Abstract
The Hspa1b (Hsp70.1) gene is one of the first genes expressed after fertilization, with expression occurring during the minor zygotic genome activation in the absence of stress. This expression can take place in the male pronucleus as early as the one-cell stage of embryogenesis. The importance of HSPA1B for embryonic viability during times of stress is supported by studies showing that depletion of this protein results in a significant reduction in embryos developing to the blastocyte stage. Recently we began addressing the mechanism responsible for allowing expression of Hspa1b during the minor ZGA and found that HSF1 and HSF2 bind the Hspa1b promoter during late spermatogenesis. In this report, we have extended those studies using western blots and chromatin immunoprecipitation assays and found that RNA Polymerase II is present in epididymal spermatozoa and bound to the Hspa1b promoter. These present results, in addition to our previous results, support a model in which the binding of HSF1, HSF2, Sp1, and RNA Polymerase II to the promoter of Hspa1b would allow the rapid formation of a transcription-competent state during the minor ZGA, thereby allowing Hspa1b expression.
INTRODUCTION
The zygotic genome activation (ZGA) is a process where maternal control of development is shifted to the zygote (reviewed in (Minami et al. 2007)). This transition can be divided into a minor ZGA where a small subset of genes including Hspa1b are expressed as early as the one-cell stage, and the major ZGA, which occurs during the two-cell stage and is characterized by a significant burst in both transcription and translation (Bellier et al. 1997; Flach et al. 1982; Howlett et al. 1985; Latham et al. 1991; Nothias et al. 1995; Taylor & Piko 1987) with more stringent transcriptional regulation (Majumder et al. 1993; Martinez-Salas et al. 1989; Rothstein et al. 1992; Wiekowski et al. 1991). During the minor ZGA, transcription in the one-cell embryo appears to be relatively promiscuous and opportunistic (Choo et al. 2002; Ma et al. 2001) with the majority of transcription occurring in the male pronucleus (Aoki et al. 1997; Worrad et al. 1994).
The Hspa1b gene encodes one of two known 70 kDa stress-inducible heat shock proteins (HSP70) and is one of the first genes expressed following fertilization, with spontaneous expression taking place as early as the one-cell stage of embryogenesis in the absence of stress (Bensaude et al. 1983; Christians et al. 1995; Fiorenza et al. 2004). Studies using inducible Hsp70 promoter driven reporter genes confirms this embryonic pattern of expression (Thompson et al. 1995). Spontaneous expression of Hspa1b mRNA increases to peak levels at the 2 cell stage and then decreases until becoming stress-inducible by the blastocyte stage (Morange et al. 1984). In bovine embryos, HSP70 appears to be stress-inducible at the 2 cell stage (Edwards & Hansen 1996).
The importance of stress-inducible HSP70 proteins for embryonic viability is supported by a number of studies. In one study HSP70 proteins were immunodepleted (Matwee et al. 2001) while another study used antisense oligonucleotides to inhibit stress-induced HSP70 expression in (Dix et al. 1998). In both studies there was a significant reduction in embryos developing to the blastocyte stage, indicating that expression of inducible HSP70 proteins is important for preimplantation embryogenesis. Although HSPA1B is important for embryonic viability, the mechanism responsible for expression of the Hspa1b gene during the minor ZGA is not known.
In somatic cells, the promoters of a number of genes, including those of the Hspa1b and C-myc genes, remain uncompacted and accessible during mitosis (Christova & Oelgeschlager 2002; John & Workman 1998; Martinez-Balbas et al. 1995; Michelotti et al. 1997; Sarge & Park-Sarge 2005). The lack of compaction of promoter regions in mitotic cells is referred to as “bookmarking” and is believed to function to permit genes that existed in a transcription-competent state prior to entry into mitosis to be maintained in a form that can be rapidly reassembled into the active state in G1. Previous research in our laboratory found that the Hspa1b promoter was bookmarked during mitosis by the binding of heat shock transcription factor 2 (HSF2) to the heat shock element (HSE) of the Hspa1b promoter (Xing et al. 2005). Bookmarking Hspa1b during mitosis allows the rapid expression of this cytoprotective gene in early G1 if the cell encounters stress. Recently we extended those studies from tissue culture to spermatogenic cells and found that HSF1, HSF2, and SP1 are present in mature spermatozoa and bound to the Hspa1b promoter (Wilkerson et al. 2008). Other reports have found that in mice HSF1 and HSF2 transcripts are present in preimplantation embryos as early as the one cell stage, while HSF1 and HSF2 proteins have been identified in 2 cell embryos (Christians et al. 1997). Interestingly, a heat-inducible DNA binding activity has been observed in one and two cell embryos (Mezger et al. 1994). Furthermore, the HSE promoter element, which is a promoter binding site for HSF1 and HSF2, plays a role in expression of Hspa1b during the early stages of embryogenesis (Christians et al. 1997).
In this report we show that RNA Polymerase II is present in mature spermatozoa and bound to the Hspa1b promoter. This observation, in addition to our previous report showing that HSF1, HSF2, and SP1 are also bound to the Hspa1b promoter in mature spermatozoa clearly supports a unique scenario where the promoter of this particular gene is maintained in a form that is ready for rapid activation in an environment where most transcription has ceased (Monesi 1964; Monesi et al. 1978; Zheng et al. 2008), chromatin has been reorganized and highly compacted (Ward & Coffey 1991), and numerous basal transcription factors, transcriptional regulators, and architectural factors are displaced from chromatin (Zheng et al. 2008). These results suggest a mechanism by which Hspa1b could be expressed during the earliest stages of embryogenesis including expression in the male pronucleus of the one-cell embryo.
RESULTS
RNA Polymerase II is Present in Mouse Sperm Nuclei
Our previous observations showed that HSF1, HSF2, and SP1 are bound to the Hspa1b gene in mature sperm. These results suggested the intriguing possibility that RNA Polymerase II may also remain associated with this promoter in mature spermatozoa. As a first test of this hypothesis, we sought to determine if RNA Polymerase II was present in mature spermatozoa using western blot analysis. We isolated and purified mature spermatozoa from the caudal epididymus of adult CD-1 mice. To minimize any extranuclear protein contamination, the purified sperm were treated with the detergent CTAB in conjunction with a 3–5 second sonication on ice to further disrupt the sperm membrane and facilitate tail removal. Light microscopy was used to confirm the removal of sperm tails and purity of the samples which was determined to be greater than 98% (data not shown). Protein extracts were prepared from sperm nuclei and a mouse C2C12 cell line (as a control) and assayed by western blot using monoclonal antibodies against the largest subunit (Rpb1) of RNA Polymerase II (Figure 1). The images presented in Figure 1 are from the same gel, only separated by 2 empty lanes to eliminate the possibility of cross-contamination. The results are representative of the banding pattern observed from western blot analysis of two independent mouse sperm protein isolations, and indicate that RNA Polymerase II is present in mature spermatozoa. In both the sperm lane (left image) and C2C12 lane (right image) we observed one predominant band migrating at approximately 220kDa that is consistent with the size of RNA Polymerase II detected in previous studies (Luthi-Carter et al. 2002; Wilkerson et al. 2007).
Figure 1. RNA Polymerase II is present in mouse sperm nuclei.
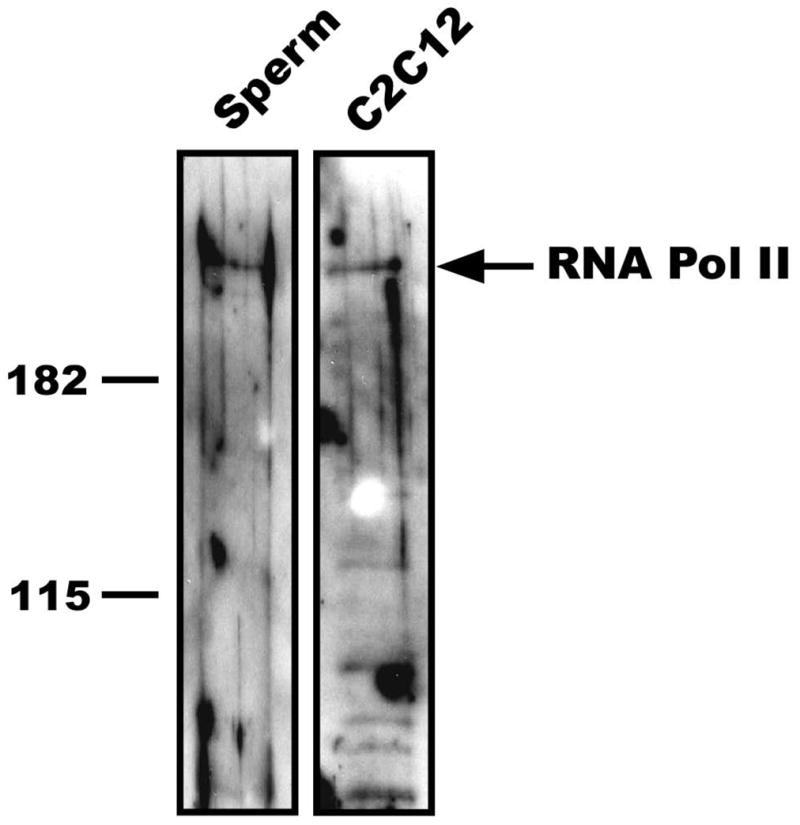
Protein extracts prepared from mouse sperm nuclei or a mouse C2C12 cell line (as a control) were separated on a 8% SDS polyacrylamide gel and assayed by western blot using mouse monoclonal antibodies against the largest subunit (Rpb1) of RNA Polymerase II (Covance). Prestained protein markers were used to indicate molecular weight (kDa). Images shown are from the same gel but separated by 2 empty lanes to eliminate cross contamination. Images are representative of results obtained from extracts prepared from two different groups of mice (two mice per group).
Chromatin Immunoprecipitation Analysis of RNA Polymerase II in Mature Sperm
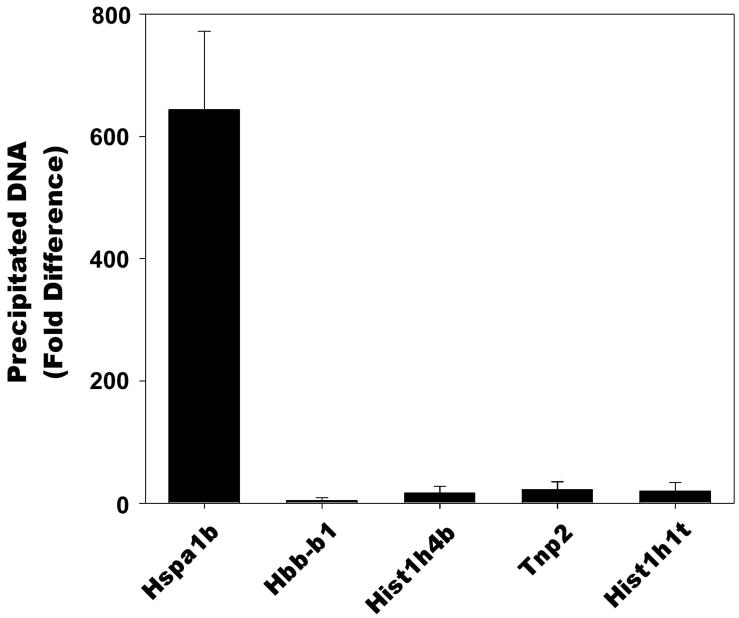
In light of our western blot results shown in Figure 1, we hypothesized that RNA Polymerase II, a factor necessary for transcription, could also be bound to the Hspa1b promoter in mature spermatozoa as part of a mechanism for allowing the preferential expression of Hspa1b in the male pronucleus of the one-cell embryo. To test this hypothesis, we performed chromatin immunoprecipitation assays on spermatozoa obtained from the caudal region of mouse epididymides using monoclonal antibodies against RNA Polymerase II and mouse IgG as a negative control antibody (Figure 2). We assayed the binding of RNA Polymerase II to the promoter of the Hspa1b gene as well as the β-globin (Hbb-b1) promoter, a negative control gene that is not expressed in spermatogenic cells and lacks a recognizable HSE promoter element. To ensure that the observed binding was not due to residual occupancy on genes that were expressed earlier in spermatogenesis, we assayed the binding of RNA Polymerase II to the promoters of histone H4 (Hist1h4b), transition protein 2 (Tnp2), and testis-specific histone H1 (Hist1h1t). DNA fragments precipitated by the indicated antibodies were assayed by quantitative real time PCR. The results indicate that binding of RNA Polymerase II to the Hspa1b promoter in mature spermatozoa is significantly higher than to the promoters of Hbb-b1, Hist1h4b, Tnp2, and Hist1h1t. There was no statistical difference (P>0.05) in the binding of RNA Polymerase II and the promoters of Hbb-b1, Hist1h4b, Tnp2, and Hist1h1t.
Figure 2. Chromatin IP analysis of RNA Polymerase II binding in mature spermatozoa.
Purified mouse cauda epididymal spermatozoa were crosslinked, lysed, sonicated, and subjected to immunoprecipitation using mouse monoclonal antibodies against the largest subunit of RNA Polymerase II or mouse IgG as a negative control antibody. Data is represented as fold difference relative to IgG. The precipitated DNA fragments were subjected to quantitative PCR using primers that amplified the proximal promoters of the Hspa1b, Hbb-b1, Hist1h4b, Tnp2, and Hist1h1t genes.
DISCUSSION
A wide range of species utilize members of the HSP70 family to promote cell survival through expression of stress-induced HSPs including HSPA1A and HSPA1B. Due to the potential role of HSPA1B in promoting embryonic viability during times of stress, we initiated studies to better understand the mechanisms regulating transcription of this gene. In this report we show that RNA Polymerase II is present in mature spermatozoa and bound to the Hspa1b promoter.
Recently, we found that that HSF1, HSF2, and SP1 are bound to the Hspa1b promoter in mature spermatozoa (Wilkerson et al. 2008). The presence of these transcription factors on the Hspa1b promoter in sperm chromatin is surprising given the fact that transcription has ceased and the chromatin has been reorganized and compacted to a level that is approximately 6-fold more compact than chromatin found in mitotic cells (Ward & Coffey 1991). It is generally thought that transcriptional silence in mature sperm is due to the high level of sperm chromatin compaction which likely excludes most sequence-specific transcription factors from binding. Due to this high level of compaction, it is unlikely that HSF1, HSF2, and SP1 are actively binding to the Hspa1b promoter at the later stages of spermiogenesis. Our hypothesis is that these factors are binding prior to, or during the condensation phase of spermatogenesis and remains bound to the Hspa1b promoter even after fertilization. The presence of RNA Polymerase II on the Hspa1b promoter provides an interesting mechanism by which the Hspa1b gene could be rapidly and preferentially expressed during the earliest stages of embryogenesis including the male pronucleus. With the opportunistic and essentially unregulated transcription in the one-cell embryo (Choo et al. 2002; Ma et al. 2001), the presence of HSF1, HSF2, SP1, and RNA Polymerase II on the Hspa1b promoter would allow the rapid transcription of this gene versus other genes that are not bound by transcription factors. In support of this hypothesis, a previous study showed that when first round replication is blocked by aphidicolin there was an increase in expression of a select number of genes including Hspa1b (Sonehara et al. 2008). It is thought that the first round of replication disrupts the compacted chromatin and allows transcription factors to assemble on promoter regions of genes (Wolffe 1991; Wolffe 1991). The transcriptional activity of Hspa1b prior to the first round of replication supports our hypothesis that the Hspa1b promoter is already poised for transcription. The observed drop in expression of Hspa1b following the first round of replication (Sonehara et al. 2008) could be due to the removal of transcription factors that were present prior to replication.
The observation that RNA Polymerase II, a factor essential for transcription, can remain bound to specific regions of DNA after compaction is unique, although not exclusive to mature spermatozoa. Previously, we observed in Jurkat cells blocked in mitosis that the binding of RNA Polymerase II to a number of HSE containing promoters was reduced, but not completely eliminated (Wilkerson et al. 2007). There was clear and reproducible binding of RNA Polymerase II to the promoters of Hsp90aa1 (Hsp90), fos, and Hspb1 (Hsp27) in mitotic cells.
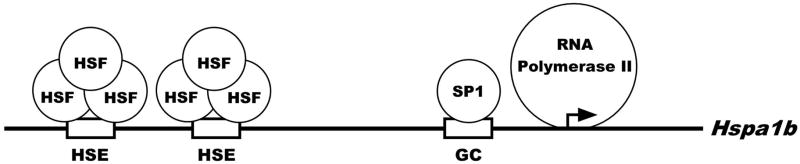
Our findings, shown schematically in Figure 3, indicate that in mature mouse spermatozoa the Hspa1b promoter is bound by a number of transcription factors important for expression of this gene. The regulatory region of the inducible HSP70 is 85% homologous between human and mouse sequences. The two heat shock elements (HSEs) are known binding sites for HSF1 and HSF2. Although HSF1 and HSF2 bind DNA as trimers and are capable of forming heterotrimers (Ostling et al. 2007; Sandqvist et al. 2009), it is not clear if these factors bind the Hspa1b promoter in mature spermatozoa as homotrimers or heterotrimers. HSF1 and HSF2 are highly homologous with their DNA binding domains being 72% identical (Schuetz et al. 1991). There is also a report suggesting an interplay between HSF1 and HSF2 (Ostling et al. 2007). The GC box is a consensus binding site for SP1, a transcription factor that is necessary for transcription of the Hspa1b gene. Our previous result showing that SP1 is bound to this GC box in mature spermatozoa suggests that SP1 has a role in transcription of Hspa1b in the one-cell embryo. Interestingly, a previous report has shown that SP1 is required for transcription of Hspa1b in the two-cell embryo (Bevilacqua et al. 2000). The mouse Hspa1b promoter contains a TATA box located at −30 which helps serve as the site for assembly of the pre-initiation complex (PIC) which most likely occurs prior to, or during the condensation phase of spermatogenesis. Based on the location of the primers used for the ChIP assays, our results suggest that RNA Polymerase II is binding at, or near the transcription start site. The culmination of our findings suggests that Hspa1b is in a state of transcriptional readiness in mature spermatozoa and ultimately the male pronucleus.
Figure 3. In vivo Promoter Occupancy of the Hspa1b gene in Mature Spermatozoa.
In mature spermatozoa, the promoter of the mouse Hspa1b gene is bound by a number of transcription factors known to be important for expression of this gene. There are two heat shock elements (HSEs) located at −140 and −110 that are known binding sites for HSF1 and HSF2 and a GC box located at −50 that is bound by SP1. RNA Polymerase II is also bound to the proximal promoter region of Hspa1b and further demonstrates the transcriptional readiness of this gene.
In mice there are two known heat inducible HSP70 proteins, HSP70.1 (HSPA1A) and HSP70.3 (HSPA1B), with HSPA1B the focus of the studies presented here. The Hspa1a and Hspa1b genes are found in a tandem array on mouse chromosome 17 (Hunt et al. 1993) and the resulting proteins differ at only two residues, rendering them nearly indistinguishable at the protein level. However, due to differences between these two genes at the nucleic acid level, they can be readily distinguished. The amino acid and gene sequences, and expression pattern of Hspa1a and Hspa1b are highly conserved between rodents and humans (Milner & Campbell 1990). Studies using antisense oligonucleotides showed that reduced levels of Hspa1a and Hspa1b disrupted preimplantation embryogenesis and increased embryonic sensitivity to arsenic (Dix et al. 1998). Another study, using knockout mice lacking both Hspa1a and Hspa1b (Hsp70.1/3−/−), suggested that these factors were not necessary for embryogenesis (Hunt et al. 2004). However, the KO mice were lighter in weight than wild type mice and cells derived from these KO mice displayed genomic instability that was enhanced by heat stress, underwent senescence sooner, and displayed a higher frequency of chromosomal aberrations and a significant increase in aberrant spermatocytes following heat stress. Considering these observations, it is possible that an accumulation of these genetic defects in later generations could lead to significant developmental problems.
Due to the high degree of conservation and likely redundant function, Hspa1a and Hspa1b are often referred to collectively as stress induced HSP70. Although the studies presented here focus on mouse Hspa1b, due to the high degree of similarity it is likely that the promoter of Hspa1a is also poised for transcriptional activation during the earliest stages of embryogenesis through mechanisms identified for Hspa1b.
There exist a number of epigenetic mechanisms by which paternal DNA patterns of expression are passed to daughter cells including DNA methylation (reviewed in (Suzuki & Bird 2008)) and occupancy and/or post-translational modifications of histones and other factors (reviewed in (Corpet & Almouzni 2008)). This list could be expanded to include non-coding RNA molecules (reviewed in (Mattick & Makunin 2006)). In the unique case of Hspa1b and most likely Hspa1a, our research suggests that the binding of HSF2, and possibly HSF1, to the HSE promoter element maintains the promoter in an “accessible” chromatin confirmation throughout mitosis (Xing et al. 2005) and spermiogenesis (Wilkerson et al. 2008).
Mature spermatozoa have traditionally be thought of as merely inert cells with the sole function of delivering paternal DNA to the ovum. However, an ever-increasing number of studies suggest a functional organization exists in sperm chromatin. In spermatozoa, protamines serve as the major DNA binding proteins around which chromatin is organized and compacted (reviewed in (Balhorn 2007)). Research has shown that insufficient levels of protamine leads to male infertility with sperm displaying both structural and functional defects (Cho et al. 2001). It seems that other DNA-interacting proteins can contribute to sperm chromatin organization. In humans for example, approximately 15% of sperm chromatin is bound by histones with the remaining 85% bound by protamines (Gatewood et al. 1990). The nucleohistone fraction displays hypersensitivity to nuclease cleavage suggesting less DNA compaction (Wykes & Krawetz 2003). It is thought that certain genes that may be important during early embryogenesis could be bound in this nucleohistone portion of chromatin or be directed for expression through other epigenetic mechanisms. In support of this hypothesis, a study found that the postnatally expressed Hbb-b1 gene was found in the protamine bound DNA fraction while the embryonically expressed IGF-2 gene was found in the histone bound DNA fraction (Wykes & Krawetz 2003). In addition to chromatin binding factors, it is possible that nuclear matrix attachment regions (MARs) may play a role in organizing sperm chromatin (Martins et al. 2004). An intact sperm nuclear matrix appears to be critical to the formation of the male pronucleus (Ward et al. 1999).
Although the results presented here are focused on mechanisms that could lead to expression of stress-induced HSP70 expression during the early stages of embryogenesis, there appears to be a role for inducible HSP70 in mature spermatozoa. Previous studies have shown that HSPA1A localizes to the sperm tail and the equatorial region of the sperm head in human spermatozoa (Mitchell et al. 2007) and in porcine spermatozoa localization was found in the equatorial region (Spinaci et al. 2005). The equatorial region of the sperm head suggests a role in sperm-oocyte fusion. When porcine spermatozoa are incubated with anti-HSP70 antibodies, there is a marked reduction in fertilization which is likely due to a disruption in sperm-ooycte fusion (Spinaci et al. 2005). It is also thought that HSPA1A has a role in human fertility based on a study showing that the presence of antibodies against HSPA1A in seminal plasma inhibits fertilization (Bohring & Krause 2003). Another study implicated reduced expression of HSPA1A in certain subtypes of male infertility (Cedenho et al. 2006).
Based on the important cytoprotective role of HSPA1B, bookmarking the Hspa1b promoter would provide a mechanism by which Hspa1b could be expressed during the earliest stages of embryogenesis in the event of cellular stress. Bookmarking the Hspa1b gene in spermatozoa makes particular sense given the need for molecular chaperones to handle the large number of proteins that are translated during embryogenesis and the various stresses that can occur, including osmotic and pH changes. The ability to rapidly express the HSPA1B protein would clearly promote embryonic viability.
MATERIALS AND METHODS
Animals
All CD-1 mice used in this study were adult males ( at least 8–10 weeks of age) obtained from Harlan (Indianapolis, IN). Animals were maintained in the Division of Laboratory Animal Resources (DLAR) and studies performed according to approved Institutional Animal Care and Use Committee (IACUC) guidelines at the University of Kentucky (Lexington, KY).
Preparation of Sperm Nuclei
Sperm nuclei were prepared as previously described (Wilkerson et al. 2008). Briefly, mature sperm was obtained from the caudal epididymides of adult male CD-1 mice by repeatedly puncturing with a 20G needle. Sperm were then gently flushed from the caudal epididymides using phosphate buffered saline (PBS). Sperm were pelleted, quickly frozen in liquid nitrogen, and then resuspended in cold PBS to lyse any red blood cells present. Centrifugation of the Percoll (Sigma Chemical Co., USA) gradient was performed according to established protocols (Hishinuma & Sekine 2004) with the following modifications. Sperm, washed three times in cold PBS then resuspended in 1mL of PBS, were loaded on a discontinuous Percoll gradient created by layering 1.5mL of 35%, 45%, 75% Percoll (v/v) in a 15mL conical tube. Centrifugation was performed at 700xg for 30 minutes. Sperm nuclei were prepared using cetyltrimethylammonium bromide (CTAB) according to published protocols (Balhorn et al. 1977) with minor modifications (Maione et al. 1997). Immediately prior to the CTAB incubation, the sperm suspension was sonicated for 3–5 sec at 20% power to dissolve clumps and assist in tail removal. Following CTAB incubation, the sperm suspension was passed through siliconized glass wool to remove any remaining tail debris.
Western Blots
Protein extracts were prepared from sperm nuclei using buffer C (20mM Hepes pH 7.9, 25% glycerol, 420mM NaCl, 1.5mM MgCl2, 0.2mM EDTA, 0.5mM PMSF, 0.5mM DTT) followed by brief sonication on ice. Protein extracts were separated on a 8% SDS polyacrylamide gel and western blots were performed using mouse monoclonal antibodies against the largest subunit (Rpb1) of RNA Polymerase II (Covance) according to our established protocols (Hilgarth et al. 2003; Lubert & Sarge 2003; Xing et al. 2004). The secondary antibody, α-IgG-HRP, was obtained from an enhanced chemiluminescence (ECL) kit (Amersham Life Science, Arlington Heights, IL). Samples for Western blots were prepared from two different groups of animals, with at least two animals used for each preparation.
Chromatin Immunoprecipitations
All chromatin immunoprecipitations (ChIP) were performed as previously published (Wilkerson et al. 2008; Worrad et al. 1994) with the following modifications. ChIP assays were performed at least twice with each batch of DNA. Pre-cleared chromatin was incubated with 5μg of mouse monoclonal RNA Polymerase II or control mouse IgG (Sigma), and rotated at 4°C for 16 hours. DNA was purified using a QIAquick PCR Purification Kit (Qiagen Inc.) and eluted in 50μl of 10mM Tris (pH 8.5). Immunoprecipitated DNA and input samples obtained prior to immunoprecipitation were analyzed by quantitative real time PCR with a Stratagene Mx 4000 system using Brilliant SYBR Green QPCR master mix (Stratagene) and the following primers (shown 5′ to 3′) from Integrated DNA Technologies (Coralville, IA):
Hspa1b: (+) CCGCAACAGTGTCAATAGC, (−) CCTTGAGTAATCGGAGTTGTGG
Hbb-b1: (+) TTGCTCCTCACATTTGCTTCTG, (−) ACTTCATCGGAGTTCACCTTTC
Hist1h4b: (+) ACGAAGCCCGCCATC, (−) TTGGCGTGCTCCGTGTAGGT
Hist1h1t: (+) GCAGTGAGCAGATATGCAAGA, (−) CCAACAGTGATGGGGTAGTG
Tnp2: (+) GATGTCACCGTGAGAAGGGTACAA, (−) ACAAAGCCCTGCCTCTCATCTTCA
The sequence used for the upstream Hspa1b primer differs at 4 of 19 nucleotides from the sequence of Hspa1a, including the 3′ terminal nucleotide of the primer. This sequence difference, under the stringency conditions employed during PCR amplification, would be expected to discriminate Hspa1b from Hspa1a. Samples were checked for specific amplification using dissociation curves analysis software. PCR products were also assayed on polyacrylamide gels with ethidium bromide staining to ensure they were of the expected size (data not shown). The Ct values were normalized to input DNA (DNA before immunoprecipitation step) and IgG controls using the formula 2^[(Ct IgG-Ct Input)−(Ct Ab-Ct Input)] (where Ab=RNA Polymerase II, or IgG). Data is represented as fold differences relative to IgG. The data shown represent quantitative PCR results from three independent sperm purifications and ChIP assays. Error bars represent standard error of the means. Statistical significance was determined using a two-tailed, unpaired t-test.
Acknowledgments
Funding
This work was supported by a National Institutes of Health (NIH) HD grant (F32HD050043) to D.C.W, and NIH grants GM61053 and GM64606 to K.D.S.
We would like to thank other members of our laboratory for insightful discussions during these studies.
Footnotes
Publisher's Disclaimer: Disclaimer. This is not the definitive version of record of this article. This manuscript has been accepted for publication in Reproduction, but the version presented here has not yet been copy edited, formatted or proofed. Consequently, the journal accepts no responsibility for any errors or omissions it may contain. The definitive version is available at http://dx.doi.org/10.1530/REP-09-0015.
Declaration of Interest
There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported
References
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R, Gledhill BL, Wyrobek AJ. Mouse sperm chromatin proteins: quantitative isolation and partial characterization. Biochemistry. 1977;16:4074–4080. doi: 10.1021/bi00637a021. [DOI] [PubMed] [Google Scholar]
- Bellier S, Chastant S, Adenot P, Vincent M, Renard JP, Bensaude O. Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J. 1997;16:6250–6262. doi: 10.1093/emboj/16.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude O, Babinet C, Morange M, Jacob F. Heat shock proteins, first major products of zygotic gene activity in mouse embryo. Nature. 1983;305:331–333. doi: 10.1038/305331a0. [DOI] [PubMed] [Google Scholar]
- Bevilacqua A, Fiorenza MT, Mangia F. A developmentally regulated GAGA box-binding factor and Sp1 are required for transcription of the hsp70.1 gene at the onset of mouse zygotic genome activation. Development. 2000;127:1541–1551. doi: 10.1242/dev.127.7.1541. [DOI] [PubMed] [Google Scholar]
- Bohring C, Krause W. Characterization of spermatozoa surface antigens by antisperm antibodies and its influence on acrosomal exocytosis. Am J Reprod Immunol. 2003;50:411–419. doi: 10.1034/j.1600-0897.2003.00103.x. [DOI] [PubMed] [Google Scholar]
- Cedenho AP, Lima SB, Cenedeze MA, Spaine DM, Ortiz V, Oehninger S. Oligozoospermia and heat-shock protein expression in ejaculated spermatozoa. Hum Reprod. 2006;21:1791–1794. doi: 10.1093/humrep/del055. [DOI] [PubMed] [Google Scholar]
- Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet. 2001;28:82–86. doi: 10.1038/ng0501-82. [DOI] [PubMed] [Google Scholar]
- Choo KB, Chen HH, Liu TY, Chang CP. Different modes of regulation of transcription and pre-mRNA processing of the structurally juxtaposed homologs, Rnf33 and Rnf35, in eggs and in pre-implantation embryos. Nucleic Acids Res. 2002;30:4836–4844. doi: 10.1093/nar/gkf623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E, Campion E, Thompson EM, Renard JP. Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription . Development. 1995;121:113–122. doi: 10.1242/dev.121.1.113. [DOI] [PubMed] [Google Scholar]
- Christians E, Michel E, Adenot P, Mezger V, Rallu M, Morange M, Renard JP. Evidence for the involvement of mouse heat shock factor 1 in the atypical expression of the HSP70.1 heat shock gene during mouse zygotic genome activation. Mol Cell Biol. 1997;17:778–788. doi: 10.1128/mcb.17.2.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova R, Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 2008 doi: 10.1016/j.tcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Garges JB, Hong RL. Inhibition of hsp70-1 and hsp70-3 expression disrupts preimplantation embryogenesis and heightens embryo sensitivity to arsenic. Mol Reprod Dev. 1998;51:373–380. doi: 10.1002/(SICI)1098-2795(199812)51:4<373::AID-MRD3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Edwards JL, Hansen PJ. Elevated temperature increases heat shock protein 70 synthesis in bovine two-cell embryos and compromises function of maturing oocytes. Biol Reprod. 1996;55:341–346. doi: 10.1095/biolreprod55.2.341. [DOI] [PubMed] [Google Scholar]
- Fiorenza MT, Bevilacqua A, Canterini S, Torcia S, Pontecorvi M, Mangia F. Early transcriptional activation of the hsp70.1 gene by osmotic stress in one-cell embryos of the mouse. Biol Reprod. 2004;70:1606–1613. doi: 10.1095/biolreprod.103.024877. [DOI] [PubMed] [Google Scholar]
- Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1982;1:681–686. doi: 10.1002/j.1460-2075.1982.tb01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Schmid CW, Bradbury EM. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem. 1990;265:20662–20666. [PubMed] [Google Scholar]
- Hilgarth RS, Hong Y, Park-Sarge OK, Sarge KD. Insights into the regulation of heat shock transcription factor 1 SUMO-1 modification. Biochem Biophys Res Commun. 2003;303:196–200. doi: 10.1016/s0006-291x(03)00312-7. [DOI] [PubMed] [Google Scholar]
- Hishinuma M, Sekine J. Separation of canine epididymal spermatozoa by Percoll gradient centrifugation. Theriogenology. 2004;61:365–372. doi: 10.1016/s0093-691x(03)00219-x. [DOI] [PubMed] [Google Scholar]
- Howlett SK, Webb M, Maro B, Johnson MH. Meiosis II, mitosis I and the linking interphase: a study of the cytoskeleton in the fertilised mouse egg. Cytobios. 1985;43:295–305. [PubMed] [Google Scholar]
- Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, Pandita TK. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol Cell Biol. 2004;24:899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt CR, Gasser DL, Chaplin DD, Pierce JC, Kozak CA. Chromosomal localization of five murine HSP70 gene family members: Hsp70-1, Hsp70-2, Hsp70-3, Hsc70t, and Grp78. Genomics. 1993;16:193–198. doi: 10.1006/geno.1993.1158. [DOI] [PubMed] [Google Scholar]
- John S, Workman JL. Bookmarking genes for activation in condensed mitotic chromosomes. Bioessays. 1998;20:275–279. doi: 10.1002/(SICI)1521-1878(199804)20:4<275::AID-BIES1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Latham KE, Garrels JI, Chang C, Solter D. Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one- and two-cell stages. Development. 1991;112:921–932. doi: 10.1242/dev.112.4.921. [DOI] [PubMed] [Google Scholar]
- Lubert EJ, Sarge KD. Interaction between protein phosphatase 2A and members of the importin beta superfamily. Biochem Biophys Res Commun. 2003;303:908–913. doi: 10.1016/s0006-291x(03)00434-0. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Hanson SA, Strand AD, Bergstrom DA, Chun W, Peters NL, Woods AM, Chan EY, Kooperberg C, Krainc D, Young AB, Tapscott SJ, Olson JM. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum Mol Genet. 2002;11:1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- Ma J, Svoboda P, Schultz RM, Stein P. Regulation of zygotic gene activation in the preimplantation mouse embryo: global activation and repression of gene expression. Biol Reprod. 2001;64:1713–1721. doi: 10.1095/biolreprod64.6.1713. [DOI] [PubMed] [Google Scholar]
- Maione B, Pittoggi C, Achene L, Lorenzini R, Spadafora C. Activation of endogenous nucleases in mature sperm cells upon interaction with exogenous DNA. DNA Cell Biol. 1997;16:1087–1097. doi: 10.1089/dna.1997.16.1087. [DOI] [PubMed] [Google Scholar]
- Majumder S, Miranda M, DePamphilis ML. Analysis of gene expression in mouse preimplantation embryos demonstrates that the primary role of enhancers is to relieve repression of promoters. EMBO J. 1993;12:1131–1140. doi: 10.1002/j.1460-2075.1993.tb05754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Martinez-Salas E, Linney E, Hassell J, DePamphilis ML. The need for enhancers in gene expression first appears during mouse development with formation of the zygotic nucleus. Genes Dev. 1989;3:1493–1506. doi: 10.1101/gad.3.10.1493. [DOI] [PubMed] [Google Scholar]
- Martins RP, Ostermeier GC, Krawetz SA. Nuclear matrix interactions at the human protamine domain: a working model of potentiation. J Biol Chem. 2004;279:51862–51868. doi: 10.1074/jbc.M409415200. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Matwee C, Kamaruddin M, Betts DH, Basrur PK, King WA. The effects of antibodies to heat shock protein 70 in fertilization and embryo development. Mol Hum Reprod. 2001;7:829–837. doi: 10.1093/molehr/7.9.829. [DOI] [PubMed] [Google Scholar]
- Mezger V, Renard JP, Christians E, Morange M. Detection of heat shock element-binding activities by gel shift assay during mouse preimplantation development. Dev Biol. 1994;165:627–638. doi: 10.1006/dbio.1994.1281. [DOI] [PubMed] [Google Scholar]
- Michelotti EF, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- Milner CM, Campbell RD. Structure and expression of the three MHC-linked HSP70 genes. Immunogenetics. 1990;32:242–251. doi: 10.1007/BF00187095. [DOI] [PubMed] [Google Scholar]
- Minami N, Suzuki T, Tsukamoto S. Zygotic gene activation and maternal factors in mammals. J Reprod Dev. 2007;53:707–715. doi: 10.1262/jrd.19029. [DOI] [PubMed] [Google Scholar]
- Mitchell LA, Nixon B, Aitken RJ. Analysis of chaperone proteins associated with human spermatozoa during capacitation. Mol Hum Reprod. 2007;13:605–613. doi: 10.1093/molehr/gam043. [DOI] [PubMed] [Google Scholar]
- Monesi V. Ribonucleic Acid Synthesis During Mitosis and Meiosis in the Mouse Testis. J Cell Biol. 1964;22:521–532. doi: 10.1083/jcb.22.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monesi V, Geremia R, D’Agostino A, Boitani C. Biochemistry of male germ cell differentiation in mammals: RNA synthesis in meiotic and postmeiotic cells. Curr Top Dev Biol. 1978;12:11–36. doi: 10.1016/s0070-2153(08)60592-x. [DOI] [PubMed] [Google Scholar]
- Morange M, Diu A, Bensaude O, Babinet C. Altered expression of heat shock proteins in embryonal carcinoma and mouse early embryonic cells. Mol Cell Biol. 1984;4:730–735. doi: 10.1128/mcb.4.4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothias JY, Majumder S, Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development. J Biol Chem. 1995;270:22077–22080. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- Ostling P, Bjork JK, Roos-Mattjus P, Mezger V, Sistonen L. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J Biol Chem. 2007;282:7077–7086. doi: 10.1074/jbc.M607556200. [DOI] [PubMed] [Google Scholar]
- Rothstein JL, Johnson D, DeLoia JA, Skowronski J, Solter D, Knowles B. Gene expression during preimplantation mouse development. Genes Dev. 1992;6:1190–1201. doi: 10.1101/gad.6.7.1190. [DOI] [PubMed] [Google Scholar]
- Sandqvist A, Bjork JK, Akerfelt M, Chitikova Z, Grichine A, Vourc’h C, Jolly C, Salminen TA, Nymalm Y, Sistonen L. Heterotrimerization of HSF1 and HSF2 Provides a Transcriptional Switch in Response to Distinct Stimuli. Mol Biol Cell. 2009 doi: 10.1091/mbc.E08-08-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK. Gene bookmarking: keeping the pages open. Trends Biochem Sci. 2005;30:605–610. doi: 10.1016/j.tibs.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Schuetz TJ, Gallo GJ, Sheldon L, Tempst P, Kingston RE. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc Natl Acad Sci U S A. 1991;88:6911–6915. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonehara H, Nagata M, Aoki F. Roles of the first and second round of DNA replication in the regulation of zygotic gene activation in mice. J Reprod Dev. 2008;54:381–384. doi: 10.1262/jrd.20053. [DOI] [PubMed] [Google Scholar]
- Spinaci M, Volpe S, Bernardini C, De Ambrogi M, Tamanini C, Seren E, Galeati G. Immunolocalization of heat shock protein 70 (Hsp 70) in boar spermatozoa and its role during fertilization. Mol Reprod Dev. 2005;72:534–541. doi: 10.1002/mrd.20367. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Taylor KD, Piko L. Patterns of mRNA prevalence and expression of B1 and B2 transcripts in early mouse embryos. Development. 1987;101:877–892. doi: 10.1242/dev.101.4.877. [DOI] [PubMed] [Google Scholar]
- Thompson EM, Legouy E, Christians E, Renard JP. Progressive maturation of chromatin structure regulates HSP70.1 gene expression in the preimplantation mouse embryo. Development. 1995;121:3425–3437. doi: 10.1242/dev.121.10.3425. [DOI] [PubMed] [Google Scholar]
- Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–574. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- Ward WS, Kimura Y, Yanagimachi R. An intact sperm nuclear matrix may be necessary for the mouse paternal genome to participate in embryonic development. Biol Reprod. 1999;60:702–706. doi: 10.1095/biolreprod60.3.702. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML. Regulation of gene expression in preimplantation mouse embryos: effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Dev Biol. 1991;147:403–414. doi: 10.1016/0012-1606(91)90298-h. [DOI] [PubMed] [Google Scholar]
- Wilkerson DC, Murphy LA, Sarge KD. Interaction of HSF1 and HSF2 with the Hspa1b promoter in mouse epididymal spermatozoa. Biol Reprod. 2008;79:283–288. doi: 10.1095/biolreprod.107.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson DC, Skaggs HS, Sarge KD. HSF2 binds to the Hsp90, Hsp27, and c-Fos promoters constitutively and modulates their expression. Cell Stress Chaperones. 2007;12:283–290. doi: 10.1379/CSC-250.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP. Activating chromatin. Curr Biol. 1991;1:366–368. doi: 10.1016/0960-9822(91)90195-3. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Implications of DNA replication for eukaryotic gene expression. J Cell Sci. 1991;99(Pt 2):201–206. doi: 10.1242/jcs.99.2.201. [DOI] [PubMed] [Google Scholar]
- Worrad DM, Ram PT, Schultz RM. Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development. 1994;120:2347–2357. doi: 10.1242/dev.120.8.2347. [DOI] [PubMed] [Google Scholar]
- Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- Xing H, Mayhew CN, Cullen KE, Park-Sarge OK, Sarge KD. HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J Biol Chem. 2004;279:10551–10555. doi: 10.1074/jbc.M311719200. [DOI] [PubMed] [Google Scholar]
- Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge OK, Sarge KD. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- Zheng J, Xia X, Ding H, Yan A, Hu S, Gong X, Zong S, Zhang Y, Sheng HZ. Erasure of the paternal transcription program during spermiogenesis: The first step in the reprogramming of sperm chromatin for zygotic development. Dev Dyn. 2008 doi: 10.1002/dvdy.21499. [DOI] [PubMed] [Google Scholar]