Abstract
In this study, we demonstrated that the surface-expressed enolase from diarrheal isolate SSU of Aeromonas hydrophila bound to human plasminogen and facilitated the latter's tissue-type plasminogen activator-mediated activation to plasmin. The bacterial surface-bound plasmin was more resistant to the action of its specific physiological inhibitor, the antiprotease α2-antiplasmin. We found that immunization of mice with purified recombinant enolase significantly protected the animals against a lethal challenge dose of wild-type (WT) A. hydrophila. Minimal histological changes were noted in organs from mice immunized with enolase and then challenged with WT bacteria compared to severe pathological changes found in the infected and nonimmunized group of animals. This correlated with the smaller bacterial load of WT bacteria in the livers and spleens of enolase-immunized mice than that found in the nonimmunized controls. We also showed that the enolase gene could potentially be important for the viability of A. hydrophila SSU as we could delete the chromosomal copy of the enolase gene only when another copy of the targeted gene was supplied in trans. By site-directed mutagenesis, we altered five lysine residues located at positions 343, 394, 420, 427, and 430 of enolase in A. hydrophila SSU; the mutated forms of enolase were hyperexpressed in Escherichia coli, and the proteins were purified. Our results indicated that lysine residues at positions 420 and 427 of enolase were crucial in plasminogen-binding activity. We also identified a stretch of amino acid residues (252FYDAEKKEY260) in the A. hydrophila SSU enolase involved in plasminogen binding. To our knowledge, this is the first report of the direct involvement of surface-expressed enolase in the pathogenesis of A. hydrophila SSU infections and of any gram-negative bacteria in general.
Aeromonas hydrophila is an emerging human pathogen that causes gastroenteritis and other extraintestinal infections, such as septicemia, meningitis, pneumonia, cellulitis, bullous lesions, and ecthyma gangrenosum (8, 43). The increasing isolation of this organism from water and food, as well as its resistance to water chlorination specifically in biofilms and to various antibiotics, could be a public health threat (16, 43).
Aeromonas spp. produce an array of virulence factors that are associated with bacterial virulence (8). In terms of gastroenteritis, three enterotoxins have been identified in our laboratory from a clinical isolate, SSU, of A. hydrophila (33). In addition, other virulence and regulatory genes that coded for the glucose-inhibited division protein (GidA), DNA adenine methyltransferase (Dam), hemolysin (Hly), ToxR-regulated lipoprotein (TagA), and the cold shock exoribonuclease R (VacB) were recently identified and characterized from the above-mentioned isolate (12-15, 29, 34). Further, we identified type III and VI secretion systems, as well as their effectors in A. hydrophila SSU, and their roles in the pathogenesis of A. hydrophila infections were established (16, 36-39).
While searching for new virulence factors in A. hydrophila SSU, we previously used a murine peritoneal culture model in which bacteria were grown in a dialysis bag implanted in the peritoneal cavity of mice (32). By restriction fragment differential display PCR using RNA as a template, we identified a glycolytic enzyme enolase in A. hydrophila SSU, which was differentially expressed under in vivo but not in vitro growth conditions (32). Enolase is a ubiquitous enzyme that catalyzes the reversible conversion of 2-phosphoglycerate (2-PGE) to phosphoenolpyruvate (PEP) (26). In addition to its metabolic role, enolase has been implicated for its contribution to several biological and pathophysiological processes by acting as a heat shock protein and in modulating gene transcription, as well as for its involvement in microbial diseases and autoimmunity (26). Although mainly localized in the cytoplasm, enolase has also been identified on the surface of neuronal, cancer, and some hematopoietic cells, as well as on the cell surfaces of several bacterial species, where it binds plasminogen (6, 17, 20, 21, 23, 25).
Plasminogen is a single-chain glycoprotein that is abundant in human plasma and extracellular fluid (20). Its conversion to a proteolytic, active two-chain form, namely plasmin, can be mediated either by eukaryotic activators such as the host-derived tissue plasminogen activator (tPA) and urokinase or by the prokaryotic activators staphylokinase and streptokinase (17, 20). tPA is a principal activator of plasminogen in plasma and intercellular fluid. The immobilization of plasminogen with its receptors dramatically enhances tPA-mediated activation of plasminogen, and the generated plasmin that continues to bind to its receptors is more resistant to the action of its specific physiological inhibitors (e.g., the antiprotease α2-antiplasmin [α2AP]) (17, 20). Plasmin is a trypsin-like serine protease with broad substrate specificity; it is physiologically involved in fibrinolysis by degradation of fibrin and also degrades noncollagenous glycoproteins of mammalian extracellular matrices and basement membranes. Thus, surface-expressed plasminogen receptors (e.g., enolase) provide a niche for host cells or bacteria to interact with the mammalian proteolytic plasminogen-plasmin system, which presumably facilitates the metasis process or promotes microbial pathogens to migrate within the host to fulfill their nutritional demands (18, 19, 21, 23, 25). Indeed, several recent studies demonstrated that enolase in certain pathogenic bacteria (e.g., Streptococci and Pneumococci) and microbiota (e.g., Lactobacilli and Bifidobacteria) not only is essential for bacterial viability because of its enzymatic activity but also promotes pathogen-host interactions and contributes to bacterial colonization and pathogenesis by means of its surface or extracellular location (1, 4, 6, 17, 19, 20, 22, 26, 27). In our previous study, we reported the presence of surface-expressed enolase in the gram-negative bacterium A. hydrophila SSU (32) and demonstrated that crude, whole-bacterial-cell lysates of A. hydrophila could bind human plasminogen (32). However, the direct role of enolase in Aeromonas infections has not yet been established.
In our present study, we provided direct evidence that the washed whole bacterial cells of A. hydrophila SSU bound human plasminogen/plasmin and facilitated tPA-mediated activation of plasminogen. We also found that this plasminogen/plasmin-bacterial cell interaction was, in part, mediated by enolase. We immunized mice with purified recombinant enolase and showed that such immunized animals were protected against mortality from a lethal challenge dose of wild-type (WT) A. hydrophila. However, we were unable to develop an enolase-negative mutant of A. hydrophila SSU, which suggests that this enzyme might be essential for the viability of this organism. Further, we studied enolase's plasminogen-binding activity in A. hydrophila SSU and identified lysine residues at positions 420 and 427 that contributed to this activity. We also identified an internal stretch of amino acid residues (252FYDAEKKEY260) in the enolase (which has 434 amino acid residues) of A. hydrophila SSU that was responsible for this plasminogen-binding activity. This is the first detailed study illustrating the role of enolase in the pathogenesis of A. hydrophila SSU infections.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and DNA techniques.
The bacterial strains and plasmids (10, 24, 33) used in this study are listed in Table 1. A. hydrophila SSU, a diarrheal isolate, was obtained from the Centers for Disease Control and Prevention, Atlanta, GA. SSU-R is a spontaneous rifampin-resistant (Rifr) strain of SSU that was prepared previously in our laboratory (35). Luria Bertani (LB) broth (Difco Laboratories, Detroit, MI) was used to grow A. hydrophila and Escherichia coli cultures at 37°C with aeration (180 rpm). The antibiotics ampicillin (Ap), tetracycline (Tc), kanamycin (Km), streptomycin (Sp), spectinomycin (Sm), and Rif were obtained from Sigma (St. Louis, MO) and were used at concentrations of 100, 15, 50, 25, 25, and 40 μg/ml, respectively. Restriction endonucleases were purchased from Promega (Madison, WI). All of the basic molecular biology techniques used in this study were previously described (33).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) and/or construction | Source or reference |
|---|---|---|
| Strains | ||
| A. hydrophila SSU | CDC, Atlanta, GA | |
| SSU-R | Rifr strain of A. hydrophila SSU | Laboratory stock |
| WT/pBReno | WT A. hydrophila SSU-R transformed with pBReno plasmid; Rifr Tcr | This study |
| Enolase mutant | The chromosomal enolase gene was deleted from WT A. hydrophila SSU-R that was transformed with the plasmid pBReno; Sm/Spr Rifr Tcr | This study |
| E. coli | ||
| SM10 | Kmr λpir | 24 |
| JM109 | endA1 recA1 gyrA96 thi-1 hsdR17(rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Promega |
| ES1301 mutS | lacZ53 mutS201::Tn5 thyA36 rha-5 metB1 deoC | Promega |
| HMS174 (DE3) | RecA-mutated K-12 background E. coli strain that carries a chromosomal copy of the T7 RNA polymerase gene under the control of the lacUV5 promoter; used as the host strain for the pET expression system | Novagen |
| Plasmids | ||
| pHPΩ45 | Contains a 2.0-kb Sm/Spr gene cassette | 33 |
| pET30a | pBR322-derived expression vector with T7 lac promoter and up- and down-stream His tags; Kmr | Novagen |
| pDMS197 | A suicide vector; R6K ori sacB Tcr | 10 |
| pALTER-1 | Vector for the site mutagenesis; Tcr Aps | Promega |
| pBReno | The coding region of A. hydrophila enolase gene was cloned in pBR322 at the ScaI/PstI sites and under the control of the Ap promoter of the vector; Tcr | This study |
| pETUD | pET30a vector containing up- and downstream DNA flanking sequences to the enolase gene; Kmr | This study |
| pETUDSm/Sp | The Sm/Sp cassette was ligated with the up- and downstream DNA flanking sequences to the enolase gene in pET30a vector; Sm/Spr Kmr | This study |
| pBUDSm/Sp | The Sm/Sp cassette was ligated with the up- and downstream DNA flanking sequences to the enolase gene in pBluescript vector; Sm/Spr Apr | This study |
| pDMSUDSm/Sp | Suicide vector pDMS197 containing the Sm/Sp cassette ligated with up- and downstream DNA flanking sequences to the enolase gene; used to generate the enolase gene deletion mutant of A. hydrophila; Kmr Tcr | This study |
| pALTER-1/eno | The native enolase gene of A. hydrophila was cloned into pALTER-1 vector at the BamHI/HindIII sites for the site mutagenesis; Tcr Aps | This study |
| pALTER/eno343K/Q | pALTER-1 vector containing the mutated A. hydrophila enolase gene eno(K343Q); Tcs Apr | This study |
| pALTER/eno394K/M | pALTER-1 vector containing the mutated A. hydrophila enolase gene eno(K394M); Tcs Apr | This study |
| pALTER/eno420K/L | pALTER-1 vector containing the mutated A. hydrophila enolase gene eno(K420L); Tcs Apr | This study |
| pALTER/eno427K/N | pALTER-1 vector containing the mutated A. hydrophila enolase gene eno(K427N); Tcs Apr | This study |
| pALTER/eno430K/R | pALTER-1 vector containing the mutated A. hydrophila enolase gene eno(K430R); Tcs Apr | This study |
| pET30aeno | The native enolase gene of A. hydrophila was cloned in pET30a and fused with the downstream His tags of the vector for hyperexpression; Kmr | This study |
| pET30aeno343K/Q | The mutated enolase gene of A. hydrophila eno(K343Q) was cloned in pET30a and fused with the downstream His tags of the vector for hyperexpression; Kmr | This study |
| pET30aeno394K/M | The mutated enolase gene of A. hydrophila eno(K394M) was cloned in pET30a and fused with the downstream His tags of the vector for hyperexpression; Kmr | This study |
| pET30aeno420K/L | The mutated enolase gene of A. hydrophila eno(K420L) was cloned in pET30a and fused with the downstream His tags of the vector for hyperexpression; Kmr | This study |
| pET30aeno427K/N | The mutated enolase gene of A. hydrophila eno(K427N) was cloned in pET30a and fused with the downstream His tags of the vector for hyperexpression; Kmr | This study |
| pET30aeno430K/R | The mutated enolase gene of A. hydrophila eno(K430R) was cloned in pET30a and fused with the downstream His tags of the vector for hyperexpression; Kmr | This study |
Plasminogen binding to A. hydrophila SSU cells and activation of plasminogen.
The tPA-mediated activation of plasminogen in the presence of bacteria was measured as described by Lähteenmäki et al. (20). Briefly, 4 × 108 log-phase-grown, phosphate-buffered saline (PBS)-washed A. hydrophila SSU cells were incubated with 4 μg of human plasminogen (Sigma), 2 ng of tPA (Molecular Innovations, Inc., Novi, MI) and a 0.45 mM concentration of the chromogenic plasmin substrate S-2251 (DiaPharma Group, Inc., West Chester, OH) in a final volume of 200 μl in PBST buffer (PBS containing 0.02% Tween 80). In some reaction mixture groups, 2 mM ɛ-aminocaproic acid (EACA; Sigma) or purified immunoglobulin G (IgG) at concentrations of 0.125 mg/ml and 0.025 mg/ml from enolase-immunized and preimmune mouse serum (as stated in the animal experiment section below) was added. The reaction mixture groups that did not contain either the bacterial cells or the tPA served as negative controls. The reaction mixtures were incubated at 37°C with slow rotation, and the increased plasmin activity was assessed at a 30-min interval for a total of 3 h by measuring absorbance at 405 nm with a VERSAmax tunable microplate reader (Molecular Devices Corporation, Sunnyvale, CA).
For the binding assay, 8 × 108 A. hydrophila SSU cells treated similarly as above were incubated with either 4 μg of human plasminogen plus 2 ng of tPA or 4 μg of human plasmin only (Sigma) in a final volume of 200 μl in PBST buffer. The reaction mixtures were incubated at 37°C with slow rotation for 3 h. In some reaction mixture groups, 2 mM EACA or purified IgG (0.125 mg/ml and 0.025 mg/ml) from enolase-immunized and preimmune mouse serum was added into the reaction mixture. The reaction mixture groups that contained only bacterial cells without the addition of plasminogen and plasmin served as negative controls. After incubation, the bacterial cells were collected and washed twice with PBST buffer and were finally resuspended in 300 μl of PBST buffer containing 0.45 mM S-2251. We incubated the reaction mixtures at 37°C for 2 h and measured plasmin activity at 405 nm. The plasmin activity levels in the negative control reaction mixture groups were used as blanks in the measurement.
For analyzing the protection of plasmin from α2AP in the presence of bacterial cells, plasmin (0.25 μg or 2.5 μg) was incubated with or without 4 × 108 log-phase-grown, PBS-washed A. hydrophila SSU cells for 30 to 60 min at room temperature prior to the addition of 0.45 mM S-2251 and α2AP in increasing concentrations (1, 8, and 16 μg). Plasmin activity was measured after 1 h of incubation at 37°C. The reaction mixture groups that were omitted from either the α2AP or both bacterial cells and α2AP cultures were used as controls. The viability of bacterial cells was monitored by serial dilution and plating at the end of each of the above experiments.
Generation and characterization of enolase gene mutant of A. hydrophila SSU.
Two pairs of primers, enoup5/enoup3 and enodn5/enodn3 (Table 2), were synthesized and used to amplify the upstream (796 bp) and downstream (718 bp) flanking DNA sequences to the enolase gene of A. hydrophila. These fragments were then ligated together through the introduced common BglII site and cloned into a pET30a vector (37) at the NotI/XbaI restriction sites, resulting in the recombinant plasmid pETUD (Table 1). An Sm/Sp gene cassette flanked by the BamHI site was removed from the plasmid pHP45Ω (33) and inserted at the BglII site (compatible with the BamHI site) of pETUD to generate the recombinant plasmid pETUDSm/Sp (Table 1). After digestion with NotI/XbaI restriction enzymes, the DNA fragment (3.5 kb) from the above plasmid was removed and ligated to a pBluescript vector (33) at the compatible restriction enzyme sites to generate the recombinant plasmid pBUDSm/Sp (Table 1). By using KpnI/XbaI restriction enzymes, the DNA fragment (3.5 kb) was removed from the plasmid pBUDSm/Sp and ligated to the pDMS197 suicide vector at the compatible restriction enzyme sites, and the resulting plasmid (pDMSUDSm/Sp) (Table 1) was transformed into E. coli SM10 (24, 33). The recombinant E. coli [pDMSUDSm/Sp] was conjugated with the parental Rifr A. hydrophila strain (33). The transconjugants were selected based on their resistance to appropriate antibiotics and sucrose and subjected to further analysis (33).
TABLE 2.
Sequences of the primers used in this study
| Primer name | Sequence (restriction enzyme)a | Purpose |
|---|---|---|
| enoup5 | 5′-TTGCGGCCGCTCAAGCACGGCGGTCTG-3′ (NotI) | PCR amplification of the upstream flanking DNA fragment of the enolase gene from A. hydrophila |
| enoup3 | 5′-CCAGATCTATGTGTATTTCCTCAGGT-3′ (BglII) | |
| enodn5 | 5′-GTAGATCTATCGTCGCCGGTTCTCTTG-3′ (BglII) | PCR amplification of the downstream flanking DNA fragment of the enolase gene from A. hydrophila |
| enodn3 | 5′-TTTCTAGAGGATCCTCGGATCGGCGG-3′ XbaI | |
| eno5 | 5′-GTAGTACTATGTCCAAGATCGTTAAAGTG-3′ (ScaI) | PCR amplification of the DNA fragment encoding the enolase gene from A. hydrophila for cloning into plasmid pBR322 |
| eno3 | 5′-TCCTGCAGTTAAGCCTGGTTCTTCACTTC-3′ (PstI) | |
| Sm5 | 5′-ATGCGCTCACGCAACTGGTC-3′ | Identification of the deletion of the enolase gene on the chromosome of A. hydrophila |
| Sm3 | 5′-TTATTTGCCGACTACCTTGG-3′ | |
| ENT5 | 5′-CCTACAAGTCCGTCAACGAG-3′ | |
| ENT3 | 5′-ACGTGCAGCGCATTGAGCAC-3′ | |
| enoF | 5′-CGCGGATCCATGTCCAAGATCGTTAAAGTGATCGG-3′ (BamHI) | Cloning of the native eno gene into pALTER-1 vector |
| enoR | 5′-CCCAAGCTTTAAGCCTGGTTCTTCACTTCTTTCAG-3′ (HindIII) | |
| enoK-Q343 | 5′-GCCAACTCCATCCTGATC(A)CAGTTCAACCAGATCGG-3′ | Site-directed mutagenesis reactions |
| enoK-M394 | 5′-CCGCTGCTGGCCAGATCA(A)TGACCGGTTCCATGAGC-3′ | |
| enoK-L420 | 5′-AAGCCCTGGGTGCC(AA)TTGGCTCCGTTCCGCGGTCTG 3′ | |
| enoK-N427 | 5′-GGGTGCCAAGGCTCCGTTCCGCGGTCTGAA(A)TGAAG-3′ | |
| enoK-R430 | 5′-TCCGCGGTCTGAAAGAAGTGA(A)GGAACCAGGCTTAA-3′ | |
| enoN | 5′-GGGTTTCATATGTCCAAGATCGTTAAAGTGATCGGTCGTG 3′ (NdeI) | Cloning of the enolase gene (native or mutated) into pET-30a(+) vector for hyperexpression |
| enoC | 5′-CCCCTCGAGAGCCTGGTTCTTCACTTCTTTCAGACCGCGG 3′ (XhoI) | |
| enoC427 | 5′-CCCCTCGAGAGCCTGGTTCTTCACTTC(T)ATTCAGACCGCGG-3′ (XhoI) | |
| enoC430 | 5′-CCCCTCGAGAGCCTGGTTC(T)CTCACTTCTTTCAGACCGCGG-3′ (XhoI) |
The underlining indicates the restriction endonuclease site. Boldface indicates mutated nucleotide(s) in the enolase gene of A. hydrophila; the original nucleotide is shown in parentheses. All primers were developed for this study.
Supplying an additional copy of the native enolase gene into WT A. hydrophila SSU.
Primers eno5 and eno3 (Table 2) were used to PCR amplify the coding region of the enolase gene from A. hydrophila. The amplified DNA fragment (1,313 bp) was cloned into the pBR322 vector at the ScaI/PstI restriction enzyme sites, generating the recombinant plasmid pBReno (Table 1). In this plasmid, the enolase gene was under the control of an Ap resistance gene promoter of the vector, and, therefore, it was constitutively expressed. The recombinant plasmid pBReno was subsequently electroporated into WT A. hydrophila (14). The presence of recombinant plasmid pBReno in WT A. hydrophila was verified by plasmid isolation and digestion with ScaI and PstI restriction enzymes.
Generation of various mutated forms of the enolase gene by site-directed mutagenesis.
An Altered Sites in vitro Mutagenesis System (Promega) was used for site-directed mutagenesis as we described previously (12). Briefly, the coding region of the native enolase gene was PCR amplified from the genomic DNA of WT A. hydrophila with the primer set enoF/enoR (Table 2) and subsequently cloned into the plasmid pALTER-1 (Tcr Aps), generating the recombinant plasmid pALTER-1/eno (Tcr Aps) (Table 1). Subsequently, E. coli JM109 cells containing the pALTER-1/eno recombinant plasmid were infected with the R408 helper bacteriophage, and the phagemid single-stranded DNA was isolated as described by the manufacturer.
This single-stranded DNA was used as a template for the mutagenesis reactions with the individual mutagenic primers enoK-Q343, enoK-M394, enoK-L420, enoK-N427, and enoK-R430, respectively (Table 2). For example, the primer enoK-Q343 was used to replace the lysine residue (K) at position 343 with a glutamine residue (Q) in the enolase. Each of the mutagenesis reactions generated a specific mutated enolase gene in plasmid pALTER-1 and was transformed into E. coli ES1301 mutS competent cells. The plasmid DNA isolated from these bacterial strains was then transformed into E. coli JM109, and the transformants were screened for Tcs and Apr. Mutations within the enolase gene in plasmid pALTER-1 were confirmed by DNA sequence analysis, and the mutated enolase genes were designated eno(K343Q), eno(K394M), eno(K420L), eno(K427N), and eno(K430R) (Table 1). Different amino acids were used to replace the lysine residues in enolase as our intent was to achieve a reasonable expression level of the mutated enolases in the E. coli system while maintaining their solubility for easy purification and biological/functional activity measurements.
Hyperexpression and purification of different forms of A. hydrophila SSU enolase.
By using the primer set enoN/enoC (Table 2), we PCR amplified native and mutated enolase genes [eno(K343Q), eno(K394M), and eno(K420L)] of A. hydrophila SSU from template plasmid DNA generated from the site-directed mutagenesis step above (Table 1). Similarly, the mutated enolase genes eno(K427N)and eno(K430R)were PCR amplified with the primer sets enoN/enoC427 and enoN/enoC430, respectively (Table 2). These amplified enolase genes were cloned into a bacteriophage T7 polymerase/promoter-based pET-30a(+) expression vector (Novagen, San Diego, CA) at the NdeI/XhoI restriction enzyme sites, and their downstream sequences were fused in frame with the His tag DNA sequence of the vector (Table 1). The resulting recombinant plasmids were transformed into the E. coli HMS174 (DE3) strain for hyperexpression. Each of these expressing strains was grown in 250 to 500 ml of LB medium with shaking (180 rpm) to an optical density at 600 nm of 0.6 before induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 30°C.
The IPTG-induced bacterial cells were harvested, and the cell lysates from these enolase-expressing strains were first passed through a ProBond nickel chelating column (Invitrogen, Carlsbad, CA) and eluted according to the manufacturer's instructions. The eluted fractions were monitored for the presence of enolase by performing sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining. Fractions containing the protein of interest (42 to 45 kDa) were pooled and dialyzed (25- to 30-kDa molecular-mass-cutoff tubing) for 12 h at 4°C in a buffer containing l20 mM Tris, 50 mM NaCl, 5 mM MgSO4, and 1 mM dithiothreitol (DTT), pH 6.5. The dialyzed fractions were filtered through a 22-μm-pore-size syringe filter (Corning, Lowell, MA) and loaded onto an ion exchange Resource Q column (6 ml; GE Healthcare, Piscataway, NJ) connected to the fast protein liquid chromatography ÄCTA Purifier 10/100. The proteins were eluted from the column by using a salt gradient (20 mM Tris, 1 M NaCl, 5 mM MgSO4, and 1 mM DTT, pH 6.5) at a flow rate of 2 ml/min. The protein peaks were analyzed by Unicorn, version 4.00.16, software (GE Healthcare) and monitored for the presence of pure enolase by SDS-PAGE and Coomassie blue staining of the gel. Fractions containing enolase were pooled and concentrated using a Centriprep instrument fitted with an Ultracel YM-10 membrane (Millipore, Billerica, MA), and the protein concentration was determined by using a Bradford dye protein assay (Bio-Rad, Hercules, CA) (5).
Enzymatic activity of various mutated forms of recombinant enolase.
The enzymatic activity of different forms of recombinant enolase was measured by their ability to catalyze the reversible conversion of 2-PGE to PEP (26, 32). In our experiments, we measured the reverse reaction (PEP to 2-PGE) due to the availability of the substrate 2-PGE. The reactions were started by the addition of 2 μg of each of the purified recombinant enolase proteins into 100 μl of the reaction buffer (100 mM HEPES, pH 7.0, 10 mM MgSO4, and 7.7 mM KCl) containing different concentrations (0.2, 0.4, 0.6, 0.8 and 1.0 mM) of PEP (Sigma). The reactions were monitored by measuring the reduction of PEP absorbance at A240 nm as a result of conversion of PEP to PGE by spectrophotometry (Ultropec 2000; GE Healthcare) every 5 s for a total of 3 min per reaction.
Plasminogen-binding activity of various mutated forms of recombinant enolase.
A sandwich enzyme-linked immunosorbent assay (ELISA) was used to detect binding of enolase to human plasminogen, as described previously (32). Briefly, microtiter plates (Evergreen Scientific, Los Angeles, CA) were coated with 2 μg per well of purified and different forms of recombinant enolase overnight at 4°C. For the positive control, the plate wells were coated with 2 μg of human plasminogen (Sigma) while wells coated with PBS only were used as negative controls. Eight wells were used for each tested recombinant enolase and control. The overnight coated plates were washed and then blocked overnight again at 4°C with buffer A (PBS containing 0.05% Tween 20, 1 mM EDTA, and 0.25% gelatin) (40). An aliquot (2 μg) of plasminogen in 50 μl of PBS was added to the coated wells and incubated for 2 h at room temperature. In some experiments, EACA or peptides (see below) were mixed with 2 μg of human plasminogen before being added to the microtiter plate wells to allow us to study their abilities to block enolase binding to the plasminogen. We used EACA at concentrations of 2, 4, 6, and 8 mM and the three peptides Seno (FYDKERKVY), Aeno (FYDAEKKEY), and AenoM (FYDAGKLEY), representing regions within the Streptococcus pneumoniae and A. hydrophila SSU enolases, at concentrations of 100 to 200 μg/well. The AenoM peptide had two amino acid substitutions (underlined) compared to the peptide Aeno.
Peptide Seno represented an internal plasminogen-binding motif (amino acid residues 248 to 256) identified in the enolase of S. pneumoniae (3, 11), while peptide Aeno (amino acid residues 252 to 260) represented a corresponding stretch of Seno in the enolase of A. hydrophila SSU (11). Peptide AenoM was the mutated form of Aeno, and the substituted amino acid residues were indicated by underlining in the sequence shown above. These peptides were synthesized by the Biomatik Corporation (Toronto, Ontario, Canada) and reconstituted in water at a concentration of 5 mg/ml. After a 2-h incubation, the plates were washed three times with buffer A and incubated for 1 h with goat antiplasminogen antibodies (primary antibody; 1:1,500 dilution) followed by horseradish peroxidase-labeled mouse, anti-goat IgG (secondary antibody; 1:2,000 dilution) (Santa Cruz Biotechnology, Inc.) (32). TMB (3,3′, 5,5′-tetramethyl-benzidine; Sigma) substrate was used for color development, and the optical density (subtracted from the blanks, i.e., the wells coated with PBS and used as the negative control) was read at 370 nm by using a VERSAmax tunable microplate reader.
Animal experiments.
Groups of 15 Swiss Webster mice (Taconic Farms, Germantown, NY) were first immunized with purified native (unmutated) recombinant enolase produced from E. coli. Briefly, animals were injected intraperitoneally (i.p.) with 10 μg of purified enolase mixed with monophosphoryl lipid A-trehalose dicorynomycolate-cell wall skeleton adjuvant(Sigma). The animals were bled before immunization, boosted twice with the antigen (10 μg) at 15-day intervals, and finally bled after 1 month. The antibody titers in the pooled sera were determined by using an ELISA having recombinant enolase as the antigen source (33). The immunized and nonimmunized mice (given the adjuvant alone without the antigen) were challenged i.p. with the WT A. hydrophila SSU at two to three times the 50% lethal dose ([LD50] 5 × 107 to 1 × 108 CFU) after 1 month of immunization. Organs (lungs, spleen, and liver) from five mice in each group were removed at day 3 of the challenge and subjected to analyses for histopathology and bacterial load (liver and spleen), as we recently described (31). Five noninfected mice were used as controls for the histopathological study. The deaths of mice were recorded, and the animals were monitored for 3 weeks postinfection.
The IgGs from the enolase-immunized and preimmune mouse sera collected from the above animal experiment were purified by using Nab Spin Kits (Thermo Fisher Scientific Inc., Rockford, IL) and further desalted with a Zeba Desalt Spin column (Thermo). The enolase antibody titers in the purified IgGs were further evaluated by ELISA using purified native recombinant enolase as the antigen source. The amount of purified IgG in both preimmune and immune sera was determined by using a Bradford dye protein assay (5). The purified IgGs were stored at 4°C and employed in the phagocytosis assay and plasminogen-binding and activation assays as described above.
Phagocytosis assay.
An aliquot (1 × 108) of log-phase-grown, PBS-washed A. hydrophila SSU cells was incubated with purified IgGs from either enolase-immunized or preimmune mouse sera at concentrations of 0.125 mg/ml and 0.025 mg/ml in a final volume of 500 μl in PBS. The bacterial cells incubated with PBS served only as negative controls. After a 1-h incubation at 37°C, the bacterial cells were washed three times with PBST buffer, and a portion of the bacterial cells was serially diluted and plated to examine viability of the bacterial cells after they were coated with the antibodies. The rest of the bacterial cells were used to infect RAW 264.7 murine macrophages (in six-well tissue culture plates) at a multiplicity of infection of 1 (37). The wells were done in triplicate for each infection group. After 3 h of incubation at 37°C with 5% CO2, the infected macrophages were washed twice with PBS and then incubated an additional hour in fresh Dulbecco's modified essential medium containing 200 μg/ml gentamicin. The gentamicin treatment killed extracellular bacteria but did not affect the viability of intracellular organisms. The macrophages were then washed three times with PBS to remove gentamicin and lysed with 200 μl of 1% (vol/vol) Triton X-100 to release intracellular bacteria. A 10-fold serial dilution was made, and bacteria were enumerated on LB agar plates in duplicate.
Statistical analysis of the data.
Where applicable, a minimum of three independent experiments were performed, and the data were analyzed using either a Student's t test or Fisher's exact test.
RESULTS
Immobilization and conversion of plasminogen to plasmin on the cell surface of A. hydrophila SSU cells.
In our previous study, we detected strong enolase activity associated with the intact A. hydrophila cells and demonstrated that crude, whole-bacterial-cell lysates of A. hydrophila could bind human plasminogen (32). To further investigate whether plasminogen indeed binds to the bacterial surface, the log phase and PBS-washed A. hydrophila SSU cells were first incubated with human plasminogen. After the unbound plasminogen was washed away, the surface-bound plasminogen was further converted to the active form of plasmin by tPA, and the plasmin activity was measured by adding the plasmin-specific chromogenic substrate S-2251.
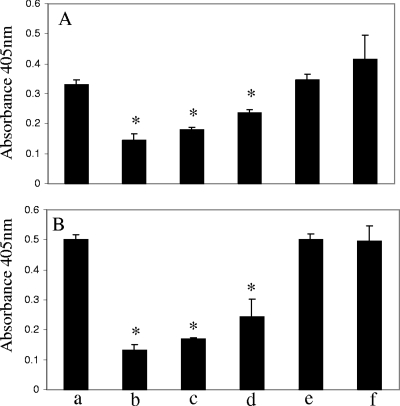
As shown in Fig. 1A, plasmin activity was detected on the cell surface of A. hydrophila SSU cells (column a), and addition of the lysine analog EACA decreased bacterial surface-associated plasmin activity by 56% (column b). These data indicated that plasminogen binding to the bacterial surface was specific and that lysine residues were involved. Importantly, we observed a dose-dependent decrease in plasmin activity (29 to 46%) in the reaction mixture groups in which purified enolase-specific antibodies were added (columns c and d). As expected, there was no decrease in plasmin activity compared to that of the control (column a) when purified IgGs from preimmune sera were added to the reaction mixtures (columns e and f). Overall, these data indicated to us that the bacterial surface-plasminogen interaction was mediated in part by enolase. A similar pattern was noticed when human plasmin was directly incubated with A. hydrophila SSU cells (Fig. 1B). No significant changes in the number of bacterial cells were observed before or after the experiment in each group and among the groups, which suggested to us that the viability of A. hydrophila SSU was not affected during the experiment.
FIG. 1.
Plasminogen and plasmin binding to the surface of A. hydrophila SSU. The log phase and PBS-washed A. hydrophila SSU cells were first incubated with 4 μg of either plasminogen (A) or plasmin (B). After washing away the unbound plasminogen/plasmin, we converted the surface-bound plasminogen to the active form plasmin by tPA and measured the plasmin activity by adding the plasmin-specific chromogenic substrate S-2251. The reaction mixtures that contained only bacterial cells without plasminogen and plasmin served as negative controls and were set as blanks when the absorbance at 405 nm was recorded. Columns a, bacterial cells incubated with plasminogen (A) or plasmin (B); columns b, addition of 2 mM EACA to the binding reaction mixture; columns c and d, addition of 0.125 mg/ml and 0.025 mg/ml, respectively, of the purified IgGs from enolase-immunized mouse serum; columns e and f; addition of 0.125 mg/ml and 0.025 mg/ml, respectively, of purified IgGs from preimmune mouse serum. The asterisk denotes statistically significant differences (P < 0.05) compared to columns a by a Student's t test.
Plasminogen binding on the surface of A. hydrophila SSU cells promoted its tPA-mediated activation and protected plasmin from its physiological inhibitor α2AP.
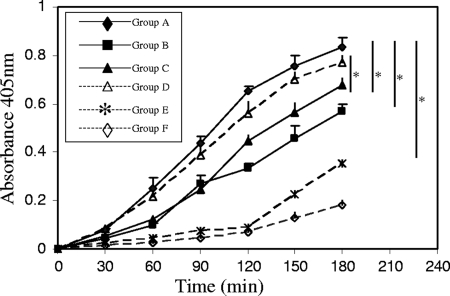
As shown in Fig. 2, there was a significant, time-dependent (30 to 180 min) formation of plasmin from plasminogen mediated by tPA (group A) in the presence of A. hydrophila SSU cells. In contrast, only marginal plasmin activity was observed in the group in which no bacterial cells were added (group E). Plasmin formation was greatly inhibited (32 to 61%) over a period of 3 h of incubation in the presence of EACA (group B) compared to that in the control group A, a finding that further emphasized lysine residue involvement and the importance of plasminogen's binding to the bacterial surface prior to its activation.
FIG. 2.
A. hydrophila SSU cells promoted tPA-mediated plasminogen activation. An aliquot of 4 × 108 CFU of log-phase-grown, PBS-washed A. hydrophila SSU cells was incubated with 4 μg of human plasminogen, 2 ng of tPA, and a 0.45 mM S-2251, a chromogenic substrate of plasmin. The reaction mixtures were incubated at 37°C with slow rotation, and the increased plasmin activities were assessed at 30-min intervals for 3 h by measuring absorbance at 405 nm. Group A, bacterial cells incubated with plasminogen and tPA; group B, addition of 2 mM EACA; group C, addition of 0.125 mg/ml of purified IgGs from enolase-immunized mouse serum; group D, addition of 0.125 mg/ml of purified IgGs from preimmune mouse serum; group E, reaction mixture containing only plasminogen and tPA and no bacterial cells; group F, reaction mixture containing only plasminogen and bacterial cells without tPA. The asterisk denotes statistically significant differences (P < 0.05) between the two groups, identified by the vertical lines, using a Student's t test.
Compared to the groups of reaction mixtures with the addition of purified IgGs from preimmune sera (group D), the presence of enolase-specific antibodies in the reaction mixtures significantly and dose-dependently decreased (12 to 43%) tPA-mediated plasmin formation (group C), with a maximum decrease (43%) occurring at the 60-min time point. In Fig. 2, data represent only experiments with high (0.125 mg/ml) enolase antibody concentrations. When a lower concentration of enolase-specific antibodies (0.025 mg/ml) was used in the reaction mixtures, the decrease in plasmin activity was in the range of 8 to 31% over a period of 3 h, with statistical differences compared to the preimmune sera only at 30- and 60-min time points (data not shown). These data further confirmed that enolase was involved in plasminogen binding and its activation. Minimal plasmin formation was noticed in the reaction mixture in which no tPA was added (group F), suggesting to us that there was no apparent plasminogen activator secreted by A. hydrophila SSU.
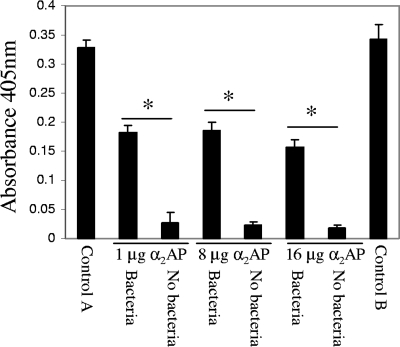
To test if the converted plasmin on the surface of A. hydrophila SSU cells was protected from the action of its specific inhibitor, bacterial cells were incubated with 0.25 μg of plasmin for 30 to 60 min before different amounts of the plasmin inhibitor α2AP were added. As shown in Fig. 3, compared to the control groups in which α2AP (control A) or both the bacterial cells and α2AP (control B) were omitted, groups receiving α2AP at various concentrations showed a statistically significant 46 to 54% decrease in plasmin activity, even in the presence of A. hydrophila cells. However, in the groups of reaction mixtures in which no bacterial cells were added, α2AP abrogated plasmin activity (above 90% inhibition), which indicated that A. hydrophila SSU cells provided partial protection to plasmin from the action of its physiological inhibitor α2AP. A similar pattern was observed when a larger amount of plasmin (2.5 μg) was used in this protection assay (data not shown).
FIG. 3.
A. hydrophila SSU cells protected plasmin from its physiological inhibitor α2AP. Human plasmin (0.25 μg) was incubated with or without 4 × 108 CFU of log-phase-grown, PBS-washed A. hydrophila SSU cells for 30 to 60 min at room temperature prior to the addition of 0.45 mM S-2251 and of α2AP in increasing concentrations (1, 8, and 16 μg). Plasmin activity was measured after 1 h of incubation at 37°C. The groups of reaction mixtures that either lacked α2AP (control A) or contained both the bacterial cells and α2AP (control B) were used as controls. The asterisk denotes statistically significant differences (P < 0.05) between the two groups (indicated by the horizontal lines) using a Student's t test.
Role of lysine residues in the enzymatic and plasminogen-binding activities of A. hydrophila enolase.
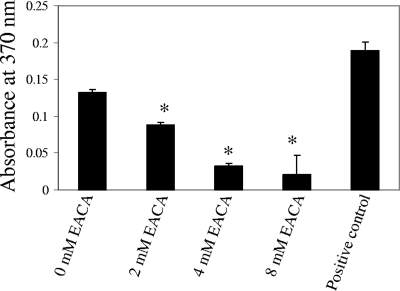
To further study the plasminogen-binding activity of enolase, we used purified recombinant A. hydrophila SSU enolase from E. coli. As shown in Fig. 4, the recombinant enolase-bound plasminogen was detected by the antiplasminogen antibody in ELISA. Importantly, this plasminogen-binding activity could also be blocked by adding EACA (lysine analog) in a dose-dependent manner. These data with the purified enolase corroborate the results shown in the plasminogen-binding and activation experiments (Fig. 1 to 3) with whole bacterial cells and confirmed the role of lysine residues of enolase in plasminogen binding.
FIG. 4.
EACA blocked plasminogen binding of recombinant enolase of A. hydrophila SSU hyperproduced in E. coli. In a sandwich ELISA, 2 μg of purified recombinant enolase of A. hydrophila was precoated in each well, and plasminogen (2 μg) with or without EACA (2 to 8 mM) was then added to the wells for a 2-h incubation at room temperature. After a washing step, we detected plasminogen binding to the coated enolase by using antiplasminogen antibody. The wells coated with 2 μg of plasminogen were used as positive controls for the system while those coated with PBS only were used as negative controls and set as blanks when the absorbance readings were taken. A total of eight wells were used per group. The asterisk denotes statistically significant differences compared to the group without the addition of EACA (Student's t test).
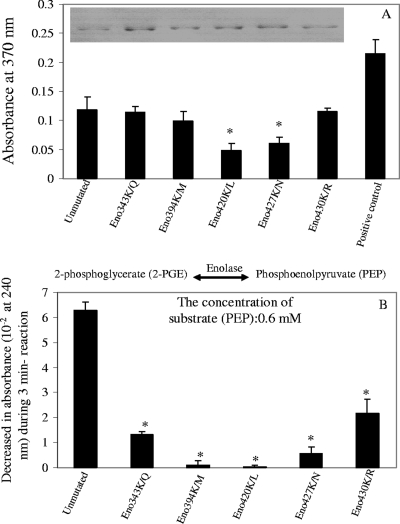
To confirm this finding, we mutated lysine residues of the A. hydrophila SSU enolase at positions 343, 394, 420, 427, and 430 by site-directed mutagenesis and generated five mutated forms of enolase, designated Eno(K343Q), Eno(K394M), Eno(K420L), Eno(K427N), and Eno(K430R), respectively. These lysine residues were chosen either because their positions were toward the carboxyl terminus of A. hydrophila SSU enolase, which has 434 amino acid residues (e.g., lysine residues 420, 427, and 430), or because their corresponding lysine residues (e.g., lysine residues 343 and 394) were important to the enolase activity of Streptococcus pyogenes (26). As with the native form of recombinant enolase, the mutated enolase genes were hyperexpressed, and the resulting recombinant proteins were purified. The integrity and purity of the recombinant enolase were examined by SDS-12% PAGE and Coomassie blue staining (Fig. 5A, inset). As shown in Fig. 5A, compared to the native recombinant enolase, the plasminogen-binding activities of Eno(K420L) and Eno(K427N) were decreased by 49 to 60%, respectively, while the binding activity remained similar to that of the native recombinant enolase in Eno(K343Q), Eno(K394M), and Eno(K430R). Interestingly, the enzymatic activity in all of the mutated forms of enolase was severely affected, with minimal activities (21%, 10%, and 35% of the native recombinant enolase activity, respectively) detected in Eno(K343Q), Eno(K427N), and Eno(K430R) (Fig. 5B). Importantly, Eno(K394M)and Eno(K420L) exhibited no detectable enolase activity. These findings were interesting as Eno(K394M)affected enolase activity but not the plasminogen-binding activity of enolase, while Eno(K420L) affected both of these activities.
FIG. 5.
Role of lysine residues in plasminogen-binding and enzymatic activities of A. hydrophila SSU enolase. (A) Sandwich ELISA showing plasminogen-binding activities of mutated forms of Aeromonas enolase (with mutations of the indicated lysine residues). Wells of the microtiter plates were precoated with either native recombinant enolase (unmutated) or mutated forms of enolase [Eno(K343KQ), Eno(K394M), Eno(K420L), Eno(K427N), and Eno(K430R)]. An aliquot (2 μg) of plasminogen was then added to the wells for a 2-h incubation at room temperature. After a washing step, binding of plasminogen was detected by using antiplasminogen antibody. Wells coated with 2 μg of plasminogen were used as positive controls for the system while the wells coated with PBS only were used as negative controls and were set as blanks when the absorbance readings were taken. A total of eight wells were used for each group. The integrity and purity of these recombinant enolases were examined by SDS-12% PAGE with Coomassie blue staining, shown here in the inset. (B) Enzymatic activity of different forms of recombinant enolase was evaluated by measuring their ability to convert PEP to 2-PGE. Different concentrations of substrate PEP were used, and the results of a typical reaction at a 0.6 mM concentration of PEP are shown. The asterisk denotes statistically significant differences compared to the native enolase group (Student's t test).
Plasminogen-binding motif of enolase in A. hydrophila SSU.
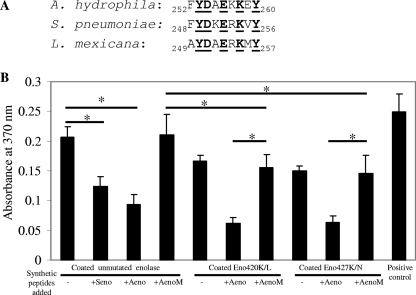
It has been recently reported that a 9-amino-acid stretch of enolase (248FYDKERKVY256) in S. pneumoniae is also responsible for plasminogen-binding activity (3). By sequence comparison, a corresponding stretch (FYDAEKKEY) was identified at amino acid residues 252 to 260 in the enolase of A. hydrophila SSU (11). The latter shared a 60% identity with similar stretches identified in S. pneumoniae and Leishmania mexicana enolase (Fig. 6A) (11, 40). To delineate the role of this stretch in the binding of enolase to plasminogen, peptides corresponding to these stretches (for both S. pneumoniae and A. hydrophila) were synthesized. These peptides were Seno (FYDKERKVY), Aeno (FYDAEKKEY), and AenoM (FYDAGKLEY). Peptide AenoM is a mutated form of Aeno, and in the forging sequence, the two amino acid substitutions are underlined.
FIG. 6.
Identification of the plasminogen-binging motif in the enolase of A. hydrophila SSU. (A) Comparison of the plasminogen-binding motif in the enolase of S. pneumoniae with the corresponding strectches identified in the enolase of A. hydrophila and L. mexicana. The conserved amino acid residues are in bold and underlined. (B) Sandwich ELISA was used for measuring the plasminogen-binding activity. Wells of the microtiter plates were precoated with either native recombinant enolase (unmutated) or mutated forms of enolase [Eno(K420L) and Eno(K427N)]. An aliquot (2 μg) of plasminogen with or without the synthetic peptides (Seno, Aeno, and AenoM) was added to the wells for a 2-h incubation at room temperature. After a washing step, binding of plasminogen was detected by using antiplasminogen antibody. The wells coated with 2 μg of plasminogen were used as positive controls for the system while the wells coated with PBS only were used as negative controls and were set as blanks when the absorbance readings were taken. A total of eight wells were used for each group. The asterisk denotes statistically significant differences between the two groups, identified by the horizontal lines (Student's t test).
As shown in Fig. 6B, when wells were coated with the native recombinant A. hydrophila SSU enolase, the addition of peptides Seno and Aeno to the reaction mixtures led to a 40 to 54% reduction in the enolase-plasminogen-binding activities compared to the reaction mixtures without the addition of peptides. These data indicated that the peptides Seno and Aeno competed with the coated enolase, thus inhibiting enolase-plasminogen-binding activity. However, when the mutated peptide (AenoM) was used, no such blocking effects were noticed, thus showing that the blocking effects of peptides Seno and Aeno were specific and that the corresponding amino acid stretch (positions 252 to 260) in the enolase of A. hydrophila SSU had a function similar to that of the plasminogen-binding motif identified in the enolase of S. pneumoniae (3, 11).
Interestingly, when peptide Aeno was added to the wells coated with the mutated forms of enolase [i.e., Eno(K420L) and Eno(K427N)], we noted a further 57 to 60% reduction in plasminogen binding to mutated enolase in comparison to the wells receiving the mutated peptide AenoM or no peptides (Fig. 6B). These data indicated that the lysine residue at position 420 or 427, along with the amino acid stretch FYDAEKKEY of A. hydrophila SSU enolase, had an additive effect on plasminogen binding.
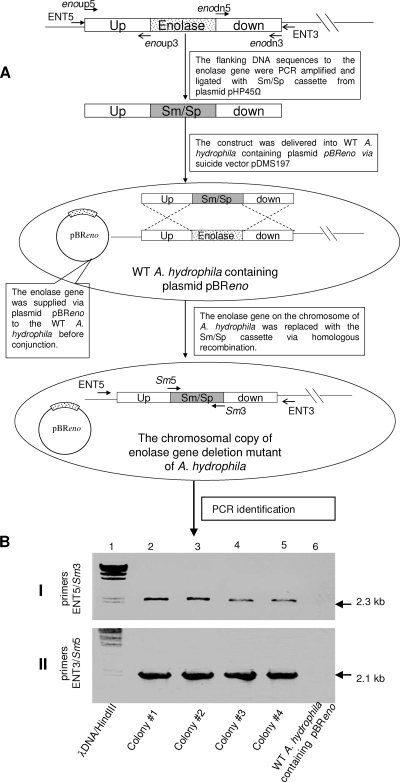
Potential role of the enolase gene in the viability of A. hydrophila SSU.
To study the contribution of enolase in the pathogenesis of Aeromonas infections, we first attempted to generate an enolase-negative isogenic mutant of A. hydrophila. However, after several attempts, we failed to obtain the enolase knockout mutant of A. hydrophila SSU, thus raising the possibility that enolase might be essential for the viability of A. hydrophila. To test this, an additional copy of the A. hydrophila enolase gene was supplied via plasmid pBR322 to the WT A. hydrophila before conjugation (Fig. 7). Consequently, we obtained four colonies on the selected LB agar plates (containing 5% sucrose, 25 μg of Sp/Sm, and 200 μg of Rif). To confirm the deletion of the chromosomal copy of the enolase gene in these A. hydrophila mutant clones, two pairs of primers were synthesized. One set was Sm5/Sm3 (Table 2 and Fig. 7), which corresponded to the coding region of the Sm/Sp gene cassette, and another was ENT5/ENT3 (Table 2 and Fig. 7), representing regions of the genomic DNA just outside of the sequences to which primers enoup5 and enodn3, respectively, were designed. Therefore, primers ENT5 and ENT3 were not within the flanking DNA sequences used for homologous recombination to generate the enolase mutant of A. hydrophila.
FIG. 7.
Construction of the chromosomal enolase gene deletion mutant of A. hydrophila SSU. (A) The flow diagram showing construction of the mutant. The primer pairs enoup5/enoup3 and enodn5/enodn3 (Table 2) were used to PCR amplify the flanking DNA fragments (open boxes) to the enolase gene (dotted box). The Sm/Sp gene cassette was removed from the plasmid pHP45Ω. The suicide vector pDMS197 was used for delivering the construct into WT A. hydrophila that contained an additional copy of the enolase gene on the plasmid pBReno. By homologous recombination, the enolase gene on the chromosome of A. hydrophila was replaced with the Sm/Sp cassette. The arrows represent the direction and position of different primers. (The figure is not drawn to scale). (B) PCR identification of the potential enolase gene deletion mutants of A. hydrophila. In the experiment shown in panel I, the primer set ENT5/Sm3 was used while primer set ENT3/Sm5 was employed for the experiment shown in panel II. Lanes 2 to 5, potential enolase mutant colonies. Other lanes are as indicated.
By using primers Sm5/Sm3, we PCR amplified a 1.0-kb segment of the Sm/Sp gene-coding region from the tested colonies, which was evidence of the integration of the Sm/Sp cassette in these clones (data not shown). The primers ENT5/Sm3 and ENT3/Sm5 were used to examine the DNA sequences adjacent to the Sm/Sp integration site after homologous recombination. As the corresponding primer binding sequences for ENT5 and ENT3 existed only on the chromosome of A. hydrophila (Fig. 7) and neither the plasmid pBReno nor pDMSUDSm/Sp contained the binding sequences for ENT5/ENT3, we expected specific PCR amplifications from the chromosome of the tested colonies. As shown in Fig. 7B, lanes 2 to 5, 2.3-kb and 2.1-kb DNA fragments were PCR amplified from the chromosomes of these tested clones when primer sets ENT5/Sm3 (panel I) and ENT5/Sm3 (panel II) were used. No band was amplified from WT A. hydrophila transformed with pBReno (Fig. 7B, lane 6), which indicated to us the correct location of the integration, which resulted in the replacement of a chromosomal copy of the enolase gene with an Sm/Sp cassette. The integrity of the native enolase gene on the plasmid pBReno in these colonies was confirmed by plasmid isolation and restriction enzyme analysis (data not shown).
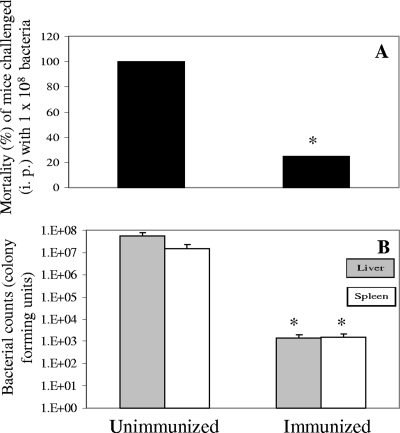
Role of enolase in the pathogenesis of A. hydrophila SSU infection.
To study enolase's role in the pathogenesis of A. hydrophila infections, we immunized mice with the purified recombinant enolase and subsequently challenged them with a lethal dose (5 × 107 to 1 × 108 CFU) of WT A. hydrophila. At 3 weeks postinfection, we observed 100% mortality in the nonimmunized group of mice compared to only 20% in the immunized group (Fig. 8A). The average bacterial load (per organ) of the nonimmunized mice at day 3 postchallenge was 5.5 × 107 CFU for the liver and 1.5 × 107 CFU for the spleen while the corresponding numbers in the immunized group of mice dropped to 1.4 × 103 and 1.6 × 103 CFU, respectively (Fig. 8B).
FIG. 8.
Role of enolase in a mouse model of A. hydrophila SSU infection. The recombinant enolase-immunized Swiss Webster mice as well as the nonimmunized mice were intraperitoneally challenged with a lethal dose (2 to 3 LD50s) of WT A. hydrophila. The deaths were recorded, and mice were observed for 3 weeks after challenge. The mortality rates from both the groups (immunized and nonimmunized) are shown in panel A while the bacterial loads in the livers and spleens of both the groups are shown in panel B. The asterisk denotes statistically significant differences between immunized and nonimmunized groups by a Fisher's exact test (A) or a Student's t test (B).
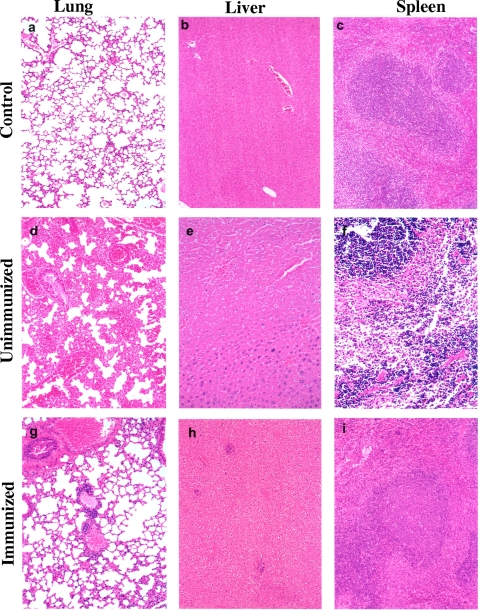
The mouse organs (lungs, liver, and spleen) from both the immunized and nonimmunized groups were histopathologically analyzed. The noninfected mice were used as controls. As shown in Fig. 9, the noninfected control mice had normal organ architecture with no lesions in the lungs, liver, and spleen (Fig. 9a, b, and c). In the nonimmunized group of mice infected with the WT bacteria (Fig. 9d, e, and f), the lung section had marked vascular congestion, alveolar hemorrhage, and widening of the interstitium. The liver section (panel e) showed prominent coagulative necrosis of the hepatic parenchyma. Several inflammatory cells were seen in the sinusoids. In the spleen section of nonimmunized mice (panel f), the splenic follicle exhibited necrosis and apoptotic cells in the red pulp in proximity to the lymphoid follicle.
FIG. 9.
The histopathological analysis of mouse tissues. Swiss Webster mice immunized with or without the purified recombinant enolase of A. hydrophila SSU were challenged i.p. with a lethal dose (2 to 3 LD50s) of WT A. hydrophila. Mouse organs (liver, lung, and spleen) were removed on day 3 postchallenge and stained with hematoxylin and eosin; the noninfected mice were used as controls. Tissues are as indicated on the figure. Magnifications, ×100 (a, b, c, d, h, and i) and ×200 (e, f, and g).
In the immunized group (Fig. 9g, h, and i), the lung section had focal thickening of alveolar septa with lymphohistiocytic infiltrates and neutrophils, but there was no evidence of alveolar hemorrhage or vascular congestion. In the liver section, occasional lobular and perivascular lymphohistiocytic infiltrates with focal areas of necrosis were observed. The increased macrophages in the red pulp and marked lymphoid activation in the splenic follicles, with germinal center formation, were present in the spleen section (panel i). Overall, architectural analysis revealed to us that mice infected with WT A. hydrophila without immunization with the recombinant enolase exhibited more severe pathologies.
These in vivo data pointed to a disseminative role of the A. hydrophila SSU enolase in the infected animals. We also explored whether enolase-specific antibodies altered the phagocytic ability of A. hydrophila SSU. For those studies, we precoated bacterial cells with either enolase-specific IgGs or IgGs from preimmune mouse serum. These bacterial cells were then used to infect murine RAW 264.7 macrophages. Our data indicated that macrophages engulfed similar numbers of bacteria precoated with either the purified IgGs from enolase-immunized or preimmune mouse serum (data not shown).
DISCUSSION
Enolase's pathogenic role has been linked to its surface expression and its ability to bind plasminogen in several pathogenic bacteria (1, 3, 11, 20, 26-28, 32). The subsequent activation of the fibrinolytic system promotes bacterial penetration of the barriers of the infected host (2, 3, 11, 20). Several bacterial molecules, such as enolase, the glyceraldehyde-3-phosphate dehydrogenase, and certain fimbrial and flagellar antigens, have been reported to serve as plasminogen receptors (20). The existence of surface plasminogen receptor(s) on A. hydrophila SSU cells was evidenced by the binding of plasminogen to the intact bacterial cells and by the ability of bacteria to aid in tPA-mediated plasminogen activation. Importantly, this binding and activation could be partially inhibited by enolase-specific antibodies, which indicated to us that enolase is one of the surface plasminogen receptors and correlated with our previous findings (32).
We also observed that a lysine analog, EACA, inhibited plasminogen binding and activation, which suggested to us that immobilization of plasminogen on the surface of A. hydrophila SSU might involve lysine residues of its receptors (e.g., enolase) in binding to the kringle domains of plasminogen (20). As α2AP is the primary circulating inhibitor of plasmin, it binds to the kringle domains and effectively inactivates soluble plasmin (17). Consequently, the protection provided by A. hydrophila cells to plasmin from the effect of α2AP was evidence that the converted plasmin was still in a bound form, and this protection further implicated the involvement of surface-expressed enolase in the pathogenesis of A. hydrophila SSU infections.
In addition to using eukaryotic plasminogen activators, such as tPA and urokinase, several pathogenic bacteria also produce their own activators. These may be either surface-bound proteolytic activators, such as the plasminogen activator (Pla) in Yersinia pestis and PrtA in Salmonella enterica, or nonproteolytic activators, such as staphylokinase and streptokinase, which are secreted by staphylococci and streptococci, respectively (9, 17-20). As we did not detect any significant conversion or activation of plasminogen without tPA in the assays we used, these data provided evidence that A. hydrophila SSU lacked a functional plasminogen activator of its own.
We further confirmed plasminogen binding with enolase of A. hydrophila SSU by using purified recombinant protein and observed a similar inhibitive role of the lysine analog EACA in the binding. These data strongly suggested involvement of lysine residues in Aeromonas enolase-plasminogen binding. It has been shown that the two carboxyl-terminal lysine residues located immediately before the termination codon play a role in enolase-plasminogen binding in both eukaryotic and prokaryotic enolase (1, 23, 27). Interestingly, the A. hydrophila SSU enolase did not possess such C-terminal lysine residues (11, 32). However, when the lysine residues of Aeromonas enolase (positions 420 and 427) were mutated, we observed decreased plasminogen-binding activities with the mutated forms, which suggested to us that the lysine residues near the C-terminal end of enolase played a role in plasminogen binding. Interestingly, when the lysine residue at position 430 (the closest lysine residue to the C-terminal end of enolase) was replaced with arginine, plasminogen-binding activity was not significantly altered. This could possibly be due to the similarity of arginine to a lysine residue as both are positively charged amino acid residues.
Recently, an internal stretch (amino acid residues 248 to 256) of pneumococcal enolase was identified as the plasminogen-binding motif (3, 11). In this study, we have shown that synthetic peptides corresponding to this internal stretch of S. pneumoniae and A. hydrophila competitively inhibited plasminogen binding to A. hydrophila SSU enolase, and we believe that these peptides represent a binding domain for plasminogen in both S. pneumoniae and A. hydrophila. A similar binding motif has been reported for the enolase of L. mexicana (40).
Ehinger et al. reported that the internal stretch of S. pneumoniae was located on the surface of the octameric pneumococcal enolase, whereas the C-terminal lysines were located in the interdimer groove and apparently not accessible to plasminogen binding (11). We observed some additive blocking effects by combining mutated forms of enolase (lysine residues at position 420 or 427) with the synthetic peptide Aeno in a sandwich ELISA (Fig. 6B). However, we need to determine the exact role of these lysine residues and the internal stretch in plasminogen binding in context of the oligomeric status of enolase on the surface of A. hydrophila SSU.
It has been reported that the amino acid residues Glu168, Glu211, Lys345, and Lys396 are important for the enzymatic activity of enolase in Saccharomyces cerevisiae (26). As expected, when we mutated the corresponding lysine residues of A. hydrophila SSU enolase at positions 343 and 394, the enzymatic activities were severely affected. A similar decrease in enzymatic activity was observed when we altered lysine residues at positions 420, 427, and 430 of Aeromonas enolase. The enzymatic activity of enolase is associated with its dimeric form (26, 42), and mutations of lysine residues toward the C-terminal end of Aeromonas enolase possibly affected its oligomeric form, thereby decreasing enzymatic activity. A recent report noted that replacement of the native enolase gene with the mutated enolase gene (in which both of the C-terminal lysine residues were either replaced or deleted) in the genome of S. pneumoniae did not affect the overall enzymatic activity on the surface of these mutated S. pneumoniae cells compared to activity of the native enolase on the surface (3). However, the positions of mutated lysine residues were different in A. hydrophila SSU from their positions in S. pneumoniae. Further, we used purified recombinant, mutated enolase instead of the surface-expressed mutated enolase to measure the enzymatic activities.
To further study the role of enolase, we started by generating an enolase-negative isogenic mutant of A. hydrophila SSU. However, we found that enolase might be essential for the viability of A. hydrophila, which was not surprising, as enolase is one of the key enzymes acting as a 2-phospho-d-glycerate hydrolase in the glycolytic cycle located in the cytoplasm of prokaryotes and eukaryotes (1). Further, there was only one copy of the enolase gene in the A. hydrophila genome (30, 32). Similar results were reported for S. pyogenes and S. pneumoniae, in which the enolase gene was indispensable (1, 28). Based on the DNA sequence (gene accession number AY141757), a CTP synthetase and a cell division protein (FtsB)-encoding gene were located immediately up- and downstream, respectively, of the enolase gene in A. hydrophila SSU. The polar effects are always a concern when knockout mutants are generated; however, in this study, our failure to obtain an enolase knockout mutant of A. hydrophila SSU was unlikely due to these possible polar effects as we supplied only the coding region of the enolase gene in trans and successfully knocked out the enolase gene from the chromosome of A. hydrophila SSU. However, further studies are needed to conclusively prove the role of enolase in the viability of Aeromonas species in general.
In this study, we showed that blocking the surface enolase of A. hydrophila SSU with anti-enolase-specific antibodies, generated by immunizing the mice with purified recombinant enolase, protected animals from subsequent Aeromonas infection. The immunized animals had minimal histopathological changes and lower bacterial loads in the organs examined. This correlated well with the plasminogen-binding ability of Aeromonas enolase and its surface display, which are linked to aiding dissemination of bacteria in the host. We noted that bacteria coated with antibodies to enolase were phagocytosed by macrophages to the same extent as were A. hydrophila SSU cells coated with purified IgGs from the preimmune serum at the same protein concentration. These data ruled out the possibility that enolase antibody-coated bacteria were rapidly engulfed by macrophages and killed, resulting in minimal mouse mortality and histopathology. From our data, it appeared that the surface-displayed enolase had a direct role in disseminating the bacteria to different organs.
Importantly, enolase is a mutifunctional protein (26), and it may affect bacterial virulence through different mechanism(s). Enolase, especially, has been reported to have the ability to bind cytoskeletal and chromatin structures and is a component of the RNA degradosome in E. coli involved in the degrading of mRNA (7, 26). Thus, enolase might play a role in regulating the expression of a variety of genes, including those coding for identified and unidentified virulence factors of A. hydrophila.
In addition to interaction with the mammalian proteolytic plasminogen-plasmin system, enolase from Streptococcus sobrinus has been reported to suppress the immune system in mice (41). Therefore, further studies that focus on these possibilities will certainly help us to delineate the pathogenic mechanism(s) of A. hydrophila infections that are linked to enolase and will be included in our future research. Although our current study is focused mainly on A. hydrophila SSU, considering the ubiquitous and conservative properties of enolase, its interaction on the cell surface with the mammalian proteolytic plasminogen-plasmin system may represent a universal pathogenic mechanism in Aeromonas infections in general and needs to be further explored.
Acknowledgments
This research was supported by grants to A.K.C. from the NIH/NIAID (AI041611) and the Environmental Protection Agency.
We thank Mardelle Susman for her carefully editing of the manuscript.
Footnotes
Published ahead of print on 6 March 2009.
REFERENCES
- 1.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 401273-1287. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann, S., M. Rohde, K. T. Preissner, and S. Hammerschmidt. 2005. The nine-residue plasminogen-binding motif of the pneumococcal enolase is the major cofactor of plasmin-mediated degradation of extracellular matrix, dissolution of fibrin and transmigration. Thromb. Haemost. 94304-311. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann, S., D. Wild, O. Diekmann, R. Frank, D. Bracht, G. S. Chhatwal, and S. Hammerschmidt. 2003. Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49411-423. [DOI] [PubMed] [Google Scholar]
- 4.Boyle, M. D., and R. Lottenberg. 1997. Plasminogen activation by invasive human pathogens. Thromb. Haemost. 771-10. [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Candela, M., S. Bergmann, M. Vici, B. Vitali, S. Turroni, B. J. Eikmanns, S. Hammerschmidt, and P. Brigidi. 2007. Binding of human plasminogen to Bifidobacterium. J. Bacteriol. 1895929-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpousis, A. J. 2007. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 6171-87. [DOI] [PubMed] [Google Scholar]
- 8.Chopra, A. K., and C. W. Houston. 1999. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 11129-1137. [DOI] [PubMed] [Google Scholar]
- 9.Coleman, J. L., and J. L. Benach. 1999. Use of the plasminogen activation system by microorganisms. J. Lab. Clin. Med. 134567-576. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207149-157. [DOI] [PubMed] [Google Scholar]
- 11.Ehinger, S., W. D. Schubert, S. Bergmann, S. Hammerschmidt, and D. W. Heinz. 2004. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 343997-1005. [DOI] [PubMed] [Google Scholar]
- 12.Erova, T. E., A. A. Fadl, J. Sha, B. K. Khajanchi, L. L. Pillai, E. V. Kozlova, and A. K. Chopra. 2006. Mutations within the catalytic motif of DNA adenine methyltransferase (Dam) of Aeromonas hydrophila cause the virulence of the Dam-overproducing strain to revert to that of the wild-type phenotype. Infect. Immun. 745763-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erova, T. E., V. G. Kosykh, A. A. Fadl, J. Sha, A. J. Horneman, and A. K. Chopra. 2008. Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis. J. Bacteriol. 1903467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erova, T. E., L. Pillai, A. A. Fadl, J. Sha, S. Wang, C. L. Galindo, and A. K. Chopra. 2006. DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect. Immun. 74410-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erova, T. E., J. Sha, A. J. Horneman, M. A. Borchardt, B. K. Khajanchi, A. A. Fadl, and A. K. Chopra. 2007. Identification of a new hemolysin from diarrheal isolate SSU of Aeromonas hydrophila. FEMS Microbiol. Lett. 275301-311. [DOI] [PubMed] [Google Scholar]
- 16.Fadl, A. A., C. L. Galindo, J. Sha, T. E. Erova, C. W. Houston, J. P. Olano, and A. K. Chopra. 2006. Deletion of the genes encoding the type III secretion system and cytotoxic enterotoxin alters host responses to Aeromonas hydrophila infection. Microb. Pathog. 40198-210. [DOI] [PubMed] [Google Scholar]
- 17.Hurmalainen, V., S. Edelman, J. Antikainen, M. Baumann, K. Lahteenmaki, and T. K. Korhonen. 2007. Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology 1531112-1122. [DOI] [PubMed] [Google Scholar]
- 18.Lahteenmaki, K., S. Edelman, and T. K. Korhonen. 2005. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 1379-85. [DOI] [PubMed] [Google Scholar]
- 19.Lahteenmaki, K., P. Kuusela, and T. K. Korhonen. 2001. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25531-552. [DOI] [PubMed] [Google Scholar]
- 20.Lahteenmaki, K., R. Virkola, R. Pouttu, P. Kuusela, M. Kukkonen, and T. K. Korhonen. 1995. Bacterial plasminogen receptors: in vitro evidence for a role in degradation of the mammalian extracellular matrix. Infect. Immun. 633659-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Alemany, R., P. Correc, L. Camoin, and P. Burtin. 1994. Purification of the plasmin receptor from human carcinoma cells and comparison to alpha-enolase. Thromb. Res. 75371-381. [DOI] [PubMed] [Google Scholar]
- 22.Lottenberg, R., D. Minning-Wenz, and M. D. Boyle. 1994. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends Microbiol. 220-24. [DOI] [PubMed] [Google Scholar]
- 23.Miles, L. A., C. M. Dahlberg, J. Plescia, J. Felez, K. Kato, and E. F. Plow. 1991. Role of cell-surface lysines in plasminogen binding to cells: identification of alpha-enolase as a candidate plasminogen receptor. Biochemistry 301682-1691. [DOI] [PubMed] [Google Scholar]
- 24.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1702575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakajima, K., M. Hamanoue, N. Takemoto, T. Hattori, K. Kato, and S. Kohsaka. 1994. Plasminogen binds specifically to alpha-enolase on rat neuronal plasma membrane. J. Neurochem. 632048-2057. [DOI] [PubMed] [Google Scholar]
- 26.Pancholi, V. 2001. Multifunctional alpha-enolase: its role in diseases. Cell Mol. Life Sci. 58902-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pancholi, V., and V. A. Fischetti. 1998. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 27314503-14515. [DOI] [PubMed] [Google Scholar]
- 28.Pancholi, V., P. Fontan, and H. Jin. 2003. Plasminogen-mediated group A streptococcal adherence to and pericellular invasion of human pharyngeal cells. Microb. Pathog. 35293-303. [DOI] [PubMed] [Google Scholar]
- 29.Pillai, L., J. Sha, T. E. Erova, A. A. Fadl, B. K. Khajanchi, and A. K. Chopra. 2006. Molecular and functional characterization of a ToxR-regulated lipoprotein from a clinical isolate of Aeromonas hydrophila. Infect. Immun. 743742-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seshadri, R., S. W. Joseph, A. K. Chopra, J. Sha, J. Shaw, J. Graf, D. Haft, M. Wu, Q. Ren, M. J. Rosovitz, R. Madupu, L. Tallon, M. Kim, S. Jin, H. Vuong, O. C. Stine, A. Ali, A. J. Horneman, and J. F. Heidelberg. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 1888272-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sha, J., S. L. Agar, W. B. Baze, J. P. Olano, A. A. Fadl, T. E. Erova, S. Wang, S. M. Foltz, G. Suarez, V. L. Motin, S. Chauhan, G. R. Klimpel, J. W. Peterson, and A. K. Chopra. 2008. Braun lipoprotein (Lpp) contributes to virulence of yersiniae: potential role of Lpp in inducing bubonic and pneumonic plague. Infect. Immun. 761390-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sha, J., C. L. Galindo, V. Pancholi, V. L. Popov, Y. Zhao, C. W. Houston, and A. K. Chopra. 2003. Differential expression of the enolase gene under in vivo versus in vitro growth conditions of Aeromonas hydrophila. Microb. Pathog. 34195-204. [DOI] [PubMed] [Google Scholar]
- 33.Sha, J., E. V. Kozlova, and A. K. Chopra. 2002. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 701924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sha, J., E. V. Kozlova, A. A. Fadl, J. P. Olano, C. W. Houston, J. W. Peterson, and A. K. Chopra. 2004. Molecular characterization of a glucose-inhibited division gene, gidA, that regulates cytotoxic enterotoxin of Aeromonas hydrophila. Infect. Immun. 721084-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sha, J., M. Lu, and A. K. Chopra. 2001. Regulation of the cytotoxic enterotoxin gene in Aeromonas hydrophila: characterization of an iron uptake regulator. Infect. Immun. 696370-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sha, J., L. Pillai, A. A. Fadl, C. L. Galindo, T. E. Erova, and A. K. Chopra. 2005. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect. Immun. 736446-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sha, J., S. F. Wang, G. Suarez, J. C. Sierra, A. A. Fadl, T. E. Erova, S. M. Foltz, B. K. Khajanchi, A. Silver, J. Graf, C. H. Schein, and A. K. Chopra. 2007. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila—part I. Microb. Pathog. 43127-146. [DOI] [PubMed] [Google Scholar]
- 38.Sierra, J. C., G. Suarez, J. Sha, S. M. Foltz, V. L. Popov, C. L. Galindo, H. R. Garner, and A. K. Chopra. 2007. Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila—part II. Microb. Pathog. 43147-160. [DOI] [PubMed] [Google Scholar]
- 39.Suarez, G., J. C. Sierra, J. Sha, S. Wang, T. E. Erova, A. A. Fadl, S. M. Foltz, A. J. Horneman, and A. K. Chopra. 2008. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanegas, G., W. Quinones, C. Carrasco-Lopez, J. L. Concepcion, F. Albericio, and L. Avilan. 2007. Enolase as a plasminogen binding protein in Leishmania mexicana. Parasitol. Res. 1011511-1516. [DOI] [PubMed] [Google Scholar]
- 41.Veiga-Malta, I., M. Duarte, M. Dinis, D. Tavares, A. Videira, and P. Ferreira. 2004. Enolase from Streptococcus sobrinus is an immunosuppressive protein. Cell Microbiol. 679-88. [DOI] [PubMed] [Google Scholar]
- 42.Wold, F. 1971. Enolase, p. 499-538. In P. D. Boyer (ed.), The enzymes. Academic Press, New York, NY.
- 43.Wu, C. J., J. J. Wu, J. J. Yan, H. C. Lee, N. Y. Lee, C. M. Chang, H. I. Shih, H. M. Wu, L. R. Wang, and W. C. Ko. 2007. Clinical significance and distribution of putative virulence markers of 116 consecutive clinical Aeromonas isolates in southern Taiwan. J. Infect. 54151-158. [DOI] [PubMed] [Google Scholar]