrary PCR analysis (data not shown) that a transposon harboring the insertion sequence IS1548, originally identified in Streptococcus agalactiae (16), exists directly upstream of the HSC5 mfp gene. As a result, we hypothesize that this efflux pump, carried on a transmissible element, was horizontally transferred and may be unique to this strain of S. pyogenes. Furthermore, we concluded from the results of antibiotic disk diffusion assays and PCR analysis (data not shown) that these efflux pumps are not directly involved in antibiotic resistance. Therefore, as they were identified as being attenuated in this STM screen, we postulate that they have some inherent physiological role associated with virulence other than expulsion of synthetic antibiotics from the bacterial cell, which is the mechanism that is most often ascribed to these pumps. An in-depth analysis of the ways in which the efflux pumps contribute to S. pyogenes virulence is ongoing.
An array of candidates involved in cellular metabolic processes have been identified in STM screens, such as the sugar dehydrogenase (B10-1) and the insertion that created a polar mutation in purB, which we described in this study. In fact, purine biosynthesis genes have been isolated in multiple STM screens for both gram-positive and gram-negative organisms (8, 19, 26, 36, 38, 47), highlighting the conclusion that purine is a key metabolite in virulence. Purine nucleotides perform diverse and integral functions in cellular metabolism. They are constituents of genetic material, enzymatic reactions, and signaling mechanisms and are involved in cellular energy. Accordingly, the inability of certain pathogens to biosynthesize purines leads to auxotrophy and avirulent phenotypes. These observations directly reflect the intricate responses of and adaptations by pathogens necessary to survive and proliferate in different host niches, particularly where essential nutrients are limited.
To efficiently sense the extra- and/or intracellular conditions and relay information, pathogens often utilize quorum-sensing systems. In gram-positive bacteria, the regulation of genes by quorum-sensing systems is mediated by signaling molecules that are secreted by an ABC transporter and detected by a two-component signal transduction system (39). Recently, the sil locus, homologous to a quorum-sensing system of S. pneumoniae, was identified in an in vivo mutagenesis study (23). The investigators determined that targeted mutations in the silC and silB genes resulted in severe attenuation in their murine model of necrotizing fasciitis (23). We identified two insertions in the same genes, silC and silB, in this STM study, suggesting that this locus is necessary for the establishment or maintenance of a successful infection in this disease model.
The data presented here support a role for the sil locus in the virulence of invasive S. pyogenes. Histopathologic analyses of zebrafish infected with each of the sil mutant strains revealed a profile different than that of the wild-type strain. Inflammatory cells observed at the injection site for infections with the silC and silB mutant bacteria appear to disrupt the microcolonies of bacteria normally found in a wild-type S. pyogenes infection, which typically show a lack of inflammatory infiltration. Hidalgo-Grass et al. suggest that this characteristic is due to a virulence trait of S. pyogenes, regulated at least in part by the sil locus, which interferes with host chemokine factors by preventing neutrophil migration to the site of infection (21, 22). Furthermore, tissue sections stained with antibodies against the bacteria and the alkaline phosphatase gene, under the control of the promoter upstream of the sil locus, although not quantitative, suggest that the sil locus is differentially regulated at various stages of the disease process. The differential expression of the sil locus adds validity to the hypothesis that these factors are components of a sensing regulon and that silC is a putative regulator of virulence genes.
Several lines of evidence from this study support the view that quorum-sensing genes or loci participate in stress management. The sil promoter was induced upon exposure to different stressors, such as temperature, H2O2, and agitation; thus, we may postulate that sil plays a role in conferring protection against a range of environmental stresses encountered in the host. However, a more detailed characterization of the response of the promoter to each of these stress factors is necessary to distinguish between a general response to stress and induction of the promoter due to distinct conditions. For example, the sil locus may be upregulated upon exposure to a lower external temperature, as is the case in open wounds of necrotizing fasciitis patients, or when intracellular proteins are damaged by reactive oxygen species, such as H2O2. The higher level of expression of the sil promoter exposed to agitation may be indicative of contact-dependent upregulation.
Generally, quorum-signaling molecules sense, among other factors, high cell density and respond accordingly. As a result, most quorum-sensing genes are upregulated in the stationary phase of growth, when cell numbers have peaked. The sil promoter was expressed maximally at mid-exponential phase, indicating that as in E. coli and S. enterica serovar Typhimurium, quorum sensing is important for regulating behavior prior to stationary phase, possibly to signal the cells to enter later growth stages and prepare for lower nutrient availability and metabolic activity (57).
The sil locus may represent a unique quorum-sensing system in S. pyogenes, regulating a unique profile of factors critical for a successful invasive infection. Although several transcriptional regulators have been found to control the expression of a number of virulence factors in response to environmental stimulants, many of the precise mechanisms contributing to streptococcal pathogenesis remain unclear. Although the pool of 1,200 mutants screened with the zebrafish was not large enough to completely saturate the genome and therefore may have missed important virulence genes, a genomic study such as the one described here enables us to at least begin to unravel the molecular mechanisms utilized by this accomplished pathogen. Further analysis of the sil locus, as well as other factors identified in this STM screen, should provide a better understanding of the intricate interplay between the pathogen and the host environment.
Acknowledgments
This work was supported by Public Health Service grant AI052141 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
We express our gratitude to Donna Runft for technical assistance and to Jesse Miller for useful suggestions on experimental design.
Editor: A. Camilli
Footnotes
Published ahead of print on 17 February 2009.
REFERENCES
- 1.Ashbaugh, C. D., H. B. Warren, V. J. Carey, and M. R. Wessels. 1998. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Investig. 102550-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Autret, N., and A. Charbit. 2005. Lessons from signature-tagged mutagenesis on the infectious mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 29703-717. [DOI] [PubMed] [Google Scholar]
- 3.Bakleh, M., L. E. Wold, J. N. Mandrekar, W. S. Harmsen, H. H. Dimashkieh, and L. M. Baddour. 2005. Correlation of histopathologic findings with clinical outcome in necrotizing fasciitis. Clin. Infect. Dis. 40410-414. [DOI] [PubMed] [Google Scholar]
- 4.Bonventre, P. F., H. Heeg, C. Cullen, and C. J. Lian. 1993. Toxicity of recombinant toxic shock syndrome toxin 1 and mutant toxins produced by Staphylococcus aureus in a rabbit infection model of toxic shock syndrome. Infect. Immun. 61793-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 1745693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caparon, M. G., and J. R. Scott. 1991. Genetic manipulation of the pathogenic streptococci. Methods Enzymol. 204556-586. [DOI] [PubMed] [Google Scholar]
- 7.Carapetis, J. R., A. C. Steer, E. K. Mulholland, and M. Weber. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5685-694. [DOI] [PubMed] [Google Scholar]
- 8.Chiang, S. L., and J. J. Mekalanos. 1998. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonisation. Mol. Microbiol. 27797-805. [DOI] [PubMed] [Google Scholar]
- 9.Cleary, P. P., E. L. Kaplan, J. P. Handley, A. Wlazlo, M. H. Kim, A. R. Hauser, and P. M. Schlievert. 1992. Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet 339518-521. [DOI] [PubMed] [Google Scholar]
- 10.Cockerill, F. R., III, R. L. Thompson, J. M. Musser, P. M. Schlievert, J. Talbot, K. E. Holley, W. S. Harmsen, D. M. Ilstrup, P. C. Kohner, M. H. Kim, B. Frankfort, J. M. Manahan, J. M. Steckelberg, F. Roberson, and W. R. Wilson. 1998. Molecular, serological, and clinical features of 16 consecutive cases of invasive streptococcal disease. Southeastern Minnesota Streptococcal Working Group. Clin. Infect. Dis. 261448-1458. [DOI] [PubMed] [Google Scholar]
- 11.Dale, J. B., R. G. Washburn, M. B. Marques, and M. R. Wessels. 1996. Hyaluronate capsule and surface M protein in resistance to opsonization of group A streptococci. Infect. Immun. 641495-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 1824696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efstratiou, A. 2000. Group A streptococci in the 1990s. J. Antimicrob. Chemother. 45(Suppl.)3-12. [DOI] [PubMed] [Google Scholar]
- 14.Eran, Y., Y. Getter, M. Baruch, I. Belotserkovsky, G. Padalon, I. Mishalian, A. Podbielski, B. Kreikemeyer, and E. Hanski. 2007. Transcriptional regulation of the sil locus by the SilCR signalling peptide and its implications on group A streptococcus virulence. Mol. Microbiol. 631209-1222. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, C. M., and M. G. Caparon. 2002. An alkaline phosphatase reporter transposon for analysis of protein secretion in gram-positive microorganisms. Appl. Environ. Microbiol. 68928-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granlund, M., L. Oberg, M. Sellin, and M. Norgren. 1998. Identification of a novel insertion element, IS1548, in group B streptococci, predominantly in strains causing endocarditis. J. Infect. Dis. 177967-976. [DOI] [PubMed] [Google Scholar]
- 17.Granok, A. B., A. Claiborne, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site of Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 1821529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanski, E., P. A. Horowitz, and M. G. Caparon. 1992. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 605119-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269400-403. [DOI] [PubMed] [Google Scholar]
- 20.Herman, A., J. W. Kappler, P. Marrack, and A. M. Pullen. 1991. Superantigens: mechanism of T-cell stimulation and role in immune responses. Annu. Rev. Immunol. 9745-772. [DOI] [PubMed] [Google Scholar]
- 21.Hidalgo-Grass, C., M. Dan-Goor, A. Maly, Y. Eran, L. A. Kwinn, V. Nizet, M. Ravins, J. Jaffe, A. Peyser, A. E. Moses, and E. Hanski. 2004. Effect of a bacterial pheromone peptide on host chemokine degradation in group A streptococcal necrotising soft-tissue infections. Lancet 363696-703. [DOI] [PubMed] [Google Scholar]
- 22.Hidalgo-Grass, C., I. Mishalian, M. Dan-Goor, I. Belotserkovsky, Y. Eran, V. Nizet, A. Peled, and E. Hanski. 2006. A streptococcal protease that degrades CXC chemokines and impairs bacterial clearance from infected tissues. EMBO J. 254628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hidalgo-Grass, C., M. Ravins, M. Dan-Goor, J. Jaffe, A. E. Moses, and E. Hanski. 2002. A locus of group A streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 4687-99. [DOI] [PubMed] [Google Scholar]
- 24.Hondorp, E. R., and K. S. McIver. 2007. The Mga virulence regulon: infection where the grass is greener. Mol. Microbiol. 661056-1065. [DOI] [PubMed] [Google Scholar]
- 25.Hui, F. M., L. Zhou, and D. A. Morrison. 1995. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 15325-31. [DOI] [PubMed] [Google Scholar]
- 26.Jones, A. L., K. M. Knoll, and C. E. Rubens. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 371444-1455. [DOI] [PubMed] [Google Scholar]
- 27.Kamezawa, Y., T. Nakahara, S. Nakano, Y. Abe, J. Nozaki-Renard, and T. Isono. 1997. Streptococcal mitogenic exotoxin Z, a novel acidic superantigenic toxin produced by a T1 strain of Streptococcus pyogenes. Infect. Immun. 653828-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotb, M. 1995. Bacterial pyrogenic exotoxins as superantigens. Clin. Microbiol. Rev. 8411-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotb, M., A. Norrby-Teglund, A. McGeer, H. El-Sherbini, M. T. Dorak, A. Khurshid, K. Green, J. Peeples, J. Wade, G. Thomson, B. Schwartz, and D. E. Low. 2002. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat. Med. 81398-1404. [DOI] [PubMed] [Google Scholar]
- 30.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11224-232. [DOI] [PubMed] [Google Scholar]
- 31.Lyon, W. R., and M. G. Caparon. 2003. Trigger factor-mediated prolyl isomerization influences maturation of the Streptococcus pyogenes cysteine protease. J. Bacteriol. 1853661-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an Rgg-like regulator in the transcription, secretion, and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1766263-66275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 634540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMillan, D. J., R. G. Beiko, R. Geffers, J. Buer, L. M. Schouls, B. J. Vlaminckx, W. J. Wannet, K. S. Sriprakash, and G. S. Chhatwal. 2006. Genes for the majority of group a streptococcal virulence factors and extracellular surface proteins do not confer an increased propensity to cause invasive disease. Clin. Infect. Dis. 43884-891. [DOI] [PubMed] [Google Scholar]
- 35.Meehl, M. A., J. S. Pinkner, P. J. Anderson, S. J. Hultgren, and M. G. Caparon. 2005. A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase. PLoS Pathog. 1e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26399-407. [DOI] [PubMed] [Google Scholar]
- 37.Miethke, T., C. Wahl, K. Heeg, B. Echtenacher, P. H. Krammer, and H. Wagner. 1992. T cell-mediated lethal shock triggered in mice by the superantigen staphylococcal enterotoxin B: critical role of tumor necrosis factor. J. Exp. Med. 17591-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. D., and M. N. Neely. 2005. Large-scale screen highlights the importance of capsule for virulence in the zoonotic pathogen Streptococcus iniae. Infect. Immun. 73921-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55165-199. [DOI] [PubMed] [Google Scholar]
- 40.Neely, M., J. Pfeifer, and M. G. Caparon. 2002. Streptococcus-zebrafish model of bacterial pathogenesis. Infect. Immun. 703904-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neely, M. N., W. R. Lyon, D. L. Runft, and M. Caparon. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 1855166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norrby-Teglund, A., P. Thulin, B. S. Gan, M. Kotb, A. McGeer, J. Anderssen, and D. E. Low. 2001. Evidence for superantigen involvement in severe group A streptococcal tissue infections. J. Infect. Dis. 184853-860. [DOI] [PubMed] [Google Scholar]
- 43.O'Brien, K. L., B. Beall, N. L. Barrett, P. R. Cieslak, A. Reingold, M. M. Farley, R. Danila, E. R. Zell, R. Facklam, B. Schwartz, and A. Schuchat. 2002. Epidemiology of invasive group a streptococcus disease in the United States, 1995-1999. Clin. Infect. Dis. 35268-276. [DOI] [PubMed] [Google Scholar]
- 44.Perez-Casal, J., J. A. Price, E. Maugin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8809-819. [DOI] [PubMed] [Google Scholar]
- 45.Phelps, H. A., and M. N. Neely. 2007. SalY of the Streptococcus pyogenes lantibiotic locus is required for full virulence and intracellular survival in macrophages. Infect. Immun. 754541-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Podbielski, A., J. A. Peterson, and P. P. Cleary. 1992. Surface protein-CAT reporter fusions demonstrate differential gene expression in the vir regulon of Streptococcus pyogenes. Mol. Microbiol. 62253-2265. [DOI] [PubMed] [Google Scholar]
- 47.Polissi, A., A. Ponitggia, G. Feger, M. Altieiri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 665620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Proft, T., S. L. Moffatt, C. J. Berkahn, and J. D. Fraser. 1999. Identification and characterization of novel superantigens from Streptococcus pyogenes. J. Exp. Med. 18989-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proft, T., S. Sriskandan, L. Yang, and J. D. Fraser. 2003. Superantigens and streptococcal toxic shock syndrome. Emerg. Infect. Dis. 91211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reed, L. J., and H. Muench. 1938. A simple method of estimation of fifty percent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 51.Ribardo, D. A., and K. S. McIver. 2003. amrA encodes a putative membrane protein necessary for maximal exponential phase expression of the Mga virulence regulon in Streptococcus pyogenes. Mol. Microbiol. 50673-685. [DOI] [PubMed] [Google Scholar]
- 52.Ribardo, D. A., and K. S. McIver. 2006. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol. Microbiol. 62491-508. [DOI] [PubMed] [Google Scholar]
- 53.Ruiz, N., B. Wang, A. Pentland, and M. G. Caparon. 1998. Streptolysin O and adherence synergistically modulate proimflammatory responses of kerationocytes to group A streptococci. Mol. Microbiol. 27337-346. [DOI] [PubMed] [Google Scholar]
- 54.Saenz, H. L., and C. Dehio. 2005. Signature-tagged mutagenesis: technical advances in a negative selection method for virulence gene identification. Curr. Opin. Microbiol. 8612-619. [DOI] [PubMed] [Google Scholar]
- 55.Schmitz, F. J., A. Beyer, E. Charpentier, B. H. Normark, M. Schade, A. C. Fluit, D. Hafner, and R. Novak. 2003. Toxin-gene profile heterogeneity among endemic invasive European group A streptococcal isolates. J. Infect. Dis. 1881578-1586. [DOI] [PubMed] [Google Scholar]
- 56.Sriskandan, S., D. Moyes, and J. Cohen. 1996. Detection of circulating bacterial superantigen and lymphotoxin-alpha in patients with streptococcal toxic-shock syndrome. Lancet 3481315-1316. [DOI] [PubMed] [Google Scholar]
- 57.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 957046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor, F. B. J., A. E. Bryant, K. E. Blick, E. Hack, P. M. Jansen, S. D. Kosanke, and D. L. Stevens. 1999. Staging of the baboon response to group A streptococci administered intramuscularly: a descriptive study of the clinical symptoms and clinical chemical response patterns. Clin. Infect. Dis. 29167-177. [DOI] [PubMed] [Google Scholar]
- 59.Unnikrishnan, M., D. M. Altmann, T. Proft, F. Wahid, J. Cohen, J. D. Fraser, and S. Sriskandan. 2002. The bacterial superantigen streptococcal mitogenic exotoxin Z is the major immunoactive agent of Streptococcus pyogenes. J. Immunol. 1692561-2569. [DOI] [PubMed] [Google Scholar]
- 60.Wessels, M. R., A. E. Moses, J. B. Goldberg, and T. J. DiCesare. 1991. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc. Natl. Acad. Sci. USA 888317-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, L., M. Thomas, A. Woodhouse, D. Martin, J. D. Fraser, and T. Proft. 2005. Involvement of streptococcal mitogenic exotoxin Z in streptococcal toxic shock syndrome. J. Clin. Microbiol. 433570-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]