Abstract
The repressor CodY is reported to inhibit metabolic genes mainly involved in nitrogen metabolism. We analyzed codY mutants from three unrelated Staphylococcus aureus strains (Newman, UAMS-1, and RN1HG). The mutants grew more slowly than their parent strains in a chemically defined medium. However, only codY mutants were able to grow in medium lacking threonine. An excess of isoleucine resulted in growth inhibition in the wild type but not in the codY mutants, indicating that isoleucine plays a role in CodY-dependent repression. Prototypic CodY-repressed genes including the virulence regulator agr are repressed after up-shift with isoleucine. The CodY-dependent repression of agr is consistent with the concomitant influence of CodY on typical agr-regulated genes such as cap, spa, fnbA, and coa. However, some of these virulence genes (e.g., cap, fnbA, and spa) were also regulated by CodY in an agr-negative background. Microarray analysis revealed that the large majority of CodY-repressed genes were involved in amino acid metabolism; CodY-activated genes were mainly involved in nucleotide metabolism or virulence. In summary, CodY in S. aureus not only acts as a repressor for genes involved in nitrogen metabolism but also contributes to virulence gene regulation by supporting as well as substituting for agr function.
Staphylococcus aureus asymptomatically colonizes the nares of healthy individuals but also causes a variety of infections in humans. Regulatory loci are necessary for the adaptation of the organism to the different nutrient limitations and stress conditions encountered in vivo. This allows the pathogen to survive and/or multiply in different compartments during colonization and infection processes. However, knowledge of the environmental conditions encountered in vivo is still incomplete, and the interaction of regulatory circuits leading to metabolic adaptation and differential expression of virulence factors remains poorly understood. The in vitro expression of most virulence factors is tightly related to the growth phase. For instance protein A (encoded by spa), fibronectin-binding proteins (encoded by fnbA and fnbB), and coagulase (encoded by coa) are expressed during the exponential growth phase, whereas most secreted proteins (e.g., hemolysins, enterotoxins, and proteases) and the capsule (enzymes encoded by the capA-capP operon) are expressed mainly during the postexponential phase (22, 37). In many bacteria, the transition to postexponential growth is accompanied by a profound reprogramming of gene expression. Several underlying mechanisms are thought to be involved in such a transition. Quorum sensing allows the bacteria to detect their own density. Basically, bacteria secrete small diffusible molecules (autoinducers) which are also effectors of their own synthesis. Upon passing a critical concentration threshold, the autoinducers activate specific transcriptional regulators, leading to the differential expression of target genes. The agr locus of S. aureus is a prototypic quorum-sensing system mainly involved in the regulation of virulence genes (37). At high cell densities, the regulatory RNAIII is expressed, leading to the inhibition of spa, for instance, and to the activation of genes encoding secreted virulence factors and the capsular polysaccharide. Besides quorum sensing, additional mechanisms have to be triggered for the growth phase transition in S. aureus since the expression of certain virulence factors remains growth phase dependent in agr mutants (42, 51, 53).
In Bacillus subtilis, the CodY repressor has been described as a central regulator important for the transition to stationary phase and sporulation (40, 46). Homologs of codY could be identified in the genome of most gram-positive species, including pathogenic staphylococci and streptococci (23, 46). In B. subtilis, GTP reaches its highest concentration during the exponential phase, when it binds to CodY and leads to the repression of late gene expression. This mechanism is linked to the stringent response since the synthesis of (p)ppGpp by Rel leads to a lowering of the GTP pool (28). The central role of GTP in gene regulation was further proven by the use of decoyinine, an inhibitor of GMP synthetase (40). Depletion of the GTP pool by this inhibitor leads to the activation of typical late genes already in the exponential phase. CodY represses genes which are primarily involved in nitrogen metabolism (proteases, oligopeptide transporters, and genes for amino acid synthesis) and activates transcription of genes of the carbon overflow pathway (46, 47). Interestingly, in Lactococcus lactis CodY does not bind GTP. Here, branched-chain amino acids (BCAAs) are bound by CodY and constitute the primary signals for codY repression (23, 39). The difference between the two species is probably due to sequence variations in the proposed GTP binding domain of CodY (39). It has been shown that CodY of B. subtilis can also use BCAAs in addition to GTP as signaling molecules (39, 45). A conserved CodY-binding site (AATTTTCWGAAATT) was first described for L. lactis (12, 24) and later confirmed in B. subtilis (5).
The metabolic regulatory cascades for gram-positive pathogens are only partly understood, and knowledge about nitrogen metabolism and amino acid availability during infection remains limited. However, there is growing evidence that CodY is an important regulatory link between metabolism and virulence gene expression in pathogenic bacteria (6, 14, 26, 32, 35, 46). The complete set of genes necessary for amino acid synthesis has been predicted to be present in the S. aureus N315 genome (25). Nevertheless, S. aureus usually requires a complex mixture of amino acids for growth, probably because of repression of the corresponding pathways (25). It can be assumed that during infection the bacterium recruits at least some of the amino acids from the host. Interestingly, in a whole-genome screen, genes coding for oligopeptide transporters were shown to be essential for infection or to be specifically activated during infection (10, 36). S. aureus is also equipped with several genes coding for proteases, whose activity could provide amino acids or peptides during infection. The analysis of other gram-positive organisms together with our own preliminary data led to the hypothesis that in S. aureus CodY may play a central role not only in metabolic adaptation but also in virulence gene expression. Indeed, it was recently shown that a codY mutant shows enhanced expression of the agr effector molecule RNAIII (34). Additionally, codY influences biofilm formation, albeit with opposite effects depending on the strain analyzed (16, 34). Our results obtained at the transcriptome level clearly demonstrate that CodY is involved in the regulation of metabolic genes and also influences gene expression of virulence genes in an agr-dependent and -independent manner.
MATERIALS AND METHODS
Strains and growth conditions.
Strains and plasmids are listed in Table 1. S. aureus strains were grown in tryptic soy broth, CYPG (10 g/liter Casamino Acids, 10g/liter yeast extract, 5 g/liter NaCl, 20% glucose, and 1.5 M phosphoglycerate), or in a chemically defined medium (CDM). For strains carrying resistance genes, antibiotics were used only in precultures at the following concentrations: kanamycin, 50 μg/ml; erythromycin, 10 μg/ml; and tetracycline, 5 μg/ml. CDM was composed as follows (final concentration in mg/liter in brackets): group 1 amino acids from a 10× stock consisting of l-tryptophan (100), l-tyrosine (100), and l-phenylalanine (100); group 2 amino acids from a 10× stock consisting of l-cysteine (50), l-histidine (100), and l-methionine (100); group 3 amino acids from a 100× stock consisting of l-glutamine (200), l-glutamic acid (100), glycine (100), and l-proline (100); group 4 amino acids from a 100× stock consisting of l-isoleucine (100), l-leucine (100), l-threonine (200), and l-valine (100); group 5 amino acids from a 100× stock consisting of dl-alanine (100), l-arginine (100), l-aspartic acid (100), l-lysine (100), hydroxy-l-proline (100), and l-serine (100); group 6 vitamins from a 50× stock consisting of p-aminobenzoic acid (0.2), biotin (0.2), folic acid (0.8), niacinamide (1), β-NAD (2.5), pantothenate calcium salt (2), pyridoxal (1), pyridoxamine dihydrochloride (1), riboflavin (2), thiamine hydrochloride (1), and vitamin B12 (0.1); group 7 nucleotides from a 100× stock (predissolved in 2 N HCl) consisting of adenine (20), guanine hydrochloride (20), and uracil (20); group 8 and 9 salts from a 50× stock consisting of K2HPO4 (200) and KH2PO4 (1,000); group 9 consisting of NaH2PO4 (3,195), MgSO4 (700), and CaCl2 (10); group 10 consisting of a 100× stock of Na2HPO4 (9,214); and group 11 carbohydrate from a 20× stock of glucose (10,000). Single amino acids were omitted in some experiments, as indicated in the figure legends. The pH of the medium was buffered to 7.0.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| TOP10 | Competent E. coli for plasmid transformation | Invitrogen, Karlsruhe. Germany |
| CYL316 | RN4220 harboring pYL112Δ19 and L54 int gene; r− | 31 |
| RN6112 | RN6390 agrA::Tn551 | Richard Novick |
| RN4220 | Restriction-deficient S. aureus strain | 29 |
| RN4220-21 | RN4220 ΔcodY::tet(M) | This work |
| Newman | Wild type | 15 |
| Newman-21 | Newman ΔcodY::tet(M) | This work |
| Newman-agr | Newman agrA::Tn551 | This work |
| Newman-21/-agr | Newman agrA::Tn551 ΔcodY::tet(M) | This work |
| UAMS-1 | Osteomyelitis isolate | 19 |
| UAMS-1-21 | UAMS-1 ΔcodY::tet(M) | This work |
| UAMS-1-agr | agrA::Tn551 | 17 |
| UAMS-1-21/-agr | UAMS-1 agrA::Tn551 ΔcodY::tet(M) | This work |
| RN1HG | rsbU restored RN1 | This work |
| RN1 | 8325 | NARSA strain collectiona |
| RN1HG-21 | RN1HG ΔcodY::tet(M) | This work |
| Plasmids | ||
| pALC2073 | E. coli-S. aureus shuttle vector with tetracycline-inducible promoter | 3 |
| pMAD | Shuttle vector for gene replacement mutagenesis | 1 |
| pCG29 | pMAD with cloned clpY-tet(M)-rpsB fragment for codY mutagenesis | This work |
| pCG30 | pALC2073 with codY integration via EcoRI restriction site | This work |
NARSA, Network on Antimicrobial Resistance in Staphylococcus aureus.
Bacteria from an overnight culture were diluted to an initial optical density at 600 nm (OD600) of 0.05 in fresh medium without antibiotics and grown with shaking (222 rpm) at 37°C to the indicated OD600. In the complemented strains, codY was induced with anhydrotetracycline (IBA GmbH, Göttingen) (0.05 μg/ml). For up-shift experiments, strains were grown in CDM without isoleucine to an OD600 of 0.4. The cultures were divided into aliquots and supplemented with isoleucine and grown for 1 h.
Construction of mutant strains and complementation.
The codY locus was replaced by a tetracycline resistance cassette [tet(M)]. Briefly, two fragments flanking codY and the tet(M) gene were amplified and annealed by overlapping PCR using oligonucleotides (see Table S2 in the supplemental material). The amplicon was cloned into pMAD using the BamHI/BglII restriction sites of pMAD to gain pCG29. Mutagenesis of strain RN4220 was performed as described previously (1). The obtained codY gene replacement mutant strain (RN4220-21) was verified by PCR (for oligonucleotides, see Table S1 in the supplemental material). codY and agr mutants of different S. aureus strains were obtained by transduction using φ11 lysates of strains RN4220-21 and RN6112, respectively. Transductants were verified by PCR and pulsed-field gel electrophoresis. All agr mutants were negative for δ-hemolysin and RNAIII expression. For complementation, codY was amplified with oligonucleotides (see Table S1 in the supplemental material) containing EcoRI sites and cloned in the EcoRI site of the tetracycline-inducible vector pALC2073, yielding plasmid pCG30. The plasmid was used to transform strain RN4220, from which it was transduced, into the codY mutant strains. Strain RN1HG is an rsbU-restored derivative of S. aureus strain RN1 (8325) obtained by site-directed mutagenesis using pMAD rsbU as described previously (50).
RNA isolation, Northern blot hybridization, and real-time reverse transcription-PCR.
RNA isolation and Northern blot analysis were performed as described previously (20). Briefly, bacteria were lysed in 1 ml of Trizol reagent (Invitrogen Life Technologies, Karlsruhe, Germany) with 0.5 ml of zirconia-silica beads (0.1 mm-diameter) in a high-speed homogenizer (Savant Instruments, Farmingdale, NY). RNA was isolated as described in the instructions provided by the manufacturer of Trizol. Digoxigenin-labeled probes for the detection of specific transcripts were generated using a DIG-Labeling PCR Kit following the manufacturer's instructions (Roche Biochemicals, Mannheim, Germany). Oligonucleotides used for probe generation are as described previously (48) or are listed in Table S1 in the supplemental material.
Microarray manufacturing and microarray design.
The microarray was manufactured by in situ synthesis of 60-base-long oligonucleotide probes (Agilent, Palo Alto, CA), selected as previously described (8). The array covers >98% of all open reading frames (ORFs) annotated in strains N315 and Mu50 (30), MW2 (2), COL (18), NCTC8325 and USA300 (13), and MRSA252 and MSSA476 (27), including their respective plasmids.
Preparation of labeled nucleic acids for expression microarrays.
Total RNA was purified from bacteria grown in CDM to an OD600 of 0.5. For each strain RNA of three independently grown cultures was analyzed. After additional DNase treatment, the absence of remaining DNA traces was confirmed by quantitative PCR (SDS 7700; Applied Biosystems, Framingham, MA) with assays specific for 16S rRNA (41, 43). Batches of 5 μg of total S. aureus RNA were labeled by Cy3-dCTP using SuperScript II (Invitrogen, Basel, Switzerland) following the manufacturer's instructions. Labeled products were then purified onto QiaQuick columns (Qiagen).
Purified genomic DNA from the different sequenced strains used for the design of the microarray was extracted (DNeasy; Qiagen), labeled with Cy5 dCTP using the Klenow fragment of DNA polymerase I (BioPrime, Invitrogen, Carlsbad, CA) (8), and used for the normalization process (49). Cy5-labeled DNA (500 ng) and a Cy3-labeled cDNA mixture were diluted in 50 μl of Agilent hybridization buffer and hybridized at a temperature of 60°C for 17 h in a dedicated hybridization oven (Robbins Scientific, Sunnyvale, CA). Slides were washed, dried under nitrogen flow, and scanned (Agilent, Palo Alto, CA) using 100% photon multiplier tube power for both wavelengths.
Microarray analysis.
Fluorescence intensities were extracted using Feature Extraction software (version 8; Agilent). Local background-subtracted signals were corrected for unequal dye incorporation or unequal load of the labeled product. The algorithm consisted of a rank consistency filter and a curve fit using the default LOWESS (locally weighted linear regression) method. Data consisting of two independent biological experiments were expressed as log 10 ratios and analyzed using GeneSpring, version 8.0 (Silicon Genetics, Redwood City, CA). A filter was applied to select oligonucleotides mapping ORFs in the Newman genome, yielding approximately 92% coverage. Statistical significance of differentially expressed genes was calculated by analysis of variance (9, 43) using GeneSpring, including the Benjamini and Hochberg false discovery rate correction of 5% (P value cutoff, 0.05) and an arbitrary cutoff of twofold for expression ratios.
Microarray data accession number.
The complete microarray data set has been posted on the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE12340 for the platform design and GPL7137 for the original data set.
RESULTS
Molecular organization of the codY operon.
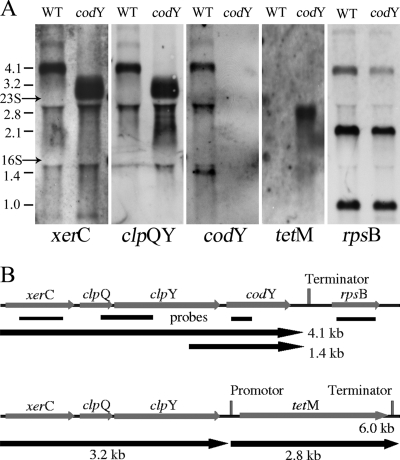
To characterize the CodY regulon, S. aureus gene replacement mutants were constructed in three clonally distinct S. aureus strains (Newman, RN1HG, and UAMS-1). In the mutant strains, codY was replaced by a tet(M) resistance cassette, as verified by Southern hybridization and PCR. To evaluate whether the mutation exerted a polar effect on the surrounding genes, Northern analysis was performed using probes specific for codY as well as for the neighboring genes (Fig. 1A). codY is part of a polycistronic operon encompassing xerC, clpQ (hslV), clpY (hslU), and codY (4.1 kb) (Fig. 1B). An additional 1.4-kb transcript codes for codY only. In the mutant, codY was replaced by tet(M), leading to termination of the transcript (xerC and clpQY) in front of tet(M). The expression levels of xerC and clpQY in the mutant were similar to those in the wild-type strains. However, there was a slight, but reproducible increase in the xer and clpQY transcript levels in the codY mutant. Microarray analysis also suggests that xerC is slightly affected by the mutation although the difference was not significant. rpsB located downstream of the codY operon was not affected in the mutant. Overall, the codY mutation showed minimal polar effects on the surrounding genes. Thus, for complementation analysis only, the codY ORF was cloned into an inducible vector (pCG30).
FIG. 1.
Genetic organization of the codY locus. (A) Total RNA from strain Newman and its codY mutant Newman-21 was hybridized with probes specific for xerC, clpQY, codY, tet(M), and rpsB. Transcript size was estimated by comparison with an RNA marker. Note that slight hybridization bands below the ribosomal rRNA are probably due to comigration of the mRNA with rRNA and were not taken as distinct mRNA species. (B) Scheme of the genetic organization of the codY operon. Probes used for Northern blot analysis are indicated with black lines. Putative transcriptional units are indicated by black arrows in the wild type (upper part) and the codY mutant (lower part). WT, wild type.
Effect of codY on selected genes with putative CodY binding motifs.
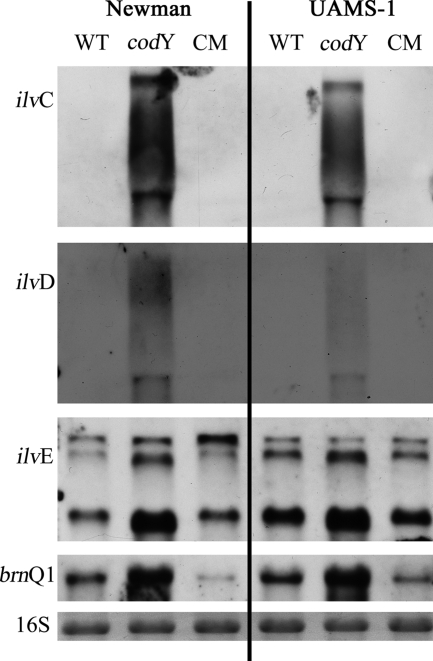
For other organisms, it has been shown that CodY functions as a repressor via binding to a proposed CodY-binding motif (AATTTTCWGAAAATT) (5, 12, 24). We performed a stringent search for the occurrence of the proposed motif (two mismatches accepted) within 1,000 bp upstream of putative ORFs in three S. aureus genomes (Col, N315, and Newman) (http://xbase.bham.ac.uk/pattern.pl?id=1327). A total of 68, 76, and 71 genes preceded by a putative CodY box were identified in strains Col, N315, and Newman, respectively. The CodY boxes were mainly localized 50 to 300 bp upstream of the translational start site (see Fig. S1 in the supplemental material). Many of the identified genes code for enzymes involved in amino acid metabolism or transport. We selected four genes containing CodY boxes for Northern blot analysis: ilvC and ilvD (part of the ilvDBC-leuABC-ilvA operon, with the CodY box in front of ilvD), ilvE, and brnQ1 (MWMN_0180, branched-chain amino acid transport system II carrier protein). It could be shown that all four genes were strongly upregulated in the codY mutants of strains Newman and UAMS-1 in comparison to the wild-type or codY-complemented mutant strains (Fig. 2). These results suggest that CodY in S. aureus binds to the same conserved motif as described for B. subtilis and L. lactis.
FIG. 2.
Detection of codY-regulated operons: ilvDBC-leuABC-ilvA (CodY box in front of ilvD, detected with ilvD and ilvC), ilvE, and brnQ1 (MWMN_0180, BCAA transport system II carrier protein). RNA was isolated from strains Newman and UAMS-1, their codY mutants (Newman-21 and UAMS-1-21), and the complemented strains ([CM] Newman-21/pCG30 and UAMS-1-21/pCG30). Bacteria were grown to an OD600 of 0.5 in CDM. The 16S rRNAs detected in the ethidium bromide-stained gels are indicated as loading controls in the lower lane. WT, wild type.
Effect of codY on growth in CDM.
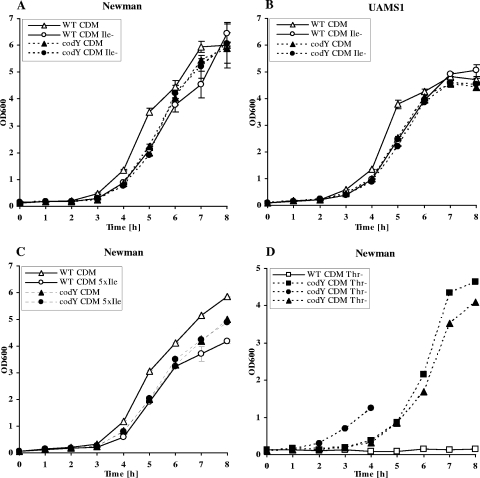
In a first attempt to characterize the codY phenotype, we performed growth analysis in CDM containing all three BCAAs (100 mg/liter each) and threonine (200 mg/liter). In this medium S. aureus strains did not grow without glucose, suggesting that amino acids were not being used as the primary carbon source. This is further supported by the lowering of the pH after prolonged growth, which also is indicative of glucose consumption. No growth of wild-type or mutant strains occurred if any of the five groups of amino acids was omitted from the medium. Growth analysis revealed that codY mutants grew more slowly in complete CDM than the wild-type strains (Fig. 3A and B). The codY mutants were also delayed in growth in complex medium such as CYPG (data not shown). Thus, one may assume that gene repression by CodY might be favorable for the organism under conditions of amino acid surplus.
FIG. 3.
Growth analysis of wild-type and codY mutants in CDM. (A and B) Growth of S. aureus strain Newman and the codY mutant Newman-21 (A) and of UAMS-1 and the codY mutant UAMS-1-21 (B) in CDM and in CDM lacking isoleucine (Ile−). (C) Growth of S. aureus strain Newman and the codY mutant Newman-21 in CDM and CDM with an excess of 500 μg/ml isoleucine (5× Ile). (D) Newman-21 was grown in medium lacking threonine (Thr−; triangle). For analysis of the observed lag phase, the codY mutant was subcultured from threonine-depleted CDM. Bacteria from CDM without threonine grown to the exponential phase (OD600 of 1) were then subcultured in medium lacking threonine (square). In addition, the bacteria were recultured in complete CDM overnight and again inoculated in CDM without threonine (circle). The lag phase was omitted after preadaptation of the bacteria to threonine depletion but was restored after intermittent growth in medium with threonine. WT, wild type.
Next, growth in CDM that had been depleted either of one of the BCAAs (valine, leucine, or isoleucine) or of threonine was analyzed. Wild-type strains and codY mutants failed to grow in medium lacking either valine or leucine (data not shown). In medium lacking isoleucine, there was no growth difference between the wild type and codY mutants in contrast to growth in CDM containing 100 mg/liter of isoleucine. We hypothesized that isoleucine may be a major signal for CodY regulation. If isoleucine is indeed a natural ligand of CodY, then maximal repression of CodY-regulated genes should occur under isoleucine-rich conditions. Thus, an excess of isoleucine may lead to growth inhibition, e.g., if some amino acids become limiting but the biosynthetic pathways remain repressed. In fact, a high concentration of isoleucine (500 mg/liter) in the medium resulted in diminished growth in the wild type in comparison to growth in medium with moderate isoleucine (100 mg/liter) (Fig. 3C). This growth inhibition is CodY dependent since in the codY mutant no effect of isoleucine excess on growth was observed. Similar results were obtained with strain UAMS-1 and strain RN1HG (data not shown). A predominant role of isoleucine in comparison to other BCAAs for signaling is emphasized by the observation that an excess of valine or leucine did not result in growth inhibition of the wild-type strains (data not shown).
Interestingly, in medium without threonine the codY mutants were able to grow but not the wild type (Fig. 3D). This indicates that, under these growth conditions, CodY represses genes necessary for threonine synthesis. The growth of the mutant in medium lacking threonine was characterized by a typical lag phase. Thus, the mutant is obviously able to adapt to the CDM conditions. When the codY mutant was subcultured from the exponential growth phase (OD600 of 0.5) into fresh medium, growth continued without lag (Fig. 3D), indicating adaptation to the medium. However, when the mutants were subcultured in complete CDM and again inoculated into medium without threonine, the typical lag phase was again apparent. Thus, the adaptation of the codY mutants to threonine-depleted medium seems to be due a regulatory adaptation in metabolism and is not mediated by the generation of suppressor mutations in genes of the biochemical pathways.
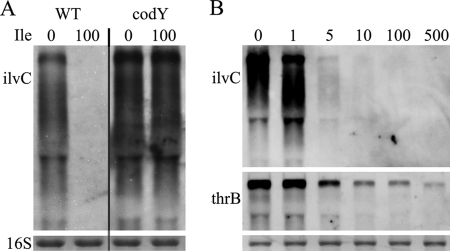
Influence of isoleucine on CodY target genes.
To gain further insight into putative CodY ligands in S. aureus, we analyzed whether isoleucine in the medium affects the transcription of codY target genes (Fig. 4). Strains were precultured in CDM without isoleucine and then supplemented with isoleucine for 1 h. Isoleucine resulted in a dose-dependent repression of the CodY target gene ilvC (Fig. 4B). The addition of 5 μg/ml isoleucine was already sufficient to cause a severe downregulation of ilvC. The repression is mediated by CodY since the codY mutants were not responsive to isoleucine under the tested conditions (Fig. 4A). However, the ilvC level observed in the wild type grown without isoleucine was lower than that in the codY mutant. This may be due to a baseline level of isoleucine produced by the bacteria or by other putative signaling molecules such as GTP. Since only the codY mutant was able to grow in medium lacking threonine, we also tested whether genes involved in threonine biosynthesis are similarly repressed via CodY. We could show that thrB transcription is also dependent on the isoleucine concentration. However, repression of thrB by isoleucine was less pronounced than that of ilvC.
FIG. 4.
Influence of isoleucine on codY target genes. (A) Bacteria were grown in CDM without isoleucine to an OD600 of 0.4. Aliquots were then supplemented with isoleucine (200 mg/liter) and grown for another 1 h. (B) Strain Newman was grown without isoleucine to an OD600 of 0.4. Aliquots were then supplemented with increasing concentrations of isoleucine and further grown for 1 h. RNA were hybridized with a probe specific for ilvC or thrB. The 16S rRNA detected in the ethidium bromide-stained gels is indicated as a loading control in the lowest panel. WT, wild type.
CodY as a virulence regulator in S. aureus.
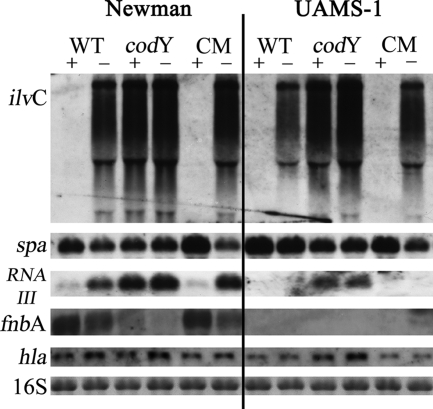
The repressor CodY may be essential not only for the regulation of metabolic genes but also for the fine-tuning of virulence-associated genes, as recently proposed by Majerczyk et al. (34). Indeed, the transcriptional pattern of RNAIII of the virulence regulator agr was identical to that of the prototypic CodY-repressed ilvC in strain Newman: repression after growth was observed with isoleucine in the wild-type and complemented strains but not in the codY mutant (Fig. 5). In contrast, genes coding for the cell surface proteins fnbA and spa were upregulated by isoleucine in a codY-dependent manner. No significant effect on hla expression was observed in strain Newman under these growth conditions. The diminished expression of spa and enhanced expression of RNAIII in the codY mutant compared to the wild type were also evident in strain UAMS-1. However, there were some differences between the results obtained from strain UAMS-1 and those from strain Newman. First, fnbA transcription was not detectable, which agrees with the results from genome sequencing showing that fnbA is not present in strain UAMS-1 (unpublished observation). Second, the effects of isoleucine in the wild-type and complemented strains were less pronounced. In addition, in this genetic background, the expression of the agr-regulated gene hla was enhanced in the codY mutant compared to the wild-type and complemented strains.
FIG. 5.
Influence of isoleucine on virulence gene expression. RNA from strain Newman and UAMS-1, their codY mutants (Newman-21 and UAMS-1-21, respectively) and the complemented strains ([CM] Newman-21/pCG30 and UAMS-1-21/pCG30, respectively) isolated from bacteria grown to an OD600 of 0.5 in CDM with (+) or without (−) isoleucine. They were then hybridized with probes specific for ilvC, spa, agr (RNAIII), fnbA, and hla. The 16S rRNA detected in the ethidium bromide-stained gels is indicated as a loading control in the lowest panel. WT, wild type.
Effect of codY on virulence gene expression in agr-negative background.
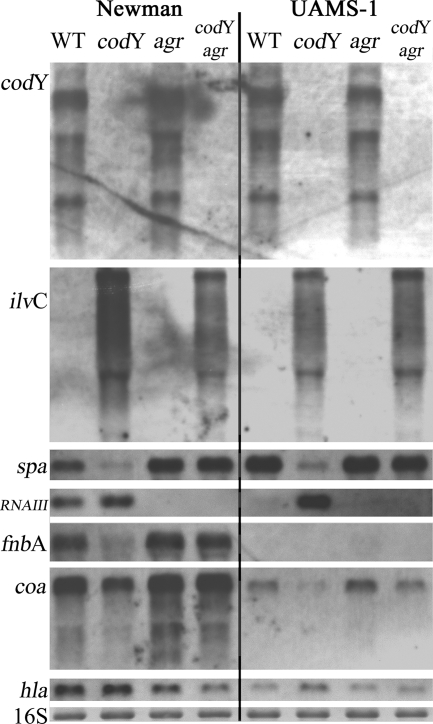
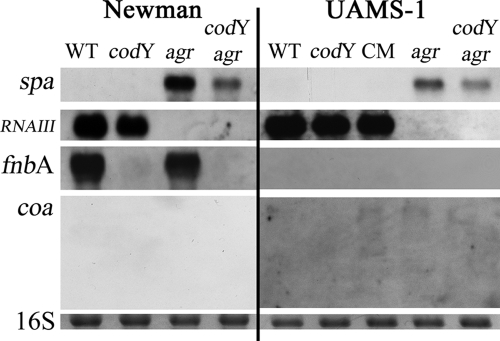
The virulence gene hla is known to be activated by RNAIII, whereas spa, coa, and fnbA are inhibited by RNAIII (37, 52, 53). To clarify whether the effect of CodY on virulence genes is solely due to RNAIII upregulation in the codY mutant, we analyzed virulence gene expression in agr and codY agr double mutants (Fig. 6). Interestingly, codY expression was not affected in the agr mutants of S. aureus. This is in contrast to Staphylococcus epidermidis, in which agr leads to elevated codY transcription (4). As expected in the agr-negative background, increased levels of spa, fnbA, and coa transcripts were detectable. In the agr-negative background, no significant effect of CodY was observed on these genes in bacteria during the exponential growth phase. However, when we analyzed bacteria from the postexponential phase, it could be shown that fnbA and spa were activated by CodY, independently of agr (Fig. 7). The lack of spa transcription in the agr-positive background is due to the strong repressive effect of RNAIII on spa transcription. Surprisingly, growth in CDM also allowed transcription of fnbA in the late growth phase. This is usually not seen using complex medium, in which fnbA, like coa, is repressed during postexponential growth independently of agr (42, 53).
FIG. 6.
Influence of CodY and agr on virulence gene expression in bacteria from exponential growth phase. RNA from the wild type (WT), codY, agr, and codY agr mutants was isolated from bacteria grown in CDM to the exponential (OD600 of 0.5) growth phase. The 16S rRNA detected in the ethidium bromide-stained gels is indicated as loading control in the lowest panel.
FIG. 7.
Influence of CodY and agr on virulence gene expression in bacteria from postexponential growth phase. RNA from wild-type (WT) strains (Newman or UAMS-1) and their codY, agr, and codY agr mutants was isolated from bacteria grown in CDM with an excess of isoleucine (500 mg/liter) to the postexponential phase. The 16S rRNA detected in the ethidium bromide-stained gels is indicated as loading control in the lowest panel.
Global effect of codY on gene expression in agr-positive and agr-negative backgrounds.
In order to acquire a more comprehensive understanding of the codY regulon, we performed microarray analysis. On the basis of the results obtained up to that point, we expected to detect the most pronounced effects of CodY in bacteria grown with isoleucine during the exponential phase. Under these conditions, CodY should be saturated with its ligand(s) and thus display full repression.
First, we determined the codY regulon by comparing gene expression in the parental strain with that in the codY mutant. A total of 124 genes (5% of the genome) showed differential expression in the codY mutant compared to its parent (Table 2). These genes were predicted to be contained in 71 operons (operon structure was predicted by the public database http://www.microbesonline.org). For each gene with significant difference in gene expression, the neighboring genes within the operons were also analyzed (see Table S3 in the supplemental material). In most cases genes that were predicted to be located in an operon were coregulated although this correspondence did not always reach the level of significance.
TABLE 2.
The codY regulon of S. aureus
| Newman ORFa | Description | Gene | Differential expression in the indicated strains (n-fold)b
|
Category | |
|---|---|---|---|---|---|
| Newman vs codY mutant | agr mutant vs agr codY mutant | ||||
| NWMN_0128 | N-Acetylglutamate gamma-semialdehyde dehydrogenase | argC | 0.39 | NS | Amino acid transport/metabolism |
| NWMN_0130 | Branched-chain amino acid transport system II carrier protein | brnQ1 | 0.33 | 0.44 | Amino acid transport/metabolism |
| NWMN_0144 | Oligopeptide ABC transport. permease | 0.03 | 0.15 | Amino acid transport/metabolism | |
| NWMN_0145 | Peptide ABC transporter. permease | 0.15 | 0.33 | Amino acid transport/metabolism | |
| NWMN_0146 | RGD-containing lipoprotein | rlp | 0.31 | 0.4* | Amino acid transport/metabolism |
| NWMN_0147 | Gamma glutamyltranspeptidase | ggt | 0.12 | 0.21 | Amino acid transport/metabolism |
| NWMN_0348 | 5-Methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | metE | 0.06 | 0.06 | Amino acid transport/metabolism |
| NWMN_0349 | Methylenetetrahydrofolate reductase protein | metH | 0.03 | 0.04 | Amino acid transport/metabolism |
| NWMN_0350 | trans-Sulfuration enzyme family protein | 0.36 | 0.06 | Amino acid transport/metabolism | |
| NWMN_0351 | Cys/Met metabolism PLP-dependent enzyme | 0.02 | 0.06 | Amino acid transport/metabolism | |
| NWMN_0425 | Cystathionine gamma-synthase | metB | 0.36 | 0.48 | Amino acid transport/metabolism |
| NWMN_0436 | Glutamate synthase large subunit | gltB | 0.08 | 0.14 | Amino acid transport/metabolism |
| NWMN_0437 | NADH-glutamate synthase small subunit | gltD | 0.09 | 0.07 | Amino acid transport/metabolism |
| NWMN_0516 | Branched-chain amino acid aminotransferase | ilvE | 0.47 | 0.52* | Amino acid transport/metabolism |
| NWMN_0831 | Argininosuccinate lyase | argH | 0.24 | 0.46 | Amino acid transport/metabolism |
| NWMN_0855 | Oligopeptide transport system permease protein | oppB | 0.15 | 0.15 | Amino acid transport/metabolism |
| NWMN_0858 | Oligopeptide transport ATP-binding protein | oppD | 0.16 | 0.19 | Amino acid transport/metabolism |
| NWMN_0860 | Hypothetical protein | 0.12 | 0.17 | Amino acid transport/metabolism | |
| NWMN_0883 | Na+/alanine symporter family protein | 0.03 | 0.04 | Amino acid transport/metabolism | |
| NWMN_1239 | Aspartate kinase | 0.06 | 0.06 | Amino acid transport/metabolism | |
| NWMN_1240 | Homoserine dehydrogenase | metL | 0.05 | 0.06 | Amino acid transport/metabolism |
| NWMN_1241 | Threonine synthase | thrC | 0.06 | 0.07 | Amino acid transport/metabolism |
| NWMN_1245 | Amino acid permease | 0.5* | 0.5* | Amino acid transport/metabolism | |
| NWMN_1277 | Prephenate dehydrogenase | tyrA | 0.50 | 0.44 | Amino acid transport/metabolism |
| NWMN_1279 | Anthranilate synthase component I | 0.11 | 0.23 | Amino acid transport/metabolism | |
| NWMN_1280 | Anthranilate synthase component II | trpG | 0.15 | 0.21 | Amino acid transport/metabolism |
| NWMN_1281 | Anthranilate phosphoribosyltransferase | trpD | 0.16 | 0.16 | Amino acid transport/metabolism |
| NWMN_1282 | Indole-3-glycerol phosphate synthase | trpC | 0.10 | 0.11 | Amino acid transport/metabolism |
| NWMN_1283 | Phosphoriborylanthranilate isomerase | trpF | 0.08 | 0.16 | Amino acid transport/metabolism |
| NWMN_1284 | Tryptophan synthase subunit beta | trpB | 0.10 | 0.09 | Amino acid transport/metabolism |
| NWMN_1304 | Aspartate kinase | lysC | 0.12 | 0.19 | Amino acid transport/metabolism |
| NWMN_1305 | Aspartate semialdehyde dehydrogenase | asd | 0.12 | 0.09 | Amino acid transport/metabolism |
| NWMN_1306 | Dihydrodipicolinate | dapA | 0.09 | 0.09 | Amino acid transport/metabolism |
| NWMN_1307 | Dihydrodipicolinate reductase | dapB | 0.15 | 0.07 | Amino acid transport/metabolism |
| NWMN_1308 | Tetrahydrodipicolinate acetyltransferase | dapD | 0.10 | 0.11 | Amino acid transport/metabolism |
| NWMN_1311 | Diaminopimelate decarboxylase | lysA | 0.29 | 0.23 | Amino acid transport/metabolism |
| NWMN_1348 | Threonine dehydratase | ilvA | 0.12 | 0.13 | Amino acid transport/metabolism |
| NWMN_1616 | Aminotransferase, class V | 0.08 | 0.10 | Amino acid transport/metabolism | |
| NWMN_1617 | d-3-Phosphoglycerate dehydrogenase | serA | 0.07 | 0.10 | Amino acid transport/metabolism |
| NWMN_1749 | Glutamine transport ATP-binding protein | 0.30 | 0.55* | Amino acid transport/metabolism | |
| NWMN_1750 | Extracellular glutamine-binding protein | 0.5* | NS | Amino acid transport/metabolism | |
| NWMN_1960 | Dihydroxy acid dehydratase | ilvD | 0.03 | 0.09 | Amino acid transport/metabolism |
| NWMN_1961 | Acetolactate synthase large subunit | ilvB | 0.02 | 0.03 | Amino acid transport/metabolism |
| NWMN_1962 | Ketol acid reductoisomerase | ilvC | 0.01 | 0.02 | Amino acid transport/metabolism |
| NWMN_1963 | 2-Isopropylmalate synthase | leuA | 0.01 | 0.02 | Amino acid transport/metabolism |
| NWMN_1964 | 3-Isopropylmalate dehydrogenase | leuB | 0.02 | 0.04 | Amino acid transport/metabolism |
| NWMN_1965 | Isopropylmalate isomerase large subunit | leuC | 0.02 | 0.04 | Amino acid transport/metabolism |
| NWMN_1966 | 3-Isopropylmalate dehydratase small subunit | leuD | 0.03 | 0.02 | Amino acid transport/metabolism |
| NWMN_2347 | Glycine betaine/l-proline transport | opuCA | 0.47 | 0.27 | Amino acid transport/metabolism |
| NWMN_2500 | Amino acid permease family protein | 0.10 | 0.11 | Amino acid transport/metabolism | |
| NWMN_2501 | 4-Aminobutyrate aminotransferase | 0.09 | 0.13 | Amino acid transport/metabolism | |
| NWMN_2571 | Imidazole glycerol phosphate synthase subunit | hisF | 0.20 | 0.21 | Amino acid transport/metabolism |
| NWMN_2572 | Phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase | hisA | 0.18 | 0.14 | Amino acid transport/metabolism |
| NWMN_2573 | Imidazole glycerol phosphate synthase | hisH | 0.20 | 0.31 | Amino acid transport/metabolism |
| NWMN_2574 | Imidazoleglycerol phosphate dehydratase | hisB | 0.11 | 0.03 | Amino acid transport/metabolism |
| NWMN_2577 | ATP phosphoribosyltransferase | hisG | 0.18 | 0.31 | Amino acid transport/metabolism |
| NWMN_2370 | Putative transport protein | 0.19 | 0.24 | Amino acid transport/metabolism | |
| NWMN_0859 | Oligopeptide transport ATP-binding protein | oppF | 0.33 | 0.30 | Amino acid transport/metabolism |
| NWMN_0428 | ABC transporter | 0.10 | 0.18 | Inorganic ion transport/metabolism | |
| NWMN_1950 | Ammonium transporter | nrgA | 0.23 | NS | Inorganic ion transport/metabolism |
| NWMN_0423 | Sodium-dependent symporter protein | 3.03 | 3.92 | Inorganic ion transport/metabolism | |
| NWMN_2288 | Nitrite transport protein | narK | 2.28 | NS | Inorganic ion transport/metabolism |
| NWMN_0016 | Adenylosuccinate synthase | purA | 2.65 | NS | Nucleotide transport/metabolism |
| NWMN_0379 | Xanthine permease | pbuX | 2.51 | NS | Nucleotide transport/metabolism |
| NWMN_1110 | Uracil permease | pyrP | 2.23 | 2.1* | Nucleotide transport/metabolism |
| NWMN_1111 | Aspartate carbamoyltransferase catalytic subunit | pyrB | 2.35 | 2.2* | Nucleotide transport/metabolism |
| NWMN_1112 | Dihydroorotase | pyrC | 2.09 | 2.1* | Nucleotide transport/metabolism |
| NWMN_1249 | GMP oxidoreductase | guaC | 2.33 | 2.1* | Nucleotide transport/metabolism |
| NWMN_0163 | Formate acetyltransferase activating enzyme | pflA | 0.40 | 0.12 | Energy production/conversion |
| NWMN_0162 | Formate acetyltransferase | pflB | 0.27 | 0.16 | Energy production/conversion |
| NWMN_0979 | Pyruvate carboxylase | pycA | 0.37 | 0.31 | Energy production/conversion |
| NWMN_1325 | Dihydrolipoamide acetyltransferase | sucB | 0.45 | 0.55* | Energy production/conversion |
| NWMN_2294 | Putative nitrate reductase gamma chain | narI | 2.44 | NS | Energy production/conversion |
| NWMN_0167 | Acetyl-coenzyme A acetyltransferase | fadA | 3.04 | NS | Energy production/conversion |
| NWMN_0435 | Transcription activator of glutamate synthase operon | gltC | 0.32 | NS | Regulation |
| NWMN_1943 | Accessory gene regulator B | agrB | 0.42 | NS | Regulation |
| NWMN_1946 | Accessory gene regulator A | agrA | 0.44 | NS | Regulation |
| NWMN_0077 | Superoxide dismutase | sodM | 0.26 | 0.49* | Defense/Virulence factor |
| NWMN_0095 | Capsular polysaccharide synthesis enzyme CapA | capA | 0.32 | 0.33 | Defense/Virulence factor |
| NWMN_0096 | Capsular polysaccharide synthesis enzyme CapB | capB | 0.29 | 0.19 | Defense/Virulence factor |
| NWMN_0097 | Capsular polysaccharide synthesis enzyme CapC | capC | 0.35 | 0.16 | Defense/Virulence factor |
| NWMN_0098 | Capsular polysaccharide synthesis enzyme CapD | capD | 0.30 | 0.36 | Defense/Virulence factor |
| NWMN_0099 | Capsular polysaccharide synthesis enzyme CapE | capE | 0.32 | 0.31 | Defense/Virulence factor |
| NWMN_0100 | Capsular polysaccharide synthesis enzyme CapF | capF | 0.33 | NS | Defense/Virulence factor |
| NWMN_0101 | Capsular polysaccharide synthesis enzyme CapG | capG | 0.30 | NS | Defense/Virulence factor |
| NWMN_0102 | Capsular polysaccharide synthesis enzyme CapH | capH | 0.34 | 0.45 | Defense/Virulence factor |
| NWMN_0103 | Capsular polysaccharide synthesis enzyme CapI | capI | 0.41 | 0.50 | Defense/Virulence factor |
| NWMN_0104 | Capsular polysaccharide synthesis enzyme CapJ | capJ | 0.32 | NS | Defense/Virulence factor |
| NWMN_0105 | Capsular polysaccharide synthesis enzyme CapK | capK | 0.31 | 0.64 | Defense/Virulence factor |
| NWMN_0107 | Capsular polysaccharide synthesis enzyme CapM | capM | 0.45 | NS | Defense/Virulence factor |
| NWMN_0108 | Capsular polysaccharide synthesis enzyme CapN | capN | 0.43 | NS | Defense/Virulence factor |
| NWMN_0262 | Truncated glycerol ester hydrolase | geh | 0.35 | NS | Defense/Virulence factor |
| NWMN_0525 | Bone sialoprotein-binding protein | sdrE | 0.50 | 1.88* | Defense/Virulence factor |
| NWMN_1084 | Antibacterial protein (phenole-soluble moduline) | 0.06 | NS | Defense/Virulence factor | |
| NWMN_1246 | Catalase | katA | 0.36 | 0.36 | Defense/Virulence factor |
| NWMN_1872 | MHC class II analog protein | eap | 0.48 | NS | Defense/Virulence factor |
| NWMN_nd | Delta-hemolysin | hld | 0.30 | NS | Defense/Virulence factor |
| NWMN_0166 | Staphylocoagulase precursor | coa | 2.49 | NS | Defense/Virulence factor |
| NWMN_0394 | Auperantigen-like protein 7 | set7nm | 3.34 | NS | Defense/Virulence factor |
| NWMN_2392 | Cell wall-anchored protein | sasG | 2.85 | 0.5* | Defense/Virulence factor |
| NWMN_2399 | Fibronectin binding protein | FnBPA | 2.01* | NS | Defense/Virulence factor |
| NWMN_0071 | Acetoin reductase | butA | 0.19 | 0.17 | Miscellaneous |
| NWMN_0429 | N-Acetylmuramoyl-l-alanine amidase | 0.50 | NS | Miscellaneous | |
| NWMN_0721 | Sigma 54 modulation protein | 0.45 | 0.47 | Miscellaneous | |
| NWMN_1309 | Hippurate hydrolase | 0.14 | 0.19 | Miscellaneous | |
| NWMN_1618 | Haloacid dehalogenase-like hydrolase | 0.43 | 0.31 | Miscellaneous | |
| NWMN_2097 | Tagatose-6-phosphate kinase | lacC | 0.20 | NS | Miscellaneous |
| NWMN_2448 | ATP-dependent Clp protease | clpC | 0.38 | 0.37 | Miscellaneous |
| NWMN_0028 | Metallo-beta-lactamase superfamily protein | 2.91 | NS | Miscellaneous | |
| NWMN_0322 | Ascorbate-specific phosphotransferase system enzyme IIC | ulaA | 2.35 | NS | Miscellaneous |
| NWMN_0220 | Hypothetical protein | 0.20 | 0.47 | Function unknown | |
| NWMN_0404 | Hypothetical protein | lpl2nm | 0.5* | 0.25 | Function unknown |
| NWMN_0667 | Hypothetical protein | 0.5* | 0.47 | Function unknown | |
| NWMN_0896 | Hypothetical protein | 0.21 | 0.20 | Function unknown | |
| NWMN_0901 | Hypothetical protein | 0.07 | 0.24 | Function unknown | |
| NWMN_0902 | Hypothetical protein | 0.11 | 0.07 | Function unknown | |
| NWMN_1243 | Hypothetical protein | 0.27 | 0.27 | Function unknown | |
| NWMN_2221 | Hypothetical protein | 0.30 | 0.42 | Function unknown | |
| NWMN_2222 | Hypothetical protein | 0.10 | 0.16 | Function unknown | |
| NWMN_2230 | Hypothetical protein | 0.21 | 0.14 | Function unknown | |
| NWMN_2470 | Hypothetical protein | 0.11 | 0.09 | Function unknown | |
| NWMN_2578 | Hypothetical protein | 0.06 | 0.25 | Function unknown | |
| NWMN_0027 | Hypothetical protein | 3.66 | 4.07 | Function unknown | |
| NWMN_0401 | Hypothetical protein | 2.38 | NS | Function unknown | |
Based on the publically available Newman genome sequence. Boldface, genes that are repressed in the codY mutant.
Fold changes are indicated for each comparison and displayed for genes showing statistically significant differential expression. Values correspond to expression ratios, i.e., averaged expression levels from two independent replicate experiments (P < 0.05). NS, not significantly different; *, limit of significance.
The vast majority of targets (106/124) appeared to be repressed by CodY (upregulated in the mutant). Sixteen of these genes were preceded by a CodY box as predicted above (see Table S2 in the supplemental material). Fifty-eight of the codY-repressed genes encompassed genes involved in amino acid transport and metabolism, e.g., the ilv operon and the peptide transporters opp and brnQ1 encoding proteins involved in the metabolism or the transport of BCAAs. Besides agr only one other regulatory gene was significantly affected by CodY. The transcription activator of the glutamate synthase operon gltC is repressed by CodY, suggesting that the CodY-repressed genes gltB and gltD are indirectly influenced via gltC.
Only 18 genes seemed to be activated by CodY (downregulated in the mutant). None of these genes contained a putative CodY binding motif, supporting the hypothesis that upregulation of these genes is probably not related to a direct interaction with CodY. Interestingly, six CodY-activated genes were predicted to be involved in nucleotide transport and metabolism.
Genes encoding virulence and defense factors were either up- or downregulated by CodY. For instance, MWMN_1084, which codes for phenole-soluble moduline, appeared to be strongly downregulated. The cap operon coding for enzymes in capsular biosynthesis and genes for katalase (katA) and superoxide dismutase (sodM) were also downregulated. In contrast, expression of genes coding for the cell surface proteins fnbA, sasG, and coa were positively influenced by CodY.
We next asked which of the 124 codY-dependent genes determined in the agr-positive background are still codY regulated in an agr-negative background. To determine this, we compared gene expression of the agr mutant versus the agr codY double mutant. Ninety-six of the 124 genes appeared to be regulated by codY independent of the agr background, and most of these were involved in amino acid metabolism and transport. In contrast, many of the genes categorized as virulence and defense factors were not significantly influenced by CodY in the agr-negative background. However genes within the cap operon, katA and sodM, were still codY repressed independent of agr. These results confirm the results of Northern blot analysis indicating that CodY acts in an agr-independent and agr-dependent manner on virulence genes, whereas the influence on metabolic genes is mostly agr-independent.
DISCUSSION
CodY has been described as a conserved repressor of genes involved in the biosynthesis and transport of amino acids in several gram-positive species. Here, we analyze the role of CodY in the human pathogen S. aureus. In S. aureus codY is cotranscribed with three genes located upstream of codY (xerC, clpQ, and clpY). xerC codes for tyrosine recombinase, and clpQY codes for the ATP-dependent heat shock protease HslVU. The same gene order is present in the genome of B. subtilis, Listeria monocytogenes, and Enterococcus faecalis, indicating a physiological link between CodY and the heat shock protease Hs1VU in these bacteria. This must remain speculative, however, since little is known about the function of Hs1VU in stress response or pathogenesis (7). In streptococcal species, L. lactis or Clostridium difficile, codY is located in a different genetic context. Alignments of the CodY sequences (http://www.ebi.ac.uk/Tools/clustalw/) from a selected set of gram-positive bacteria revealed two clusters: cluster one containing CodY from S. aureus, B. subtilis, L. monocytogenes, and C. difficile and cluster two with CodY from E. faecalis, streptococci, and lactococci. Thus, the evolution of the genetic context is not congruent with sequence differences within CodY. For instance, E. faecalis resembles S. aureus with respect to the gene context, whereas from the CodY amino acid sequence, E. faecalis is very similar to the streptococcal species.
From studies of other bacteria, it was postulated that BCAAs are major ligands of CodY, resulting in severe repression of target genes. We could show that in S. aureus isoleucine, although not required for growth, resulted in strong repression of CodY target genes. Furthermore, an excess of isoleucine resulted in growth inhibition in wild-type bacteria. This suggests that certain target genes that are needed for optimal growth are repressed under these conditions. Since an excess of leucine or valine had no effect on codY target gene repression or growth, we conclude that isoleucine is the major ligand in S. aureus as proposed for CodY from L. lactis (11).
Interestingly, only codY mutants but not wild-type S. aureus strains were able to grow without threonine, indicating that threonine synthesis requires enzymes whose transcription is efficiently repressed by CodY. Indeed, thrC (coding for threonine synthase) was shown to be repressed by CodY (shown by microarray analysis and Northern analysis). Since threonine auxotrophy was also seen under conditions without isoleucine, it can be assumed that besides isoleucine, other ligands (or CodY without ligand) are sufficient to mediate the CodY-dependent repression of the threonine biosynthetic genes.
Functionally, there are also clear differences between different species in the GTP-binding capacity of CodY (39). CodY from S. aureus is similar to its homolog in B. subtilis with respect to a proposed GTP-binding motif derived from structure and sequence analyses (33). For an in-depth analysis of the role of GTP as a signaling molecule, mutants with defects in internal GTP synthesis and/or the stringent control are currently being examined.
Overall, it can be assumed that CodY of S. aureus functions in a manner similar to that which has been shown for other gram-positive organisms. This is probably also true for the proposed CodY binding motif since we were able to detect CodY target genes based on a stringent motif search allowing only two mismatches within the consensus sequence. Lowering the stringency to four or five mismatches resulted in >1,000 putative sites within the genome, which appears of little informative value. However, the high stringency may have caused us to miss several real CodY boxes as a previous study in B. subtilis showed that even up to five mismatches within the CodY box consensus could result in a functional element (5). In fact, a CodY box was predicted for only a subset of the codY-dependent genes identified by our microarray analysis. When we allowed three mismatches within 200 bp upstream of ORFs, a reasonable number (222) of additional genes with a putative codY box were found. For instance, in the upstream sequence of the cap operon and the fnbA gene, a codY motif could now be identified. Interestingly, the CodY box preceding the cap operon overlapped with the mapped −10 region (38). The putative CodY box in front of fnbA was located between a sigma B binding motif and the transcriptional start site. Thus, these two virulence-associated genes may be direct targets of CodY regulation. Direct binding assays such as chromatin immunoprecipitation with microarray analysis or gel retardation assays using purified CodY are needed to clarify this topic in the future.
From microarray analysis it became clear that, overall, the codY regulon is conserved between gram-positive species with regard to profound and mostly direct repression of genes involved in amino acid biosynthesis and transport. Genes that are activated via CodY are much less conserved, and a direct interaction of CodY with this activated target gene could be shown only for ackA of B. subtilis (44). In our screen, several genes involved in nucleotide synthesis and transport were activated by codY. Similarly, in L. monocytogenes, guaA and guaB, which encode the enzymes involved in biosynthesis of GMP, which encode, were also found to be activated by codY (6). Thus, there may be at least an indirect link between CodY and the intracellular nucleotide pool.
The most comprehensive data on the CodY regulon and function are based on the analysis of the nonpathogenic bacteria B. subtilis and L. lactis. There is emerging evidence that CodY also has an impact on virulence gene expression in gram-positive pathogens (6, 14, 26, 32, 35, 46). Interestingly, in S. aureus and L. monocytogenes, the quorum-sensing agr system is affected by CodY in opposite directions: in L. monocytogenes CodY leads to agr activation, while in S. aureus it leads to agr repression (6, 34). Although a putative CodY box with three mismatches is located upstream of agrB in S. aureus, this box overlaps with hld and is not in proximity to the transcriptional start site or the binding site of the sensor histidine kinase ArgA. Thus, the mechanism leading to agr repression remains unclear. Nevertheless, repression of the agr system by CodY presumably enhances the tight growth phase-dependent activation of this quorum-sensing system which by definition is activated at higher cell densities. Thus, under conditions of isoleucine limitation (or in codY mutants), the agr system is prematurely activated. Consequently, agr-activated genes like toxins and the cap operon also become activated, whereas genes known to be downregulated by the agr system are switched off. This can be seen as an escape mechanism for the bacteria under limiting conditions. A CodY-dependent repression of hla and the ica operon (coding for the enzyme generating the polysaccharide intercellular adhesin) was shown recently by Majerczyk et al. (34). In our microarray analysis, hla and the ica operon were not significantly affected in the codY mutants. Both genes are known to be poorly expressed in strain Newman. However, increased expression of hla in the codY mutant could be confirmed for strain UAMS-1. Genes of the ica operon were below the threshold in our analysis but also showed a clear tendency toward higher expression in the codY mutant, as further confirmed by reverse transcription-PCR in both genetic backgrounds (strain Newman and UAMS-1) (data not shown).
Genes for cell-associated proteins such as fnbA, spa, and coa are activated by CodY. The role of CodY in the activation of these virulence genes might be primarily via agr. However, direct activation through CodY binding can be presumed for fnbA activation because of the presence of a putative CodY box and the observation that fnbA is still CodY dependent in an agr-negative background. Additional regulatory mechanism(s) may act primarily at the posttranscriptional level on some of the genes since, besides agr, no other virulence regulatory gene was differentially expressed between wild-type and mutant strains.
A link to the situation during infection or colonization is hard to draw since several studies have shown that agr is not activated during chronic infections and/or colonization (20, 21), and little is known about the growth conditions in vivo. However, our analysis of the codY regulon in an agr-negative background clearly shows that CodY also regulates some of the virulence factors independently of agr. We and others have shown elsewhere that hla can be transcribed independently of agr in vivo (21, 54). Thus, CodY may contribute to virulence gene regulation in vivo not only by supporting but also by replacing agr function: under limited conditions (low isoleucine) the capsule and several secreted enzymes and toxins are derepressed, whereas some of the cell surface molecules are downregulated. Overall, the impact of codY mutation on S. aureus needs to be assessed in vivo as some codY targets may contribute to cell adhesion and survival in hostile environments.
Supplementary Material
Acknowledgments
We thank Vittoria Bisanzio for her excellent technical assistance. We are grateful to Inigo Lasa for the donation of plasmid pMAD rsbU. Isolate RNA was obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NASA) Program supported under NIAID/NIH contract HHSN2722007 00055C.
This work was supported by grants to C.W. from the Deutsche Forschungsgemeinschaft (TR34) and by grants 3100A0-116075/1 (P.F.) and 3100A0-112370/1 (J.S.) from the Swiss National Science Foundation.
Footnotes
Published ahead of print on 27 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 706887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., T. Bae, O. Schneewind, F. Takeuchi, and K. Hiramatsu. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, B. T., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 697851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batzilla, C. F., S. Rachid, S. Engelmann, M. Hecker, J. Hacker, and W. Ziebuhr. 2006. Impact of the accessory gene regulatory system (Agr) on extracellular proteins, codY expression and amino acid metabolism in Staphylococcus epidermidis. Proteomics 63602-3613. [DOI] [PubMed] [Google Scholar]
- 5.Belitsky, B. R., and A. L. Sonenshein. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol. 1901224-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett, H. J., D. M. Pearce, S. Glenn, C. M. Taylor, M. Kuhn, A. L. Sonenshein, P. W. Andrew, and I. S. Roberts. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 631453-1467. [DOI] [PubMed] [Google Scholar]
- 7.Butler, S. M., R. A. Festa, M. J. Pearce, and K. H. Darwin. 2006. Self-compartmentalized bacterial proteases and pathogenesis. Mol. Microbiol. 60553-562. [DOI] [PubMed] [Google Scholar]
- 8.Charbonnier, Y., B. Gettler, P. Francois, M. Bento, A. Renzoni, P. Vaudaux, W. Schlegel, and J. Schrenzel. 2005. A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus. BMC Genomics 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchill, G. A. 2004. Using ANOVA to analyze microarray data. BioTechniques 37173-175, 177. [DOI] [PubMed] [Google Scholar]
- 10.Coulter, S. N., W. R. Schwan, E. Y. W. Ng, M. H. Langhorne, H. D. Ritchie, S. Westbrock-Wadman, W. O. Hufnagle, K. R. Folger, A. S. Bayer, and C. K. Stover. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30393-404. [DOI] [PubMed] [Google Scholar]
- 11.den Hengst, C. D., P. Curley, R. Larsen, G. Buist, A. Nauta, S. D. van, O. P. Kuipers, and J. Kok. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.den Hengst, C. D., S. A. van Hijum, J. M. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 28034332-34342. [DOI] [PubMed] [Google Scholar]
- 13.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367731-739. [DOI] [PubMed] [Google Scholar]
- 14.Dineen, S. S., A. C. Villapakkam, J. T. Nordman, and A. L. Sonenshein. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66206-219. [DOI] [PubMed] [Google Scholar]
- 15.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 695-107. [DOI] [PubMed] [Google Scholar]
- 16.Garzoni, C., P. Francois, A. Huyghe, S. Couzinet, C. Tapparel, Y. Charbonnier, A. Renzoni, S. Lucchini, D. P. Lew, P. Vaudaux, W. L. Kelley, and J. Schrenzel. 2007. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genomics 8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiger, T., C. Goerke, M. Mainiero, D. Kraus, and C. Wolz. 2008. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J. Bacteriol. 1903419-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 1872426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillaspy, A. F., S. G. Hickmon, R. A. Skinner, J. R. Thomas, C. L. Nelson, and M. S. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 633373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goerke, C., S. Campana, M. G. Bayer, G. Döring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 681304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA, and sae of Staphylococcus aureus on the induction of α-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 401439-1448. [DOI] [PubMed] [Google Scholar]
- 22.Goerke, C., and C. Wolz. 2004. Regulatory and genomic plasticity of Staphylococcus aureus during persistent colonization and infection. Int. J. Med. Microbiol. 294195-202. [DOI] [PubMed] [Google Scholar]
- 23.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 401227-1239. [DOI] [PubMed] [Google Scholar]
- 24.Guedon, E., B. Sperandio, N. Pons, S. D. Ehrlich, and P. Renault. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 1513895-3909. [DOI] [PubMed] [Google Scholar]
- 25.Heinemann, M., A. Kummel, R. Ruinatscha, and S. Panke. 2005. In silico genome-scale reconstruction and validation of the Staphylococcus aureus metabolic network. Biotechnol. Bioeng. 92850-864. [DOI] [PubMed] [Google Scholar]
- 26.Hendriksen, W. T., H. J. Bootsma, S. Estevao, T. Hoogenboezem, A. de Jong, R. de Groot, O. P. Kuipers, and P. W. Hermans. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190590-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 1019786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 2782169-2176. [DOI] [PubMed] [Google Scholar]
- 29.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 30.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 3571225-1240. [DOI] [PubMed] [Google Scholar]
- 31.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103101-105. [DOI] [PubMed] [Google Scholar]
- 32.Lemos, J. A., M. M. Nascimento, V. K. Lin, J. Abranches, and R. A. Burne. 2008. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 1905291-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levdikov, V. M., E. Blagova, P. Joseph, A. L. Sonenshein, and A. J. Wilkinson. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J. Biol. Chem. 28111366-11373. [DOI] [PubMed] [Google Scholar]
- 34.Majerczyk, C. D., M. R. Sadykov, T. T. Luong, C. Lee, G. A. Somerville, and A. L. Sonenshein. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 1902257-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malke, H., and J. J. Ferretti. 2007. CodY-affected transcriptional gene expression of Streptococcus pyogenes during growth in human blood. J. Med. Microbiol. 56707-714. [DOI] [PubMed] [Google Scholar]
- 36.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26399-407. [DOI] [PubMed] [Google Scholar]
- 37.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 38.Ouyang, S., S. Sau, and C. Y. Lee. 1999. Promoter analysis of the cap8 operon, involved in type 8 capsular polysaccharide production in Staphylococcus aureus. J. Bacteriol. 1812492-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petranovic, D., E. Guedon, B. Sperandio, C. Delorme, D. Ehrlich, and P. Renault. 2004. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol. Microbiol. 53613-621. [DOI] [PubMed] [Google Scholar]
- 40.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 151093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renzoni, A., C. Barras, P. Francois, Y. Charbonnier, E. Huggler, C. Garzoni, W. L. Kelley, P. Majcherczyk, J. Schrenzel, D. P. Lew, and P. Vaudaux. 2006. Transcriptomic and functional analysis of an autolysis-deficient, teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 503048-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saravia-Otten, P., H.-P. Müller, and S. Arvidson. 1997. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 1795259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scherl, A., P. Francois, Y. Charbonnier, J. M. Deshusses, T. Koessler, A. Huyghe, M. Bento, J. Stahl-Zeng, A. Fischer, A. Masselot, A. Vaezzadeh, F. Galle, A. Renzoni, P. Vaudaux, D. Lew, C. G. Zimmermann-Ivol, P. A. Binz, J. C. Sanchez, D. F. Hochstrasser, and J. Schrenzel. 2006. Exploring glycopeptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genomics 7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shivers, R. P., S. S. Dineen, and A. L. Sonenshein. 2006. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol. Microbiol. 62811-822. [DOI] [PubMed] [Google Scholar]
- 45.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53599-611. [DOI] [PubMed] [Google Scholar]
- 46.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in gram-positive bacteria. Curr. Opin. Microbiol. 8203-207. [DOI] [PubMed] [Google Scholar]
- 47.Sonenshein, A. L. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5917-927. [DOI] [PubMed] [Google Scholar]
- 48.Steinhuber, A., C. Goerke, M. G. Bayer, G. Döring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on the expression of virulence factors. J. Bacteriol. 1856278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talaat, A. M., S. T. Howard, W. Hale, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toledo-Arana, A., N. Merino, M. Vergara-Irigaray, M. Debarbouille, J. R. Penades, and I. Lasa. 2005. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 1875318-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandenesch, F., J. Kornblum, and R. P. Novick. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 1736313-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolz, C., D. McDevitt, T. J. Foster, and A. L. Cheung. 1996. Influence of agr on fibrinogen binding in Staphylococcus aureus Newman. Infect. Immun. 643142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolz, C., P. Pöhlmann-Dietze, A. Steinhuber, Y.-T. Chien, A. C. Manna, W. J. van Wamel, and A. L. Cheung. 2000. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36230-243. [DOI] [PubMed] [Google Scholar]
- 54.Xiong, Y. Q., J. Willard, M. R. Yeaman, A. L. Cheung, and A. S. Bayer. 2006. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J. Infect. Dis. 1941267-1275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.