Abstract
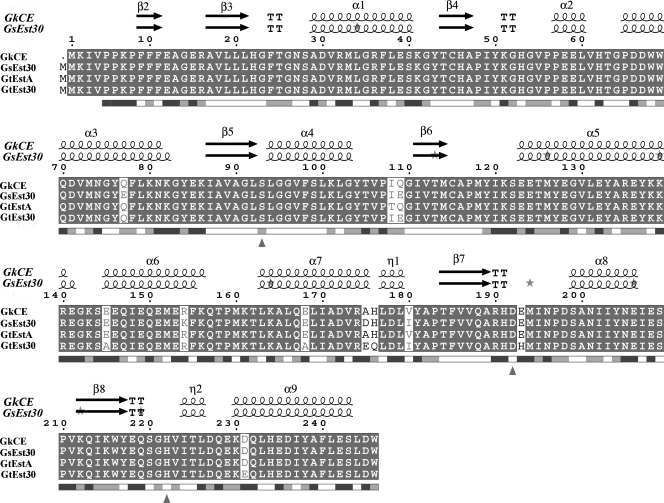
The gene GK3045 (741 bp) from Geobacillus kaustophilus HTA426 was cloned, sequenced, and overexpressed into Escherichia coli Rosetta (DE3). The deduced protein was a 30-kDa monomeric esterase with high homology to carboxylesterases from Geobacillus thermoleovorans NY (99% identity) and Geobacillus stearothermophilus (97% identity). This protein suffered a proteolytic cut in E. coli, and the problem was overcome by introducing a mutation in the gene (K212R) without affecting the activity. The resulting Est30 showed remarkable thermostability at 65°C, above the optimum growth temperature of G. kaustophilus HTA426. The optimum pH of the enzyme was 8.0. In addition, the purified enzyme exhibited stability against denaturing agents, like organic solvents, detergents, and urea. The protein catalyzed the hydrolysis of p-nitrophenyl esters of different acyl chain lengths, confirming the esterase activity. The sequence analysis showed that the protein contains a catalytic triad formed by Ser93, Asp192, and His222, and the Ser of the active site is located in the conserved motif Gly91-X-Ser93-X-Gly95 included in most esterases and lipases. However, this carboxylesterase showed no more than 17% sequence identity with the closest members in the eight families of microbial carboxylesterases. The three-dimensional structure was modeled by sequence alignment and compared with others carboxylesterases. The topological differences suggested the classification of this enzyme and other Geobacillus-related carboxylesterases in a new α/β hydrolase family different from IV and VI.
Esterases catalyze hydrolysis and synthesis of ester bonds. Even if the biological functions have not been fully described, they have been involved in catabolic pathways (3, 5). Essentially, carboxylesterases (CEs; EC 3.1.1.1) exhibit high regio- and stereospecificity, require no cofactor, and are active in organic solvents, which make them attractive in important industrial and medical roles in the synthesis and hydrolysis of stereospecific compounds, including the metabolic processing of drugs and antimicrobial agents (4, 24, 29).
Due to their importance, CEs have been identified in a wide range of organisms, and several of these have been cloned, including those from several Bacillus stearothermophilus strains (20) and from Pseudomonas sp. strain S34 (19). The elucidation of many gene sequences and the resolution of some crystal structures have permitted a structural classification of these enzymes in several families within the α/β-hydrolase fold family (2, 8).
Esterases from thermophiles have become objects of special interest for structural investigation and for a broad range of biotechnological applications. CE and lipase properties and applications have been reviewed recently by Bornscheuer (5) and Jaeger et al. (14-16).
In the search for new CEs, the gene GK3045 (741 bp) of Geobacillus kaustophilus HTA426 is of particular interest since this microorganism can grow at up to 74°C (optimally at 60°C). It was isolated from the deep-sea sediment of the Mariana Trench (41, 42) at a depth of 10,897 m. The complete genome sequence of this strain, which is composed of a 3.54-Mb chromosome and a 47.9-kb plasmid, has been determined as the first thermophilic bacillus (43).
In this paper, we describe for the first time the cloning and characterization of a thermostable CE from G. kaustophilus HTA426 (CEGk). In addition, a plausible three-dimensional structure is proposed and compared with known structures.
MATERIALS AND METHODS
Strains, plasmids, enzymes, and chemicals.
The genomic DNA of G. kaustophilus HTA426 was isolated by a previously described method (42). The Escherichia coli Rosetta (DE3) strain and a pET28a expression vector were obtained from Novagen. The DNA Taq polymerase, restriction endonucleases, and alkaline phosphatase were purchased from New England Biolabs (Beverly, MA), and T4 DNA ligase was obtained from Roche Diagnostics (Barcelona, Spain). All the substrates (p-nitrophenyl [NP] esters), isopropyl-β-d-thiogalactopyranoside (IPTG), kanamycin, chloramphenicol, and standard proteins used in molecular mass studies were obtained from Sigma. All other chemicals were of reagent grade and were obtained from commercial sources.
Cloning of the GK3045 gene and DNA manipulation.
The cloning and transformation techniques used were essentially those described by Sambrook et al. (37). The locus tag GK3045 (GeneID 3185723) was amplified by PCR (TGradient; Biometra) using oligodesoxyribonucleotide primers containing restriction enzymes sites for EcoRI in the 5′ and NotI in the 3′: 5′-GGAATTCATGAAAATTGTTCCGCCGAAGCCG-3′ and the downstream primer 5′-GGCGGCCGCTTACCAATCTAACGATTCAAGAAATGCATAAAT-3′ (restriction sites are underlined in both cases). The amplified product was digested with these restriction enzymes (2 h at 37°C), purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany), and ligated (14 h at 16°C) into the expression vector pET28a. Both strands of the resulting plasmid were automatically sequenced. This vector encodes an N-terminal six-His tag, thus enabling the expressed protein to be purified by affinity chromatography (7). This pET28a6His-GkCE expression plasmid was transferred to the host E. coli Rosetta (DE3) competent cells for protein expression.
Enzyme assays.
Esterase activity was determined spectrophotometrically by hydrolysis of different esters of p-NP. The standard reaction medium (1 ml) contained 0.8 mM p-NP caprylate, 50 mM phosphate buffer, 0.36% Triton X-100, 0.1% gum arabic, pH 8.0, and 12.5 μg of purified enzyme (34). The hydrolysis was measured at 405 nm with a ɛ405 of 16,980 M−1 cm−1, using a Shimadzu UV-2401PC spectrophotometer. One unit of activity was defined as the amount of enzyme releasing 1 μmol of p-NP per min under assay conditions.
To determine the substrate specificity, different p-NP esters such as p-NP acetate (C2), p-NP butyrate (C4), p-NP caprylate (C8), and p-NP laurate (C10), were used.
Protein analysis.
The concentration of protein was assayed by the bicinchoninic acid method (39), using bovine serum albumin as a standard. The molecular weight of the native protein was determined by gel filtration (Superdex 200 10/300GL column; GE Healthcare); the column was equilibrated in 50 mM phosphate buffer, 0.15 M NaCl, pH 8.0, at a flow rate of 0.5 ml/min. The molecular mass under denaturing conditions (sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) was determined using 12% acrylamide gel.
To determine the mass of the original protein and the proteolytic product accurately, molecular weight was also measured by liquid chromatography-mass spectrometry with electrospray ionization, using an HP/1100 LC/MSD Ion Trap System (Agilent Technologies), according to previously published methods (33, 46). Data analysis for the LC/MSD Ion Trap, version 3.2, was used to determine the molecular masses of the purified proteins in a spectrum. With this method the molecular masses obtained are accurate within about 1 Da.
Stability of CEGk.
The pH stability of the purified enzyme was studied in the pH range of 4.0 to 10. The following buffers (50 mM) were used: sodium acetate (pH 4.0 to 5.5), potassium phosphate (pH 6 to 8), sodium bicarbonate (pH 8 to 9), and boric acid (pH 10 to 11). In the pH stability study, the residual enzyme activity was measured after a 24-h incubation at 30°C with the standard reaction medium for the enzyme assay. The optimum pH toward the activity of the esterase was also determined with the same buffers.
The thermostability of the enzyme was examined using the standard assay with p-NP caprylate after incubation of the enzyme from 50°C to 80°C for different periods of time.
The stability of the enzyme against denaturing agents (detergents, organic solvents, and urea) was also studied using a standard assay with p-NP caprylate. The activity was measured at two different temperatures (30 and 60°C) immediately after each compound had been mixed with the enzyme and after 60 min of incubation, as described by Young-Jun Park et al. (32). Blank samples were prepared with the buffer solution instead of the enzyme and incubated in the same way. Each measurement was carried out with two different concentrations of the compounds: 0.1% to 5% (wt/vol) concentrations of detergents, 40% to 90% (vol/vol) concentrations of organic solvents, and 1 M to 8 M urea.
The inhibitory effect of the chemical modifiers, which are specific to particular amino acids, such as pyridoxal 5′-phosphate (PLP) to Lys, phenylglyoxal (PGO) to Arg, phenylmethylsulfonyl (PMSF) to Ser, and diethylpirocarbonate to His, was examined. Enzyme activity was measured using a standard assay with p-NP caprylate. Two different concentrations (0.5 mM and 5 mM) of each inhibitor were added to the enzyme, the mixture was incubated for 30 min at 30°C, and then the reaction was started with the addition of p-NP caprylate. Paraoxon and eserine, which are known as indicators for the classification of esterases, were also studied under the same conditions as described above.
A 5 mM concentration of divalent cations (CaCl2, CuSO4, FeSO4, MnCl2, MgCl2, and ZnSO4) were separately added to the enzyme and incubated for 30 min at 30°C to investigate the effect on the enzyme activity. In order to clarify whether the divalent cations are required for the reaction, 10 mM EDTA was added to the enzyme and incubated for 60 min at 60°C. The residual activities were measured using the standard assay with p-NP caprylate after incubation.
Expression and purification of CEGk.
The selected clone from the screening was grown in 1 liter of Terrific Broth medium supplemented with antibiotics overnight at 37°C under constant shaking. The culture was induced at an optical density at 600 nm of 5 by adding 0.5 mM IPTG for 6 h at 30°C. The culture was centrifuged at 4°C for 20 min at 6,000 × g. The cell pellet was resuspended in 250 ml of 50 mM phosphate buffer, pH 8.0, with protease inhibitor cocktail tablets (Roche).
Resuspended cells were disrupted using a mill homogenizor (Mini ZetaII; Netzsch) for 15 min at 2,000 rpm. The insoluble fraction of the lysate was removed by centrifugation at 4°C for 20 min at 6,000 × g. To remove nucleic acids, 3 U/ml DNase I (Sigma) was added, and the sample was shaken slowly for 30 min at room temperature, followed by centrifugation for 20 min at 6,000 × g and 4°C.
The enzymatic extract purification was performed in two steps: tangential ultrafiltration with a 100-kDa cutoff membrane on a QuixStand system (UFP-100-C-4MA, GE Healthcare) followed by His tag affinity chromatography (ÄKTA Purifier, GE Healthcare) with 1 ml of HisTrap FF on a 1-ml column (GE Healthcare), equilibrated with 25 mM phosphate buffer, 500 mM NaCl, and 5 mM imidazol, pH 8.0. The enzyme was eluted with a step gradient between 50 to 65 mM imidazol at a flow rate of 0.5 ml/min. Peaks of protein were pooled, and active fractions were analyzed for purity by SDS-PAGE.
Site-directed mutagenesis.
Site directed mutations were generated by a QuikChange Site-Directed Mutagenesis Kit (Stratagene) using the following primers: S93G-up (5′-GGCTGGATTGGGGCTTGGAGGCGTAT-3′), S93G-down (5′-ATACGCCTCCAAGCCCCAATCCAGCC-3′), D192V-up (5′-CCAAGCGCGCCATGATGAGATGATCAATCC-3′), D192V-down (5′-GGATTGATCATCTCATCATGGCGCGCTTGG-3′), H222L-up (5′-TATGAGCAATCAGGCCTTGTGATTACGCTTGAT-3′), and H222L-down (5′-ATCAAGCGTAATCACAAGGCCTGATTGCTCATA-3′) (mutations are underlined). Complementary primers with the wild-type gene bearing the nucleotides to be changed were used for PCR. The PCR mixtures were treated afterward with DpnI to digest wild-type DNA (methylated). The mutation sites were confirmed by DNA sequencing. For expression, plasmids were transformed into E. coli Rosetta (DE3) cells.
Mutants K212Stop and K212R were also constructed following the kit instructions. The oligonucleotides designed for the site-directed mutagenesis were the following: K212Stop-up (5′-AAATTGAATCGCCGGTCTAACAAATCAAATGGTAT-3′), derived from the 5′ end, and K212Stop-down (5′-ATACCATTTGATTTGTTAGACCGGCGATTCAATTT-3′), derived from the 3′ end; K212R-up (5′-TGAATCGCCGGTCAGACAAATCAAATGGT-3′), derived from the 5′ end, and K212R-down (5′-ACCATTTGATTTGTCTGACCGGCGATTCA-3′), derived from the 3′ end (mutations are underlined).
Sequence analysis.
Protein similarity searchers and alignment were performed using the data from CLUSTAL W (44), and only sequences previously cloned and with high homology were used. Model-building was developed by SWISS-MODEL and the Swiss-PdbViewer programs (1, 12, 38) using Est30 from Geobacillus stearothermophilus ([Est30Gs], Protein Data Bank [PDB] code 1TQH) as the template. ESPript (11) output was used to render the analysis of multiple sequence alignments. A phylogram was built using TreeView software with the neighbor-joining (36) method. Topology diagrams were prepared with the TOPS program (28).
RESULTS
Amino acid sequence comparison.
The deduced amino acid sequence of the CEGk gene showed significant identity with sequences of other Geobacillus and Bacillus species in the database. The CEGk is most closely related to EstA of Geobacillus thermoleovorans YN (DQ 288886; GI 82791235) with a sequence identity of 99% (Fig. 1). The enzyme also showed high homology to Est30Gs (PDB code 1TQH; ATCC 12980 and ATCC 7954) and to the thermostable Est30 of Geobacillus thermodenitrificans ([Est30Gt] YP_001127089), with sequence identities of 97 and 96%, respectively. All of these microorganisms are extremophiles, growing at high temperatures and under high-alkaline conditions. In addition, sequence alignment revealed that CEGk contains the typical catalytic triad composed of Ser93-Asp192-His222 and the consensus motif (Gly-X-Ser-X-Gly) around the active-site serine (Fig. 1), typical for α/β hydrolases such as lipases and esterases. Ser93Gly, Asp192Val, and His222Leu mutants were expressed as described in Materials and Methods in order to verify if they belong to the catalytic triad core. The enzymatic activities for p-NP substrates (p-NP acetate, butyrate, and caprylate) were measured for the wild type and the mutants to evaluate the effect of the changes on catalysis. No activity was found in Ser93, Asp192, or His222 mutants, showing that these amino acids are likely to be essential for the enzyme catalysis.
FIG. 1.
Multiple sequence alignment for CE from G. kaustophilus HTA426 (GkCE) and related Geobacillus CEs. EsPript outputs (11) obtained with the sequences from the SWISSPROT databank and aligned with CLUSTAL W (43). Sequences are grouped according to similarity. The enzyme showed 97% sequence identity with Est30Gs, 99% with EstA from G. thermoleovorans (GtEstA) NY, and 96% with Est30Gt. Residues strictly conserved have a solid background. Symbols above blocks of sequences represent the secondary structure, springs represent helices, and arrows represent β-strands. The residues forming the hydrophobic specificity pocket are indicated by small black asterisks. Triangles represent the location of the active site. K212 represents the protease cut.
Enzyme cloning, overexpression, and purification.
Sequence analysis of the genome revealed one major open reading frame of 741 bp, which encodes a hypothetical polypeptide of 246 residues, corresponding to a molecular mass of 30 kDa. Genomic DNA from G. kaustophilus HTA426 was used as the template to amplify the gene encoding the CE by PCR with the oligodeoxiribonucleotides described in Materials and Methods. The PCR product was purified and digested with EcoRI/NotI to ligate it into the expression vector pET28a. The recombinant plasmid was used to transform electrocompetent cells of E. coli Rosetta (DE3). The selected clone was grown in TB medium supplemented with kanamycin-chloramphenicol and induced with 0.5 mM IPTG for 6 h. The CE activity was almost totally expressed in soluble form and represented, under these conditions, about 28% of the total protein expressed, as calculated by bioinformatics analysis with Image Quant TL software (Amersham Biosciences) (Fig. 2, lane 2).
FIG. 2.

SDS-PAGE of the GK3045 gene product after 6 h of IPTG induction. Each lane contained 40 μg of protein. M, molecular size standards (P7708S; New England Biolabs). Lane 1, Rosetta (DE3) transformed with pET28a and induced; lane 2, the cell extract containing cloned CEGk after tangential ultrafiltration; lane 3, CEGk obtained by site-directed mutagenesis (K212R) without the proteolytic cut; lane 4, smaller and inactive CEGk obtained by site-directed mutagenesis (K212Stop); lane 5, His-Trap FF column pooled fraction in which a purified CEGk (of about 30 kDa) and another protein (of about 27 kDa) are shown; the latter is the result of a proteolytic cut.
The CE was purified from E. coli cells by a two-step procedure consisting of a 100-kDa ultrafiltration step and Ni2+ chelating affinity chromatography into a HisTrap FF column. After these steps, the enzyme was purified 3.36-fold with a 65% recovery and showed specific activity of 19.06 U/mg for the hydrolysis of p-NP caprylate at room temperature and pH 8.0 (data not shown). The SDS-PAGE gel of purified enzyme showed the expected band of 30 kDa (Fig. 2, lane 2) and an additional one at 27 kDa. The two bands that appeared in the gel (30 kDa and 27 kDa) were also described by Soliman et al. (40) in the closely related esterase, EstA of G. thermoleovorans NY, but with no explanation.
The molecular masses of these two bands were also confirmed by liquid chromatograph-mass spectrometry as 32,045 kDa and 27,754 kDa, respectively (data not shown), and by gel filtration chromatography (data not shown), confirming the monomeric nature of this CE. To explore the origin of these two bands, which could not be separated by a Ni2+ affinity chromatography, a proteolytic analysis of the C terminus was carried out by using the program CLC Protein Workbench (version 3.0.2) (www.clcbio.com). Bioinformatics approaches were used to identify potential peptidase cleavage sites. By scanning the amino acid sequence for patterns that matched the corresponding cleavage site for the protease, both molecular weights could be found. This showed a possible endopeptidase cut in K212-Q213 at the beginning of the β8-strand, in which K212 belongs to the hydrophobic specificity pocket (Fig. 1). This possible cleavage is also present in the sequence of EstA from G. thermoleovorans YN (Fig. 1).
Thus, two mutants were constructed, one introducing a stop codon (K212Stop) and the other introducing a similar amino acid (Arg) (K212R). The first clone resulted in an expected 27-kDa dead enzyme (Fig. 2, lane 4) since the catalytic histidine (His222) was removed. On the other hand, the second mutation resulted in an active enzyme with only one band of 30 kDa (Fig. 2, lane 3) with a similar specific activity (5.76 U/mg versus 5.84U/mg in the wild type). The latter mutant was used for the rest of this work and clearly confirmed the proteolytic attack as the origin of these two bands in the CEGk and probably explains the results obtained in EstA of G. thermoleovorans NY (40).
Biochemical characterization of the recombinant enzyme.
Substrate specificity was initially tested toward several p-NP esters at 1 mM concentrations of substrates of different chain lengths using a spectrophotometric assay. Maximum activity was observed when p-NP acetate (C2) was used as a substrate (Table 1). A decrease in activity was detected as the length of the side chain increased, which is similar behavior to that described for other esterases. This confirms the assumption that the enzyme is an esterase rather than a lipase. The activity toward p-NP myristate (C14 acyl group) and p-NP palmitate (C16 acyl group) was not detected under the specified conditions.
TABLE 1.
Kinetic parameters of recombinant CEGka
| Substrate (chain) | Km (mM) | KSI (mM)b | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| p-NP acetate (C2) | 2.77 | 280.41 | 101.23 | |
| p-NP butyrate (C4) | 1.36 | 9.07 | 6.67 | |
| p-NP caprylate (C8) | 0.40 | 2.02 | 4.79 | 11.97 |
| p-NP laurate (C10) | 14.62 | 0.02 | 70.46 | 4.82 |
| p-NP myristate (C14) | 0.98 | 0.087 | 0.09 | |
| p-NP palmitate (C16) | 1.75 | 0.025 | 0.01 | |
| α-Naphthyl acetate | 0.63 | 12.25 | 19.24 | |
| β-Naphthyl acetate | 1.22 | 44.87 | 36.77 |
The activity was assayed in the standard reaction media for enzyme assay with different substrate concentrations and 12.5 μg of protein, at pH 8.0 and room temperature.
KSI, substrate constant.
Table 1 represents the kinetic constants calculated for p-NP esters (C2 to C16) and naphthyl acetate esters at pH 8.0. Km values were in the millimolar range, with caprylate being the lowest, although it had optimal activity for p-NP acetate. The values of Km and kcat cannot be compared with other thermostable relatives of Geobacillus CEs since the kinetic constants have not been evaluated. However, it is interesting that inhibition occurs at high substrate concentrations in the case of p-NP caprylate (Table 1) and p-NP laurate, with substrate inhibition constants of 2.02 and 0.02 mM, respectively. This means that it was not possible to determine the substrate specificity of the enzyme at a unique concentration of substrate, as described previously in the bibliography (17, 27). The kcat/Km values are similar to those reported for other CEs like Rv3487c from Mycobacterium tuberculosis (48). The enzyme also showed good activity toward esters of naphthyl acetate, with kcat/Km values in the range of 19.24 and 36.77 s−1 mM−1 for α- and β-naphthyl acetate, respectively. The activity toward these substrates is similar to that shown by others esterases, like YesT from Bacillus subtilis (26), with a Km toward esters of naphthyl acetate around 1 mM and toward p-NP acetate of 2.8 mM, or like the rhamnogalacturonan acetyl esterase from Bacillus halodurans C-125, with a Km toward p-NP acetate of 14.1 mM (30).
The effect of pH on activity and stability was tested using p-NP caprylate because of its chemical stability compared with acetate and butyrate, as described by Kakugawa et al. (18). CEGk was most active at pH 8.0 (data not shown). Despite the high sequence homology, the related esterases show different pH optima: Est30Gs (10) prefers a high pH of 9.5, unlike EstA from G. thermoleovorans YN (40), for which the optimum pH is 7.5. As shown after 24 h of incubation at different pHs, the CEGk was remarkably stable in a pH range of 6.0 to 9.0 (Fig. 3), retaining 62% of activity at pH 6.0. However, the enzyme was inactivated at pH 11.0 after 1 h or after 2 h at acidic pH 4.0 (data not shown).
FIG. 3.
pH stability profiles of the purified CEGk. Samples were analyzed after 24 h of incubation in media with different pH values (50 mM sodium acetate, pH 4.0 to 5.5; 50 mM potassium phosphate, pH 6 to 8; 50 mM sodium bicarbonate, pH 8 to 9; and 50 mM boric acid, pH 10.0 to 11.0) to determine residual activity under the standard reaction conditions for the enzyme assay, using p-NP caprylate as a substrate.
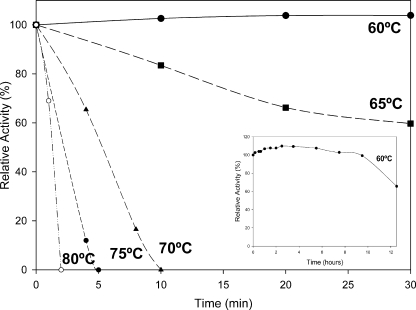
Thermal stability was determined in a temperature range of 60 to 80°C. As shown in Fig. 4, the esterase was most stable at temperatures between 60 and 65°C but strongly decreased at temperatures above 70°C. At physiological temperature (60°C) the enzyme showed some structural modifications that increase the activity by 10%, and it was also fully stable for 5 h (Fig. 4, inset). This activation by temperature is a common feature in thermostable enzymes (6, 25). After 12 h of incubation at such temperatures, the residual activity was about 60% of the initial. Activity profiling with respect to temperature was not carried out due to substrate instability at high temperatures.
FIG. 4.
Temperature stability profiles of the purified CEGk at pH 8.0. Enzyme was incubated for different periods of time at 60°C (•), 65°C (▪), 70°C (▴), 75°C (⧫), and 80°C (○), and then activity was measured under the standard reaction conditions at 25°C using p-NP caprylate as the substrate. (Inset) Expanded study of thermostability of the enzyme at the physiological temperature of G. kaustophilus (60°C) under the experimental conditions described above.
Inhibition studies.
The effect of detergents, organic solvents, and urea on CE activity was examined using a standard enzyme assay. Even after incubation for 1 h at 60°C with 5% nonionic detergents, like Tergitol or Triton X-100, the CEGk was not significantly inactivated (Table 2). The enzyme still retained 23% of residual activity after 60 min at 30°C of incubation in 2% SDS; however, it was extensively inactivated at 60°C. The activity was not significantly affected in 40% concentrations of organic solvents at 30°C but was strongly inactivated at higher concentrations (90%) or at higher temperatures (60°C). Furthermore, the enzyme was activated by 1 M urea (150%), and it was stable in 8 M urea at 30°C for 60 min. The purified CE shows thermostability in addition to high stability against various compounds that affect protein folding. CEGk is more stable in the presence of various organic solvents than EstBB1 from B. subtilis (strain RRL-BB1), where almost complete inactivation was observed in the presence of 60% (vol/vol) ethanol and 60% (vol/vol) isopropanol (23). The CE EstA1 from Bacillus sp. strain BP-7 was strongly inhibited by SDS or urea, even at 0.3% (vol/vol), whereas these compounds did not cause inhibition of the CEGk at this concentration; the esterase retained 100% of activity even in the presence of 8 M urea (35).
TABLE 2.
Effect of different denaturant compounds toward CEGk activity
| Compounda | Concn | Relative CEGk activity (%)
|
|
|---|---|---|---|
| After mixing | After incubationb | ||
| Detergents | |||
| Tween 20 | 1% | 99 ± 1 | 100 ± 3 (64 ± 3) |
| 5% | 75 ± 3 | 64 ± 4 (59 ± 5) | |
| Triton X-100 | 1% | 100 ± 3 | 105 ± 6 (95 ± 1) |
| 5% | 85 ± 5 | 94 ± 4 (88 ± 2) | |
| Tergitol | 1% | 100 ± 5 | 104 ± 4 (98 ± 6) |
| 5% | 103 ± 2 | 103 ± 2 (99 ± 3) | |
| SDS | 0.1% | 111 ± 7 | 111 ± 3 (0) |
| 2% | 23 ± 2 | 23 ± 4 (0) | |
| CTAB | 0.1% | 96 ± 3 | 96 ± 3 (3,00 ± 1) |
| 0.3% | 98 ± 4 | 99 ± 4 (2 ± 1) | |
| Organic solvents | |||
| Methanol | 40% | 103 ± 3 | 101 ± 3 (3 ± 1) |
| 90% | 73 ± 4 | 8 ± 4 (0) | |
| Ethanol | 40% | 86 ± 6 | 56 ± 8 (8 ± 2) |
| 90% | 94 ± 4 | 54 ± 5 (0) | |
| Acetone | 40% | 74 ± 2 | 64 ± 5 (2 ± 1) |
| 90% | 50 ± 4 | 49 ± 5 (2 ± 1) | |
| 2-Propanol | 40% | 88 ± 3 | 35 ± 6 (16 ± 3) |
| 90% | 87 ± 5 | 25 ± 8 (0) | |
| DMSOb | 40% | 97 ± 8 | 95 ± 7 (0) |
| 90% | 62 ± 4 | 0 (0) | |
| TBME | 40% | 44 ± 7 | 40 ± 4 (41 ± 5) |
| 90% | 33 ± 6 | 33 ± 4 (3 ± 1) | |
| Urea | 1 M | 152 ± 5 | 94 ± 3 (93 ± 5) |
| 8 M | 112 ± 3 | 91 ± 2 (4 ± 1) | |
CTAB, cetyltrimethylammonium bromide; DMSO, dimethyl sulfoxide; TBME, tert-butyl methyl ether.
The enzyme was incubated at 30°C and 60°C (numbers in parentheses) in 50 mM phosphate buffer, pH 8.0, with two different concentrations of denaturing agents. After 60 min, an aliquot of 50 μl was removed from each incubation mixture, and the residual activity was measured using the standard reaction conditions.
Addition of 5 mM concentrations of divalent cations (Mg2+, Cu2+, Co2+, Ca2+, Zn2+, and Mn2+), and subsequent incubation in such media for 30 min at 30°C did not affect enzymatic activity. This, together with the fact that EDTA (10 mM) had no effect after such incubations, indicates that the enzyme was not a metalloprotein (data not shown).
To investigate the amino acids involved in the catalytic mechanism, the inhibition effect of various chemical modifiers was also determined (Table 3). The CEGk was completely inhibited by PMSF and paraoxon at tested concentrations. However, eserine, like PGO and PLP, had no effect on the activity, even at 5 mM. These results suggested that serine and histidine are located at or near the esterase catalytic site and are involved in the activity mechanism. The fact that the esterase was inhibited by organophosphates such as paraoxon and by serine inhibitors such as PMSF demonstrated that the enzyme is a serine esterase (CE).
TABLE 3.
Effect of different inhibitors toward CEGk activity
| Inhibitor | CEGk activity (%) at the indicated concn (mM)a
|
Inhibited amino acid | |
|---|---|---|---|
| 0.5 | 5 | ||
| PMSF | 0 | 0 | Ser |
| PGO | 100 ± 2 | 94 ± 2 | Arg |
| PLP | 99 ± 2 | 95 ± 2 | Lys |
| DPCb | 45 ± 5 | 36 ± 3 | His |
| Eserine | 105 ± 2 | 98 ± 3 | |
| Paraoxon | 0 | 0 | |
The enzyme was incubated at 30°C for 30 min in 50 mM phosphate buffer, pH 8.0, with two different concentrations of the inhibitors. After 30 min, an aliquot of 50 μl was removed from each incubation mixture, and the residual activity was measured using the standard reaction conditions, with p-NP caprylate as a substrate.
Diethylpirocarbonate.
Determination of molecular structure.
The three-dimensional structure of the Est30Gs crystallized CE (PDB code 1TQH; 97% identity) (22) was used as a template to construct a model of the CEGk (Fig. 5A). In this model, the K212 was located in the external structure of the protein at the beginning of the β8 strand. Therefore, this amino acid could be susceptible to the attack of a protease of E. coli Rosetta as described above.
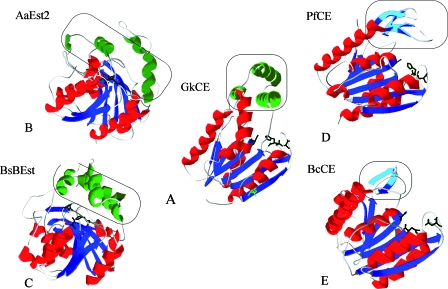
FIG. 5.
Schematic representation of tertiary structures. (A) Modeled structure of the monomeric CEGk; the possible proteolytic site (K212) is highlighted in green. (B) A. acidocaldarius CE ([AaEst2] PDB code 1EVQ). (C) B. subtilis CE ([BsBEst] PDB code 2R11). (D) CEPf (PDB code 1AUO). (E) CEBc (PDB code 2H1I). Catalytic triads are shown in black (Ser-Asp-His). β-Strands and α-helices belonging to the canonical α/β hydrolase fold are shown in blue and red, respectively. Gray parts of the structures indicate the presence of β-turns. Framed lids formed by β-strands and α-helices are shown in cyan and green, respectively. These figures were rendered using SWISS-MODEL and Swiss-PdbViewer (36).
By sequence identity with the protein 1TQH, the catalytic triad of the CEGk might consist of Ser93, Asp192, and His222. The catalytic Ser93 was located at a sharp turn between β5-strand and helix-α4 (Fig. 1), which is called the nucleophilic elbow, and it presumably forms water-mediated hydrogen bond interactions with the carboxylate side chain of Asp192 (as in Est30Gs) (Fig. 5A). The conformation of Ser93 could be stabilized by hydrogen bond interaction of the hydroxyl group with the side chain of His222, as in Est30Gs (22). All three catalytic residues are located at the top of the C-terminal β-sheet. Those residues are essential for enzyme activity and are conserved in all related enzymes: Est30Gs EstA from G. thermoleovorans, and Est30Gt (Fig. 1). Moreover, as occurs in other nonrelated CEs, such as those from Bacillus anthracis, B. halodurans, or Oceanbacillus iheyensis, the CEGk contains only one methionine at the beginning of the sequence (22). In fact, N-terminal methionine residues are often removed after translation. In the general structure, the only discrepancy in these Geobacillus related enzymes is the cap (Fig. 6C to F, boxes), which is smaller in Est30Gs, where the arginine 154 has been changed for a lysine, thus decreasing the length of the α6-helix (Fig. 1). Unfortunately, the effect of this smaller cap could not be correlated with the kinetic parameters (maximum rate and Km) since only relative activities (percentages) toward different p-NP esters have been described for Est30Gs (10).
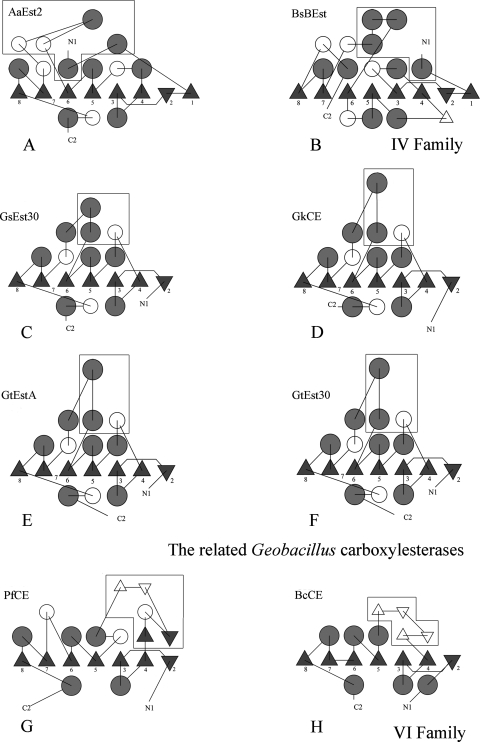
FIG. 6.
Topology diagrams of the members of the Geobacillus CE family and other members of the α/β hydrolase family. (A) Diagram of A. acidocaldarius CE (AaEst2; PDB code 1EVQ). (B) Diagram of B. subtilis CE (BsBEst; PDB code 2R11). (C) Diagram of Est30Gs (PDB code 1TQH). (D) Diagram of the recombinant CEGk. (E) Diagram of EstA from G. thermoleovorans YN (GtEstA). (F) Diagram of Est30Gt. (G) Diagram of CEPf (PDB code 1AUO). (H) Diagram of CEBc (PDB code 2H1I). α-Helices are represented by circles, and β-strands are represented by triangles. White circles represent 310-helices or other small helices in the structures. The framed figures represent those strands and loops that are not contained in the central β-sheet and that form the lids. The diagrams were made using the TOPS program (27).
The CEGk folds into two domains, with the catalytic triad and substrate-binding site located at the interface between the domains (Fig. 5A). The smaller domain of CEGk consists of three α-helices that form a cap over the active site: α2 (Pro57-Leu60), α5 (Glu123-Arg140), and α6 (Glu145-Lys156) (Fig. 1). This cap lies mainly near the N-terminal end of the central sheet. The larger domain resembles the classical α/β hydrolase fold. It contains a central seven-stranded sheet, β2 to β8, and a classical α/β hydrolase fold surrounded by six α-helices, α1 (Ala29-Ser40), α3 (Pro65-Asn81), α4 (Leu94-Gly103), α7 (Leu163-Leu179), α8 (Ser199-Glu206), and α9 (Lys 230-Ser243).
DISCUSSION
This paper describes the cloning of a bacterial CE (CEGk) with high activity and stability against various compounds and with good thermal stability, which make it very attractive for future biotechnological applications. Many clinically useful drugs contain esters, and they could be potential substrates of CEGk. These compounds include anti-inflammatory drugs like naproxen methyl ester or ketoprofen methyl ester (19) or even the anticancer prodrug campothecin-11 (47). Therefore, it would be interesting to study the catalytic activity of the enzyme toward these applied substrates.
The paper shows that the CEGk is different from other known bacterial CEs in its properties and amino acid sequence. The molecular structure, topology, and substrate specificities of CEGk and its related proteins were compared in order to assign it to a family according to the lipolytic classification of Arpigny et al. (2), which includes eight families of bacterial lipases and esterases. CEGk shows only slight homology to two different families of esterases: the hormone-sensitive lipase family (IV) and family VI.
The topology diagram (Fig. 6C to F) shows that the first β-strand of the typical α/β hydrolase fold is missing from the CEGk structure and in the other related CEs. Therefore, the central β-sheet of all these enzymes has only seven strands (β2 to β8), with the N-terminal β2-strand antiparallel to the others. The latter contrasts with the topology of CE Est2 from Alicyclobacillus acidocaldarius (PDB code 1EVQ) and brefeldin A esterase from B. subtilis (PDB code 2R11), which have been classified as family IV members of the α/β hydrolase superfamily (9, 45). Both enzymes present the canonical α/β hydrolase core (31) composed of eight β-strands, with the second antiparallel to the others (Fig. 6A and B). In addition, the lids of CEGk and its relatives lie mainly near the N-terminal ends of the central sheets (Fig. 5A, instead of surrounding the catalytic triad, as observed in A. acidocaldarius Est2 (Fig. 5B). This lid seems to be a different helical region from the core domain. At the same time, the cap of A. acidocaldarius Est2 is composed of three α-helices and two 310-helices (Fig. 6C to F) instead of the two α-helices and one 310-helix in the Geobacillus-related CEs. In addition, the B. subtilis brefeldin A esterase shows a lid formed by four α-helices (43). A. acidocaldarius Est2 and the B. subtilis brefeldin A esterase show only 16 and 6% sequence identity, respectively, with CEGk. Because of these differences, it seems that the family of related Geobacillus CEs cannot be classified in the family IV of lipolytic enzymes.
A comparison of CEGk and its related CEs with two representative structures of family VI, Pseudomonas fluorescens CE ([CEPf] PDB code 1AUO) (20) and Bacillus cereus CE ([CEBc]PDB code 2H1I), shows that they all share similar β-cores (β2 to β8) and molecular masses (about 23 to 30 kDa) (2). However, the active form in family VI is usually a dimer, in contrast to the results described here, where CEGk is monomeric. CEPf and CEBc share a different active cleft from the rest of the CEs from Geobacillus species (Fig. 6G and H). Also, the cap domains are missing, and the catalytic cores are covered by four small β-strands that appear to block the binding of long chain acid esters by covering the C-terminal strands of the β-core (the active-site cleft) close to the catalytic triad (Fig. 5D and E). The substrate specificity of CEBc has not been defined yet, but CEPf is active on acyl chains from C2 to C10 (13), whereas the Geobacillus CEs are active on chain lengths from C2 to C12. The cap domain in CEGk is adjacent to the active site of the enzyme although its role is unclear; it might participate in the ability of the CEs to hydrolyze esters of longer chain fatty acids (21). In addition, CEPf and CEBc show low sequence identity with CEGk (15 and 12%, respectively) and, in consequence, with its related esterases (Est30Gs, EstA from G. thermoleovorans NY, and Est30Gt). Thus, Geobacillus sp. CEs do not fit in terms of the topology of this family VI, or its cap domain, or its substrate specificity.
From the above, it is evident that Geobacillus sp. CEs derive from a common ancestral version but differ in topology, cap domain, and substrate specificity from families IV and VI. This is also evident when a phylogenetic study is carried out (Fig. 7). CEGk and its relatives are closely related and seem to be the common ancestors of families IV and VI separated by a long distance.
FIG. 7.
Phylogenetic analysis of the studied esterases. Family IV, A. acidocaldarius CE (AaEst2) and B. subtilis CE (BsEst); family VI, CEBc and CEPf; new Geobacillus sp. family, CEGk, Est30Gs, G. thermoleovorans YN CE (GtEstA), and Est30Gt. The phylogenetic tree was constructed using TreeView software with the neighbor-joining method.
Conclusions.
A hypothetical CE from G. kaustophilus HTA426 has been cloned in E. coli, overexpressed, and mutated to obtain a stable 30-kDa protein by removing the endopeptidase cut. This increase in expression by repairing the endopeptidase cut has not been previously described for this enzyme. The enzyme was active toward short-length p-NP esters, with high stability under alkaline pH, temperature (60°C), nonionic detergents, denaturant agents, and organic solvents, which are important characteristics required for applications in detergent formulations and biotransformations. CEGk and its relatives belong to a new bacterial CE family, described here for the first time, which is not fully characterized and which has a large cap. This new family is totally different from the other CEs from families IV and VI on the basis of comparisons of their amino acid sequences, crystal structures, and some biological properties.
Acknowledgments
This study was partially supported by MEC (BIO2007-62510) and Programa de Ayuda a Grupos de Excelencia de la Región de Murcia, de la Fundación Séneca (04541/GERM/06, Plan Regional de Ciencia y Tecnología 2007/2010). S.M.G. is a holder of a predoctoral research grant (FPU) from Fundación Séneca, Murcia, Spain. J.N.F is a holder predoctoral research (FPI) grant from MEC, Spain.
We are very grateful to Alejandro Torrecillas from Centro de Apoyo a la Investigación y Desarrollo (Murcia, Spain) for his help.
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22195-201. [DOI] [PubMed] [Google Scholar]
- 2.Arpigny, J. L., and K. E. Jaeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343177-183. [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, R., M. Hoffmann, and U. Keller. 1998. Molecular analysis of a gene encoding a cell-bound esterase from Streptomyces chrysomallus. J. Bacteriol. 1806396-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornemann, S., J. M. Cassells, J. S. Dordick, and A. J. Hacking. 1992. The use of enzymes to regioselectivity deacylate sucrose esters. Biocatalysis 71-12. [Google Scholar]
- 5.Bornscheuer, U. T. 2002. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2673-81. [DOI] [PubMed] [Google Scholar]
- 6.Costantino, H. R., S. H. Brown, and R. M. Nelly. 1990. Purification and characterization of an alpha-glucosidase from a hyperthermophilic archaeobacterium, Pyrococcus furiosus, exhibiting a temperature optimum of 105 to 115°C. J. Bacteriol. 1723654-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowe, J., H. Dobeli, R. Gentz, E. Hochuli, D. Stuber, and K. Henco. 1994. 6xHis-Ni-NTA chromatography as a superior technique in recombinant protein expression/purification. Methods Mol. Biol. 31371-387. [DOI] [PubMed] [Google Scholar]
- 8.Cygler, M., J. D. Schrag, J. L. Sussman, M. Harel, I. Silman, M. K. Gentry, and B. P. Doctor. 1993. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases and related proteins. Protein Sci. 2366-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Simone, G., S. Galdiero, M. Gluseppe, D. Lang, M. Rossl, and C. Pedone. 2000. A snapshot of a transition state analogue of a novel thermophilic esterase belonging to the subfamily of mammalian hormone-sensitive lipase. J. Mol. Biol. 10761-771. [DOI] [PubMed] [Google Scholar]
- 10.Ewis, H. E., A. T. Abdelal, and C. D. Lu. 2004. Molecular cloning and characterization of two thermostable carboxyl esterases from Geobacillus stearothermophilus. Gene 329187-195. [DOI] [PubMed] [Google Scholar]
- 11.Gouet, P., E. Courcelle, and D. I. Stuart, and F. Metoz. 1999. ESPript: multiple sequence alignment in PostScript. Bioinformatics 15305-308. [DOI] [PubMed] [Google Scholar]
- 12.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis 182714-2723. [DOI] [PubMed] [Google Scholar]
- 13.Hong, K. H., W. H. Jang, K. D. Choi, and O. J. Yoo. 1991. Characterization of Pseudomonas fluorescens carboxylesterase: cloning and expression of the esterase gene in Escherichia coli. Agric. Biol. Chem. 112839-2845. [PubMed] [Google Scholar]
- 14.Jaeger, K. E., B. W. Dijkstra, and M. T Reetz. 1999. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 53315-351. [DOI] [PubMed] [Google Scholar]
- 15.Jaeger, K. E., and T. Eggert. 2002. Lipases for biotechnology. Curr. Opin. Biotechnol. 13390-397. [DOI] [PubMed] [Google Scholar]
- 16.Jaeger, K. E., S. Ransac, B. W. Dijkstra, C. Colson, M. Van Heuvel, and O. Misset. 1994. Bacterial lipases. FEMS Microbiol. Rev. 1529-63. [DOI] [PubMed] [Google Scholar]
- 17.Job, V., G. L. Marcone, M. S. Pilone, and L. Pollegioni. 2002. Glycine oxidase from Bacillus subtilis. J. Biol. Chem. 2776987-6993. [DOI] [PubMed] [Google Scholar]
- 18.Kakugawa, S., S. Fushinobu, T. Wakagi, and H. Shoun. 2007. Characterization of a thermostable carboxylesterase from the hyperthermophilic bacterium Thermotoga maritima. Appl. Microbiol. Biotechnol. 74584-591. [DOI] [PubMed] [Google Scholar]
- 19.Kim, G. J., G. S. Choi, J. Y. Kim, J. B. Lee, D. H. Jo, and Y. W. Ryu. 2003. Screening, production and properties of a stereospecific esterase from Pseudomonas sp. S34 with high selectivity to (S)-ketoprofen ethyl ester. J. Mol. Catal. B 2229-35. [Google Scholar]
- 20.Kim, J. Y., G. S. Choi, Y. J. Kim, Y. W. Ryu, and G. J. Kim. 2002. A new isolate Bacillus stearothermophilus JY144 expressing a novel esterase with high enantioselectivity to (R)-ketoprofen ethyl ester: strain selection and gene cloning. J. Mol. Catal. B 18133-145. [Google Scholar]
- 21.Kim, K. K., H. K. Song, D. H. Shin, K. Y. Hwang, S. Choe, O. J. Yoo, and S. W. Suh. 1997. Crystal structure of carboxylesterase from Pseudomonas fluorescens, an alpha/beta hydrolase with broad substrate specificity. Structure 121571-1584. [DOI] [PubMed] [Google Scholar]
- 22.Liu, P., Y. F. Wang, H. E. Ewis, A. T. Abdelal, C. D. Lu, R. W. Harrison, and I. T Weber. 2004. Covalent reaction intermediate revealed in crystal structure of the Geobacillus stearothermophilus carboxylesterase Est30. J. Mol. Biol. 342551-561. [DOI] [PubMed] [Google Scholar]
- 23.Maqbool, Q. U., S. Johri, S. Rasool, S. Riyaz-ul-Hassan, V. Verma, A. Nargotra, S. Koul, and G. N. Qazi. 2006. Molecular cloning of carboxylesterase gene and biochemical characterization of encoded protein from Bacillus subtilis (RRL BB1). J. Biotechnol. 1251-10. [DOI] [PubMed] [Google Scholar]
- 24.Margolin, A. L. 1993. Enzymes in the synthesis of chiral drugs. Enzyme Microb. Technol. 15266-280. [DOI] [PubMed] [Google Scholar]
- 25.Martínez-Martínez, I., J. Navarro-Fernández, F. García Carmona, H. Takami, and A. Sánchez-Ferrer. 2008. Characterization and structural modelling of a novel thermostable glycine oxidase from Geobacillus kaustophilus HTA426. Proteins 701429-1441. [DOI] [PubMed] [Google Scholar]
- 26.Martínez-Martínez, I., J. Navarro-Fernández, F. García-Carmona, and A. Sánchez-Ferrer. 2008. YesT: a new rhamnogalacturonan acetyl esterase from Bacillus subtilis. Proteins 71379-388. [DOI] [PubMed] [Google Scholar]
- 27.Martínez-Martínez, I., J. Navarro-Fernández, J. D. Lozada-Ramírez, F. García-Carmona, and A. Sánchez-Ferrer. 2006. Maximization of the production of His-tagged glycine oxidase and its M261 mutant proteins. Biotechnol. Prog. 22647-652. [DOI] [PubMed] [Google Scholar]
- 28.Michalopoulus, I., G. M. Torrance, D. R. Gilbert, and D. R. Westhead. 2004. TOPS: an enhanced database of protein structural topology. Nucleic Acid Res. 1251-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moher, P., L. Rosslein, and C. Tamm. 1989. Kinetic resolution of racemic β-γ epoxy esters with pig esterase. Tetrahedron Lett. 32513-2516. [Google Scholar]
- 30.Navarro-Fernández, J., I. Martínez-Martínez, S. Montoro-García, F. García-Carmona, H. Takami, and A. Sánchez-Ferrer. 2008. Characterization of a new rhamnogalacturonan acetyl esterase from Bacillus halodurans C-125 with a new putative carbohydrate binding domain. J. Bacteriol. 1901375-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, and J. Schrag. 1992. The alpha/beta hydrolase fold. Protein Eng. 5197-211. [DOI] [PubMed] [Google Scholar]
- 32.Park, Y. J., S. Y. Choi, and H. B. Lee. 2006. A carboxylesterase from the thermoacidophilic archaeon Sulfolobus solfataricus P1; purification, characterization, and expression. Biochim. Biophys. Acta 1760820-828. [DOI] [PubMed] [Google Scholar]
- 33.Pearcy, J. O., and T. D Lee. 2001. MoWeD: a computer program to rapidly deconvolute low resolution electrospray liquid chromatography/mass spectrometry runs to determine component molecular weights. J. Am. Soc. Mass Spectrom. 12599-606. [DOI] [PubMed] [Google Scholar]
- 34.Prim, N., A. Blanco, J. Martinez, F. I. J. Pastor, and P. Diaz. 2000. EstA, a gene coding for a cell-bound esterase from Paenibacillus sp. BP-23, is a new member of the bacterial subclass of type B carboxylesterases. Res. Microbiol. 151303-312. [DOI] [PubMed] [Google Scholar]
- 35.Prim, N., F. I. Pastor, and P. Diaz. 2001. Cloning and characterization of a bacterial cell-bound type B carboxylesterase from Bacillus sp. BP-7. Curr. Microbiol. 42237-240. [DOI] [PubMed] [Google Scholar]
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modelling server. Nucleic Acids Res. 313381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 15076-85. [DOI] [PubMed] [Google Scholar]
- 40.Soliman, N. A., M. Knoll, Y. R. Abdel-Fattah, R. D. Schmid, and S. Lange. 2007. Molecular cloning and characterization of thermostable esterase and lipase from Geobacillus thermoleovorans YN isolated from desert soil in Egypt. Process Biochem. 421090-1100. [Google Scholar]
- 41.Takami, H., A. Inoue, F. Fuji, and K. Horikoshi. 1997. Microflora in the deepest sea mud of the Mariana Trench. FEMS Microbial. Lett. 152279-285. [DOI] [PubMed] [Google Scholar]
- 42.Takami, H., S. Nishi, J. Lu, S. Shimamura, and Y. Takaki. 2004. Genomic characterization of thermophilic Geobacillus species isolated from the deepest sea mud of the Mariana Trench. Extremophiles 8351-356. [DOI] [PubMed] [Google Scholar]
- 43.Takami, H., Y. Takaki, G. J. Chee, S. Nishi, S. Shimamura, H. Suzuki, S. Matsui, and I. Uchiyama. 2004. Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res. 326292-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei, Y., J. A. Contreras, P. Sheffield, T. Osterlund, U. Derewenda, R. E. Kneusel, U. Matern, C. Holm, and Z. S. Derewenda. 1999. Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase. Nat. Struct. Biol. 6340-345. [DOI] [PubMed] [Google Scholar]
- 46.Williams, J. D., B. E. Weiner, J. R. Ormand, J. Brunner, A. D. Thornquest, Jr., and J. D. Burinsky. 2001. Automated molecular weight assignment of electrospray ionization mass spectra. Rapid Commun. Mass Spectrom. 152446-2455. [DOI] [PubMed] [Google Scholar]
- 47.Yoon, K. J., E. J. Krull, C. L. Morton, W. G. Bornmann, R. E. Lee, P. M. Potter, and M. K. Danks. 2003. Activation of a camptothecin prodrug by specific carboxylesterases as predicted by quantitative structure-activity relationship and molecular docking studies. Mol. Cancer Ther. 21171-1181. [PubMed] [Google Scholar]
- 48.Zhang, M., J. D. Wang, Z. F. Li, J. Xie, Y. P. Yang, Y. Zhong, and H. H. Wang. 2005. Expression and characterization of the carboxyl esterase Rv3487 from Mycobacterium tuberculosis. Protein Expr. Purif. 4259-66. [DOI] [PubMed] [Google Scholar]