Abstract
Conjugative plasmids encode antibiotic resistance determinants or toxin genes in the anaerobic pathogen Clostridium perfringens. The paradigm conjugative plasmid in this bacterium is pCW3, a 47-kb tetracycline resistance plasmid that encodes the unique tcp transfer locus. The tcp locus consists of 11 genes, intP and tcpA-tcpJ, at least three of which, tcpA, tcpF, and tcpH, are essential for the conjugative transfer of pCW3. In this study we examined protein-protein interactions involving TcpA, the putative coupling protein. Use of a bacterial two-hybrid system identified interactions between TcpA and TcpC, TcpG, and TcpH. This analysis also demonstrated TcpA, TcpC, and TcpG self-interactions, which were confirmed by chemical cross-linking studies. Examination of a series of deletion and site-directed derivatives of TcpA identified the domains and motifs required for these interactions. Based on these results, we have constructed a model for this unique conjugative transfer apparatus.
Conjugation systems are important contributors to the dissemination of antibiotic resistance determinants and virulence factors. Extensive analysis of conjugative plasmids from gram-negative bacteria has led to the elucidation of a general mechanism of conjugative transfer (10, 22). In this process, the transferred DNA is processed by components of a relaxosome complex. Specifically, the DNA is nicked at the origin of transfer (oriT) by a relaxase, which remains covalently coupled to the transferred DNA strand. The single-stranded DNA complex then interacts with the coupling protein, a DNA-dependent ATPase that provides the energy to actively pump the DNA through the mating pair formation (Mpf) complex into the recipient cell (36). The coupling protein interacts with both DNA processing proteins and components of the Mpf complex (1, 4, 12, 35, 38). These interactions have been demonstrated using bacterial and yeast two-hybrid approaches as well as gel filtration, pull-down, and coimmunoprecipitation studies.
The mechanism of conjugative transfer has yet to be precisely determined for conjugative plasmids from gram-positive bacteria although bioinformatics analysis has identified similar gene arrangements and conservation of gene sequences within the transfer regions encoded on conjugative plasmids identified from strains of streptococcal, staphylococcal, enterococcal, and lactococcal origin (15). It was proposed that gram-positive and gram-negative conjugation systems utilize a similar transfer mechanism (15).
In the anaerobic pathogen Clostridium perfringens conjugative plasmids have been shown to encode antibiotic resistance genes or extracellular toxins (3, 8, 9, 18). Although the contribution of conjugation to disease dissemination has not been systematically evaluated, it has been proposed that transfer of the C. perfringens enterotoxin plasmid pCPF4969 to normal flora isolates of C. perfringens may contribute to the severity of disease caused by non-food-borne isolates of C. perfringens (9).
The prototype conjugative plasmid in C. perfringens is the 47-kb tetracycline resistance plasmid, pCW3. The complete sequence of pCW3 has been determined, and its unique replication protein and conjugation locus have been identified (8). Bioinformatics analysis of this C. perfringens tcp conjugation locus identified several proteins with limited similarity to proteins encoded within the transfer region of the conjugative transposon, Tn916 (8). The role of the tcp locus in the transfer of pCW3 has been confirmed by isolation of independent tcpA, tcpF, and tcpH mutants and subsequent complementation studies (8, 29). Since the region that encompasses the tcp locus is conserved in all conjugative plasmids from C. perfringens (2, 3, 8, 9, 18, 27) and since divergent tcpA homologues can complement a pCW3tcpA mutant (29), it appears that the conjugative transfer of both antibiotic resistance and toxin plasmids from this bacterium utilizes a common but poorly understood mechanism. Note that the C. perfringens tcp conjugation locus is different from the transfer regions of conjugative plasmids from other gram-positive bacteria.
We have recently shown that the essential conjugation protein TcpH, a putative membrane-associated Mpf complex component, is localized to the poles of C. perfringens cells, as is another essential conjugation protein, TcpF (37). TcpH has also been shown to interact with itself and with the pCW3-encoded TcpC protein (37). In this study we have focused on the essential conjugation protein TcpA. Since TcpA encodes an FtsK/SpoIIIE domain found in DNA translocases (8), it is proposed that TcpA is involved in the movement of DNA during conjugative transfer, fulfilling a role equivalent to that of coupling proteins in other conjugation systems. Like such proteins, TcpA encodes two N-terminal transmembrane domains (TMDs) and a C-terminal cytoplasmic region that contains three motifs predicted to be involved in ATP binding and hydrolysis (8). Our previous studies revealed that the conserved motifs, motif I (Walker A box), motif II (Walker B box), and motif III (RAAG box), are essential for the function of TcpA. The C-terminal 61 amino acids (aa), though not essential for TcpA function, were shown to be important for efficient transfer of pCW3, as were the putative TMDs (29).
To further investigate pCW3 transfer and the role of TcpA in this process, we have used bacterial two-hybrid analysis to examine protein-protein interactions involving TcpA. Using this system, interactions were observed between TcpA and itself, TcpC, TcpG, and TcpH. In addition, TcpC and TcpG were also found to self-interact. By combining these data with other data generated in this laboratory (37), we have constructed a model for the conjugative transfer of pCW3.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The Escherichia coli host strains used were DH5α (Life Technologies), BTH101 (20), and Rosetta 2 (Novagen), which were grown in 2× yeast extract-tryptone medium (26) or autoinduction medium (Invitrogen) supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml) as appropriate. Plasmids are listed in Table 1.
TABLE 1.
Plasmids
| Plasmid | Characteristic(s)a | Source or reference |
|---|---|---|
| pCW3 | Confers conjugative tetracycline resistance | 31 |
| pET28a+ | N-terminal His tag fusion expression vector; 5.3 kb; Kanr | Novagen |
| pGEX-4T | N-terminal GST fusion expression vector with thrombin cleavage site and additional linker; 5.0 kb; Apr | 16 |
| pKT25 | N-terminal T25 fusion vector for bacterial two-hybrid system; a derivative of pSU40; 3.4 kb; Kanr | 19 |
| pMAL-c2 | N-terminal MalE (MBP) fusion expression vector with an exact deletion of the malE signal sequence for cytoplasmic expression; 6.7 kb; Apr | New England BioLabs |
| pUT18C | N-terminal T18 fusion vector for bacterial two-hybrid system; a derivative of pUC19; 3.0 kb; Apr | 19 |
| pJIR3117 | pKT25 Ω(tcpH) | 37 |
| pJIR3137 | pUT18C Ω(tcpA1-530) | 37 |
| pJIR3144 | pKT25 Ω(tcpF) | 37 |
| pJIR3146 | pKT25 Ω(tcpG) | 37 |
| pJIR3168 | pKT25 Ω(tcpB) | 37 |
| pJIR3169 | pKT25 Ω(tcpD) | 37 |
| pJIR3170 | pKT25 Ω(tcpE) | 37 |
| pJIR3171 | pKT25 Ω(tcpI) | 37 |
| pJIR3175 | pKT25 Ω(tcpC) | 37 |
| pJIR3178 | pKT25 Ω(tcpJ) | 37 |
| pJIR3138 | pMAL-c2X (BamHI/SalI) harboring the product of PCR with JRP2602/JRP2603 (BamHI/SalI; 2.5 kb) and expressing MBP-TcpH | This study |
| pJIR3222 | pMAL-c2X (BamHI/SalI) harboring the product of PCR with JRP2790/JRP2791 (BamHI/SalI; 1.0 kb) and expressing MBP-TcpC | This study |
| pJIR3351 | pKT25 (Asp718/BamHI) harboring the product of PCR with JRP2617/JRP2618 (Asp718/BamHI; 1,593 bp) and expressing tcpA1-530 | This study |
| pJIR3353 | pUT18C (Asp718/BamHI) harboring the product of PCR with JRP2617/JRP2645 (Asp718/BamHI; 1,410 bp) and expressing tcpA1-469 | This study |
| pJIR3354 | pUT18C (Asp718/BamHI) harboring the product of PCR with JRP2617/JRP2824 (Asp718/BamHI; 1,098 bp) and expressing tcpA1-365 | This study |
| pJIR3355 | pUT18C (Asp718/BamHI) harboring the product of PCR with JRP2617/JRP2825 (Asp718/BamHI; 951 bp) and expressing tcpA1-316 | This study |
| pJIR3357 | pJIR3137 tcpA(K242A) | This study |
| pJIR3358 | pJIR3137 tcpA(D334A E345A) | This study |
| pJIR3359 | pJIR3137 tcpA(Q379A) | This study |
| pJIR3362 | pJIR3137 tcpAΔ46-104 | This study |
| pJIR3399 | pKT25 (Asp718/BamHI) harboring the product of PCR with JRP2870/JRP2871 (Asp718/BamHI; 801 bp) and expressing intP | This study |
| pJIR3556 | pT7Blue (EcoRV) harboring the product of PCR with JRP3589/JRP2414 (1,593 bp) and expressing tcpAΔ1-530 | This study |
| pJIR3558 | pGEX-4T (BamHI/XhoI) harboring a fragment of pJIR3556 (BamHI/XhoI; 1,593 bp) and expressing GST-TcpA1-530 | This study |
| pJIR3559 | pMAL-c2 (BamHI/XhoI) harboring a fragment of pJIR3556 (BamHI/XhoI; 1,593 bp) and expressing MBP-TcpA1-530 | This study |
| pJIR3424 | pET28a+ (SacI/XhoI) harboring the product of PCR with JRP3684/JRP3686 (SacI/XhoI; 921 bp) and expressing His-TcpGΔ1-28 | R. Bantwal, T. Bannam, and J. Rood, unpublished |
Apr, ampicillin resistance; Kanr, kanamycin resistance.
Molecular techniques.
E. coli plasmid DNA was isolated by an alkaline lysis method, according to the manufacturer's instructions (Qiagen). Purified C. perfringens DNA was obtained as described previously (30). PCR amplification used Taq DNA polymerase (Roche) and a 0.5 μM final concentration of each primer. Denaturation (94°C for 30 s), annealing (50°C for 30 s), and extension (72°C for 3 to 5 min) steps were carried out for 30 cycles. Sequence analysis of constructs was performed on an Applied Biosystems 3730S capillary sequencer. Sequence data were analyzed using Vector NTI (Invitrogen), in conjunction with the Sanger Institute freeware Artemis, release 6. For details of oligonucleotide primers, see Table S1 in the supplemental material.
Construction of vectors.
PCR products carrying the pCW3-derived wild-type tcpA gene were generated using the primer pair JRP2617 and JRP2618 and cloned into the bacterial two-hybrid vector pKT25, generating pJIR3351. Gene products expressing the TcpA deletion derivatives comprised of residues 1 to 469 (TcpA1-469), TcpA1-365, and TcpA1-316 were generated using the forward primer JRP2617 and the reverse primers JRP2645, JRP2824, JRP2825, respectively, and cloned into the bacterial two-hybrid vector pUT18c, generating pJIR3353, pJIR3354, and pJIR3355, respectively. Site-directed mutations or internal deletions of the tcpA gene were generated using a Quikchange (Stratagene) mutagenesis kit. The starting vector in these experiments was the pUT18c Ω(tcpA) vector pJIR3137. The primer pairs JRP3406 and JRP3407, JRP3408 and JRP3409, and JRP3410 and JRP3411 were used to construct vectors expressing the TcpA derivatives with the mutations K242A, D334A E335A, and Q379A, which had changes in motif I, II, or III, respectively. The TMD deletion derivative TcpAΔ46-104 was expressed from a vector generated by Quikchange mutagenesis of pJIR3137 using the primer pair JRP2831 and JRP2832. Primers JRP3589 and JRP2414 were used to generate the wild-type tcpA gene for cloning into the glutathione S-transferase (GST) expression vector pGEX-4T (GE Life Sciences) and the maltose binding protein (MBP) expression vector pMALc2 (New England Biolabs), generating pJIR3558 and pJIR3559, respectively.
Bacterial two-hybrid analysis.
The genes encoding the potential interacting proteins were cloned separately into the vectors pKT25 and pUT18C (19), and the resultant constructs were used to cotransform strain BTH101. For negative controls, strains expressing a fusion protein and carrying a corresponding empty vector (pKT25 or pUT18C, respectively), were used. Quantitative assessment of protein-protein interactions was performed as described previously (19, 37). Quantitative β-galactosidase assays (6) were performed either as described previously (37) or by using a modified method for high-throughput analysis in which cultures were grown and lysed in 96-well trays. β-Galactosidase activity (Miller units) was calculated from the initial rate of the reaction (6).
Protein expression and purification.
Derivatives of TcpA, TcpC, TcpG, and TcpH tagged with either GST (pGEX-4T), His6 (pET28a), or MBP (pMALc2) were overexpressed in Rosetta 2 cells cultured in autoinduction medium overnight at 28°C and then for an additional 24 h at 22°C. Each tagged protein was affinity purified with either GST or amylose resins. His6-tagged TcpG (His6-TcpG) was purified by the Monash University Protein Production Unit by high-throughput affinity chromatography using the ÄKTAxpress system.
Cross-linking and far-Western analysis.
Cross-linking experiments were performed using dimethyl 3,3′-dithiobispropionimidate-2HCl (DTBP) (Pierce Biotechnology, USA), a cleavable and bifunctional imidoester cross-linker, as described previously (37). Immunoblotting of cross-linked proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using the following antibodies: Penta-His (Qiagen), anti-MBP rabbit polyclonal (New England BioLabs), horseradish peroxidase-conjugated sheep anti-mouse (Chemicon) or horseradish peroxidase-conjugated sheep anti-rabbit (Chemicon). Anti-TcpC rabbit polyclonal antibodies were kindly provided by Radhika Bantwal. Far-Western analysis was performed using biotin-labeled GST-TcpA, with a biotin-labeled GST control, as previously described (37).
RESULTS
TcpA forms homooligomers.
Since coupling proteins and DNA translocases are known to form homohexamers (13, 23), it was predicted that interactions between TcpA monomers would be observed. To investigate protein-protein interactions in this study, we used a bacterial two-hybrid system based on the interaction of the T25 and T18 domains of adenylate cyclase from Bordetella pertussis (19). To determine if TcpA forms homooligomers, the wild-type tcpA gene was cloned in frame into vectors expressing the T25 (pKT25-tcpA) and T18 (tcpA-pUT18) domains. These vectors were then used to cotransform BTH101 cells, and the resultant strain was tested for expression of β-galactosidase (Fig. 1A). The results showed that significantly more β-galactosidase was detected in cells containing the complementary pair of TcpA fusion proteins than in the negative controls. This result indicated that the TcpA derivatives formed a homooligomer in the bacterial two-hybrid system.
FIG. 1.
Protein-protein interactions identified by bacterial two-hybrid analysis. (A) TcpA interacts with several proteins encoded by the tcp locus. E. coli BTH101 cells were cotransformed with TcpA-pUT18 and the pKT25 derivatives encoding IntP or TcpA to TcpJ. As negative controls, BTH101 cells were cotransformed with a vector expressing the test protein fused to either the T25 or T18 domain and a corresponding control plasmid (pUT18C or pKT25, respectively). Quantitative β-galactosidase assays were performed in triplicate, and the standard error is indicated. The activity (Miller units) is given for cells expressing Tcp proteins (striped boxes) and controls (checkered boxes). Statistical analysis was carried out using a Student's t test after analysis of variance. Statistical significance is indicated by an asterisk, where P is <0.05 by comparison to the negative control. (B) TcpC and TcpG self-interact. Bacterial two-hybrid analysis was also used to investigate TcpC self-interaction and TcpG self-interaction. The fusion derivative pair TcpC-pUT18 and pKT25-TcpC and the pair TcpG-pUT18 and pKT25-TcpG were also tested for β-galactosidase activity as for panel A.
Chemical cross-linking confirms TcpA self-interaction.
To confirm the TcpA self-interaction, a chemical cross-linking approach was used. Purified MBP-TcpA fusion protein was treated with increasing concentrations of the reversible imidoester cross-linker DTBP and examined by immunoblotting using MBP-specific antiserum. Under nonreducing conditions, even before the addition of DTBP, TcpA can be seen to form significant amounts of a very-high-molecular-weight complex (>250 kDa) (Fig. 2A). Upon the addition of increasing amounts of cross-linker, the ratio of TcpA monomer to complex increased. The cross-linking reaction mediated by DTBP was reversed under reducing conditions (24). At lower concentrations of DTBP, under reducing conditions, the TcpA complex was no longer visible. However, at higher levels of cross-linker, the reaction was no longer reversible. To control for the presence of the MBP tag, purified MBP was also subjected to chemical cross-linking under the same conditions (Fig. 2B) and shown not to self-interact. These results indicated that the formation of the higher-molecular-weight complex was enhanced by the presence of DTBP in a concentration-dependent manner. These data strongly suggested that the MBP-TcpA monomers had been cross-linked, providing physical evidence that TcpA forms homooligomers and confirming the conclusion drawn from bacterial two-hybrid analysis.
FIG. 2.
TcpA, TcpC, and TcpG form homooligomers in the presence of the chemical cross-linker DTBP. (A) Duplicate samples of purified MBP-TcpA were exposed to increasing concentrations of the chemical cross-linker DTBP. The resultant products were then separated on a 10% SDS-polyacrylamide gel in the presence and absence of DTT, which reduces the disulfide bonds created by the cross-linker. The samples were then transferred to nitrocellulose for Western blotting using commercial anti-MBP-specific antibodies. (B) To confirm that the MBP tag is not responsible for this interaction, purified MBP was also subjected to chemical cross-linking and probed using commercial anti-MBP antibodies. (C) Cross-linked samples of MBP-TcpC were detected with anti-TcpC-specific antibodies. (D) Cross-linked samples of His-TcpG were detected with commercial anti-His antibodies. Protein size standards (Std; in kDa) are indicated at left in each panel.
TcpA interacts with TcpC, TcpG, and TcpH.
Coupling proteins are known to interact with proteins involved in both DNA processing reactions and the Mpf complex export machinery (1, 4, 38). To identify additional protein-protein interactions involving TcpA, the bacterial two-hybrid analysis was expanded to investigate other proteins encoded on the tcp locus (Fig. 1A). BTH101 cells were cotransformed with vectors expressing TcpA fused to the T18 domain (TcpA-T18) and each of the T25-Tcp fusion proteins, IntP and TcpA to TcpJ. The results showed that significantly more β-galactosidase expression was detected in cells containing the fusion protein pairs T25-TcpC and TcpA-T18, T25-TcpG and TcpA-T18, and T25-TcpH and TcpA-T18 than in the corresponding negative controls. These results provide evidence that TcpA also interacts with TcpC, TcpG, and TcpH. No interactions were detected with the other proteins encoded by the tcp locus.
TcpC and TcpG also form homooligomers.
In addition to coupling proteins, other components of conjugation systems are known to form homooligomers (1, 25, 32, 38), including the pCW3 conjugation protein TcpH (37). Bacterial two-hybrid analysis also showed that TcpC and TcpG also self-interact (Fig. 1B). To confirm these interactions a chemical cross-linking approach was again used. Purified MBP-TcpC (ca. 80 kDa) and His-TcpG (ca. 33 kDa) fusion proteins were treated with increasing concentrations of DTBP and examined by immunoblotting using MBP- and His-specific antisera, respectively. Under nonreducing conditions, even before the addition of DTBP, TcpC formed moderate amounts of a multimer approximately 240 kDa in size (Fig. 2C), which corresponds to the expected size of a TcpC trimer. Upon the addition of increasing amounts of cross-linker, a much larger complex (>250 kDa) was formed. Both large-complex and trimer formation were reduced upon the addition of dithiothreitol (DTT) at lower levels of cross-linker. However, at the higher concentrations of cross-linker, the reaction was no longer completely reversible. These results indicate that the formation of the high-molecular-weight complex was dependent on the presence of DTBP in a concentration-dependent manner. Conversely, trimer formation was independent of cross-linker but still sensitive to the reducing agent DTT. Similarly, chemical cross-linking of TcpG resulted in the formation of a dimer, which could be reduced in the presence of DTT (Fig. 2D). These data provide physical evidence that both TcpC and TcpG form homooligomers and are consistent with data obtained using bacterial two-hybrid analysis.
Biochemical confirmation of TcpA-TcpH interaction.
Far-Western analysis was used to confirm TcpA-TcpH interactions as both proteins, individually, form very large homooligomers. In these experiments, partially purified MBP-TcpH was separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with biotin-labeled GST-TcpA. Interactions between TcpA and TcpH were detected with the antistreptavidin analog, ExtrAvidin (Sigma). To control for interactions involving the MBP and GST tags, MBP was resolved alongside the MBP-TcpH sample, and duplicate membranes were probed with biotin-labeled GST. The results (Fig. 3) indicate that GST-TcpA interacts with an oligomer of TcpH, as indicated by Western analysis using TcpH-specific antibodies, but does not interact with the monomer.
FIG. 3.
TcpA interacts with oligomeric MBP-TcpH. Partially purified MBP (lanes 1) and MBP-TcpH (lanes 2) were separated by 10% SDS-PAGE. (A) Coomassie blue staining of MBP and MBP-TcpH preparations. (B) Western blot probed with TcpH-specific antibodies. (C) Far-Western blot probed with biotin-labeled GST-TcpA. Protein-protein interaction between GST-TcpA and MBP-TcpH was detected using ExtrAvidin peroxidase (Sigma). (D) Far-Western blot containing MBP and MBP-TcpH preparations probed with biotin-labeled GST to control for GST-MBP interactions. Approximate protein molecular sizes are indicated (kDa). Arrows indicate the MBP-TcpH monomer (M) and oligomer (O).
The TMDs of TcpA and the Walker B motif are required for TcpA self-interaction.
Previously, a series of TcpA deletion and site-directed mutants were constructed and tested for their ability to complement a pCW3ΔtcpA mutant (29). Specifically, C-terminal deletion derivatives encompassing the last 61 aa of the 530-aa TcpA protein (TcpA1-469), then the RAAG motif (III) (TcpA1-365), and the Walker B box (motif II) (TcpA1-316) were constructed, as was an internal deletion derivative in which the region encoding the two N-terminal TMDs (TcpAΔ46-104) was removed. In addition, an N-terminal deletion derivative, TcpA358-530, was also constructed. The final derivatives of TcpA contained alanine substitutions in the Walker A motif (K242A), the Walker B motif (D334A E345A), or the RAAG motif (Q379A) (29).
To determine if loss of biological function could be attributed to decreased protein-protein interactions, these same derivatives (Fig. 4A) were constructed in the bacterial two-hybrid vector pUT18c and tested for their ability to interact with the wild-type T25-TcpA protein. The results showed that the C-terminal 61 aa of TcpA were not required for TcpA self-interaction since the β-galactosidase levels detected in cells containing this fusion protein (TcpA1-469-T18) were similar to levels in the wild-type protein (TcpA1-530-T18) (Fig. 4B). The deletion derivatives TcpA1-365-T18 and TcpA1-316-T18 were not able to interact with the wild-type protein; these cells had β-galactosidase levels indistinguishable from the negative controls. Since the region between aa 365 and 469 appeared to be required for TcpA self-interaction, a TcpA358-530 derivative was also tested in the bacterial two-hybrid system. It did not interact with full-length TcpA. Deletion of the putative TMDs of TcpA (TcpAΔ46-104) significantly reduced β-galactosidase activity, indicating that this region is also important for TcpA self-interaction. Finally, single amino acid substitutions within motif I (K242A) and motif III (Q379A) had no effect on the ability of TcpA to self-interact, but a substitution within motif II (D334A E335A) abolished TcpA self-interaction.
FIG. 4.
The role of TcpA regions in protein-protein interactions. The bacterial two-hybrid system was used to test the ability of a series of deletion and site-directed mutants of TcpA-T18 (A) to interact with T25-TcpA. Previously, plasmids expressing these derivatives were tested for their ability to complement a pCW3ΔtcpA mutant (29). Complementation efficiency is summarized as follows: ++, full complementation compared to the wild-type gene; +, reduced transfer efficiency; −, no transfer of pCW3; ND, not determined. The location of each site-directed substitution is indicated by X. (B) Domains of TcpA required for TcpA self-interaction. E. coli BTH101 cells were cotransformed with pKT25-tcpA and the various tcpA-pUT18 derivatives and the control plasmid pUT18c. Quantitative β-galactosidase assays were performed in triplicate, and the standard error is indicated. Statistical analysis was carried out using a Student's t test after analysis of variance. Statistical significance is indicated by an asterisk where P is <0.05 by comparison to the wild-type TcpA control. The domains of TcpA required for TcpA-TcpC interaction (C), TcpA-TcpG interaction (D), and TcpA-TcpH1-581 interaction (E) were determined in a similar manner.
The C-terminal 61 aa of TcpA are required for interactions with TcpC.
To determine the regions of TcpA involved in interactions with TcpC, the same TcpA derivatives were tested for their ability to interact with the wild-type T25-TcpC protein in the bacterial two-hybrid system. The results showed that the C-terminal 61 aa of TcpA were required for TcpA-TcpC interaction since β-galactosidase activity was significantly reduced compared to the wild-type TcpA protein (Fig. 4C). The deletion derivatives TcpA1-365-T18 and TcpA1-316-T18 were also unable to interact with the wild-type TcpC protein. Single amino acid substitutions within motif I and motif III had no effect on the ability of TcpA to interact with TcpC, but amino acid substitutions within motif II abolished the TcpA-TcpC interaction. Deletion of the putative TMDs of TcpA also resulted in significantly reduced β-galactosidase activity, indicating that this region is important for TcpA-TcpC interaction.
The TMDs of TcpA are required for TcpA-TcpG and TcpA-TcpH interactions.
The domains of TcpA involved in interactions with TcpG and TcpH were investigated in a similar manner. In these experiments, a smaller derivative of TcpH, TcpH1-581, was used as it had previously been shown to interact with TcpA (37). The C-terminal 61 aa of TcpA were not required for TcpA-TcpG (Fig. 4D) or TcpA-TcpH (Fig. 4E) interactions as wild-type levels of β-galactosidase activity were detected. Other deletion derivatives of TcpA were not able to interact with the wild-type TcpG or TcpH protein, and amino acid substitutions within motif II, but not motif I or III, abolished both TcpA-TcpG and TcpA-TcpH interactions. Deletion of the putative TMDs of TcpA also eliminated TcpA-TcpG and TcpA-TcpH interactions.
DISCUSSION
In this study we have investigated protein-protein interactions involving the essential pCW3 conjugation protein, TcpA. TcpA encodes domains found in DNA translocases and coupling proteins (8, 29), proteins that are known to form homooligomers (13, 23) and to interact with components of the Mpf complex (1, 4, 12, 36, 38). We have now shown that TcpA interacts with itself and with other pCW3 conjugation proteins such as TcpC, TcpG, and the essential transmembrane component TcpH. These findings are all consistent with the postulated role of TcpA as a coupling protein that links the transferred DNA to the Mpf complex.
Homooligomerization of TcpA was demonstrated in both bacterial two-hybrid and cross-linking experiments. Oligomerization of TcpA was observed even in the absence of cross-linker under nonreducing conditions. Furthermore, this SDS-stable interaction could also be reduced in the presence of DTT, indicating that disulfide bonds may be involved. Although it is postulated that TcpA will form hexamers similar to other coupling proteins and FtsK-like DNA translocases, the TcpA oligomers (>200 kDa) could not be resolved by either SDS-PAGE or size exclusion chromatography (data not shown).
Using the bacterial two-hybrid system, we identified regions of TcpA that were essential for homooligomerization. Since TcpA1-469 can interact in this system, unlike TcpA1-365, oligomerization of TcpA appears to be dependent on aa 365 to 469. This region encompasses the RAAG motif (III) that, based on comparative analysis of the structural similarity between TrwB and F1-ATPase, encodes a putative arginine finger (14). In AAA+ ATPases such as F1-ATPase, the arginine finger is thought to interact with the C-terminal phosphate of an ATP molecule that is bound to the preceding protomer in the hexameric ring, affecting oligomerization, ATP recognition, and ATP hydrolysis (11, 28). Deletion of this putative arginine finger may explain the loss of detectable self-interaction with TcpA1-365.
The conserved glutamine of the RAAG motif [TcpA(Q379A)] was also targeted in this study. This residue is predicted to be required for sensing of the triphosphate moiety of ATP, triggering its hydrolysis (21, 34). Although this amino acid has been shown to be important for the function of TcpA (29), we have now shown that it is not involved in protein-protein interactions. This finding is consistent with its role in ATP hydrolysis.
Targeting of the second conserved motif, which encompasses the Walker B box, resulted in the loss of TcpA self-interaction. Functional analysis of the Walker B motif in TcpA (29) and the TraG coupling protein from RP4 (7) has indicated that this motif is essential for conjugative transfer. The aspartate residue targeted for substitution is predicted to be required for the coordination of the Mg2+ ion involved in ATP hydrolysis, whereas the neighboring glutamate primes a water molecule for a nucleophilic attack on the γ-phosphate of the bound ATP (34). Since this region is also important for oligomerization, it is unclear if TcpA(D334A E345A) is nonfunctional due to its putative role in ATP hydrolysis or its inability to oligomerize. Furthermore, these phenotypic changes may also be the result of incorrect folding of the TcpA(D334A E345A) fusion protein. Mutagenesis of the Walker A motif (I) of TcpA [TcpA(K242A)] did not alter TcpA oligomerization, which is consistent with its putative role in ATP binding and hydrolysis.
The region of TcpA encoding the two predicted TMDs, aa 46 to 104, was required for efficient protein-protein interactions between TcpA monomers. Reduced oligomerization may account for the significant reduction in transfer frequency observed when this derivative was tested for conjugative efficiency in the pCW3ΔtcpA mutant (29). This result is consistent with the role of the TMDs of the coupling protein, TraG, which are also required for oligomerization in vitro, as well as protein-protein interactions with relaxosome components (33). By contrast, the TMDs of FtsK are not essential for its ATPase activity, multimer formation, or DNA translocase activity (5) although FtsK is significantly larger and therefore may have additional structural features that support protein-protein interactions.
TcpA was also found to interact with TcpC, TcpG, and TcpH using the bacterial two-hybrid system. TcpH is an essential membrane-located transfer protein previously shown to form homooligomers and predicted to form a key component of the Mpf complex (37). TcpH encodes an essential VirB6 domain as well as ATP binding motifs and has been localized to the poles of C. perfringens using fluorescence microscopy (37). The TcpA-TcpH interaction described here appears to occur between oligomeric forms of TcpA and TcpH, as indicated by both the far-Western analysis and the bacterial two-hybrid analysis. Derivatives of TcpA that had significantly lower levels of self-interaction in the bacterial two-hybrid system were also unable to interact with TcpH. In RP4, deletion of the equivalent TraG TMDs results in the loss of TraG oligomerization and interaction with the relaxase, TraI (33), though it is not known if the TraG-TraI interaction specifically requires the TMDs or oligomeric TraG. In the F plasmid coupling protein, purified TraD has been shown to form dimers in the absence of the other F Tra proteins; however, the coexpression of additional Tra proteins resulted in the formation of large membrane-associated complexes (17).
TcpC is most closely related to open reading frame 13 (Orf13) from the conjugative transposon Tn916 (8). Although there are no published functional data on Orf13, preliminary studies in this laboratory suggest that TcpC is involved in the conjugative transfer of pCW3 (R. Bantwal, T. Bannam, and J. Rood, unpublished results). In this study, the efficiency of TcpA-TcpC interactions was reduced upon the deletion of the C-terminal 61 aa of TcpA as well as the deletion of the TMDs. Although the C-terminal region of TcpA is not absolutely required for transfer of pCW3, deletion of this region resulted in a reduction in transfer efficiency of 3 orders of magnitude (29). Furthermore, the deletion of the C-terminal region does not affect interactions between TcpA and itself, TcpG, or TcpH, which suggests that the TcpA1-469 molecule is correctly folded. This region of TcpA, which is required for both efficient transfer and TcpA-TcpC interactions, clearly plays an important role in the conjugation process.
TcpA also interacts with TcpG, a putative peptidoglycan hydrolase (8). Peptidoglycan hydrolases, such as the lytic transglycosylase encoded by Orf7 from pIP501, are predicted to be important for the localized opening of the cell wall that is required for the assembly of the Mpf complex (1). In pIP501, Orf7 forms a dimer and also interacts with Orf10, a putative coupling protein (1). Here, we have shown that TcpG forms dimers that are able to interact with the putative coupling protein TcpA. Although TcpG belongs to a different family of peptidoglycan hydrolases and, as such, is predicted to target different linkages within the peptidoglycan layer, TcpG may play a similar functional role in pCW3 transfer.
Attempts to confirm TcpA-TcpC and TcpA-TcpG interactions using biochemical methods were not successful. Although initial attempts to identify interactions between GST-TcpA and MBP-TcpC using chemical cross-linking approaches looked promising, we were unable to distinguish between potential complexes involving homo- and heterooligomers, primarily because of the large sizes of these complexes (data not shown). Further attempts to confirm TcpA-TcpC interactions by cross-linking and far-Western analysis using a smaller derivative of TcpC in which the TMDs had been deleted (His-TcpCΔ1-98) were also unsuccessful although the role of the TMDs of TcpC in this interaction are not known. Cross-linking studies using both MBP-TcpA and GST-TcpA with His-TcpG were hindered by nonspecific interactions with the commercial anti-His antibodies. Far-Western analysis using combinations of purified MBP-TcpA, GST-TcpA, and His-TcpG both on nitrocellulose membranes and as biotin-labeled probes failed to identify reproducible TcpA-TcpG interactions. If the TcpA-TcpG interaction also occurs between oligomers, this may explain the difficulties we had confirming these interactions.
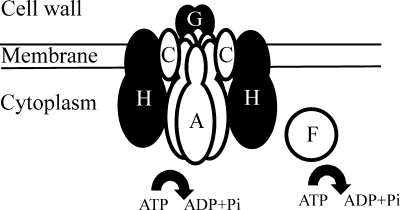
Finally, the results obtained here and in previous studies (37) have allowed us to develop a model for the interaction of the proteins involved in the conjugative transfer of pCW3. In this model (Fig. 5), oligomeric TcpH, a putative membrane-spanning Mpf protein is embedded in the cell membrane at the poles (37) and forms a key component of the mating channel. TcpA, the putative coupling protein, interacts with oligomeric TcpH. Both TcpA and TcpH also interact with the accessory protein TcpC (37), which has a putative N-terminal TMD, indicating that it may also be membrane associated (8). The interaction between TcpA and TcpC is dependent on the 61 C-terminal amino acids of TcpA, a region that is also important for conjugative transfer of pCW3. TcpA, but not TcpH, also interacts with TcpG, a putative cell wall-associated peptidoglycan hydrolase. Finally, immunofluorescence has been used to colocalize TcpF, a putative ATPase, with TcpH at the poles of C. perfringens cells (37) although we have no evidence for TcpH-TcpF interactions.
FIG. 5.
Model of the conjugative transfer apparatus of pCW3. The diagram represents a cross-section of the Tcp conjugation apparatus, which is present at the poles of C. perfringens cells (37). The relative locations of TcpA, TcpC, TcpG, TcpF, and TcpH are indicated. Proteins that have been shown to interact are shown here as overlapping. With the exception of TcpH, protein localization is based on computer predictions (37). The curved arrows indicate postulated ATP hydrolysis. Note that although not represented here, the TcpH and TcpC proteins have been shown to form homooligomers.
In this study we have identified interactions between several Tcp conjugation proteins that form the unique pCW3 Mpf apparatus. We anticipate that the resultant model will provide a valuable framework upon which further interactions can be mapped and that it will aid in the determination of the mechanism of pCW3 transfer in C. perfringens, a clinically important pathogen.
Supplementary Material
Acknowledgments
We thank Daniel Ladant for kindly providing his bacterial two-hybrid assay system, Noelene Quinsey for large-scale protein purification, Radhika Bantwal for anti-TcpC antibodies and helpful discussions, and Jason Steen for helpful discussions.
This research was supported by a grant from the Australian Research Council to the ARC Centre of Excellence in Structural and Functional Microbial Genomics and by grant AI056177-03 from the U.S. National Institute of Allergy and Infectious Diseases. Jennifer A. Steen (née Parsons) was the recipient of a postgraduate scholarship awarded by the ARC Centre of Excellence and the Department of Microbiology. W. L. Teng was the recipient of a Monash Graduate Scholarship.
Footnotes
Published ahead of print on 27 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abajy, M. Y., J. Kopec, K. Schiwon, M. Burzynski, M. Doring, C. Bohn, and E. Grohmann. 2007. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria. J. Bacteriol. 1892487-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham, L. J., and J. I. Rood. 1985. Molecular analysis of transferable tetracycline resistance plasmids from Clostridium perfringens. J. Bacteriol. 161636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abraham, L. J., A. J. Wales, and J. I. Rood. 1985. Worldwide distribution of the conjugative Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid 1437-46. [DOI] [PubMed] [Google Scholar]
- 4.Atmakuri, K., E. Cascales, and P. J. Christie. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 541199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aussel, L., F.-X. Barre, M. Aroyo, A. Stasiak, A. Z. Stasiak, and D. Sherratt. 2002. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108195-205. [DOI] [PubMed] [Google Scholar]
- 6.Baker, S. J., C. Daniels, and R. Morona. 1997. PhoP/Q. regulated genes in Salmonella typhi: identification of melittin sensitive mutants. Microb. Pathog. 22165-179. [DOI] [PubMed] [Google Scholar]
- 7.Balzer, D., W. Pansegrau, and E. Lanka. 1994. Essential motifs of relaxase (TraI) and TraG proteins involved in conjugative transfer of plasmid RP4. J. Bacteriol. 1764285-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannam, T. L., W. L. Teng, D. Bulach, D. Lyras, and J. I. Rood. 2006. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J. Bacteriol. 1884942-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brynestad, S., M. R. Sarker, B. A. McClane, P. E. Granum, and J. I. Rood. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 693483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie, P. J. 2004. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim. Biophys. Acta 1694219-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey, M. J., D. Jeruzalmi, J. Kuriyan, and M. O'Donnell. 2002. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 3826-835. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Lopez, R., M. P. Garcillan-Barcia, C. Revilla, M. Lazaro, L. Vielva, and F. de la Cruz. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30942-966. [DOI] [PubMed] [Google Scholar]
- 13.Gomis-Ruth, F. X., and M. Coll. 2001. Structure of TrwB, a gatekeeper in bacterial conjugation. Int. J. Biochem. Cell Biol. 33839-843. [DOI] [PubMed] [Google Scholar]
- 14.Gomis-Ruth, F. X., G. Moncalian, F. de la Cruz, and M. Coll. 2002. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein. Detailed structural features and mapping of the active site cleft. J. Biol. Chem. 2777556-7566. [DOI] [PubMed] [Google Scholar]
- 15.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, K., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192262-267. [DOI] [PubMed] [Google Scholar]
- 17.Haft, R. J. F., E. G. Gachelet, T. Nguyen, L. Toussaint, D. Chivian, and B. Traxler. 2007. In vivo oligomerization of the F conjugative coupling protein TraD. J. Bacteriol. 1896626-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes, M. L., R. Poon, V. Adams, S. Sayeed, J. Saputo, F. A. Uzal, B. A. McClane, and J. I. Rood. 2007. Epsilon-toxin plasmids of Clostridium perfringens type D are conjugative. J. Bacteriol. 1897531-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karimova, G., A. Ullmann, and D. Ladant. 2000. A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol. 32859-73. [DOI] [PubMed] [Google Scholar]
- 20.Karimova, G., A. Ullmann, and D. Ladant. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 373-82. [PubMed] [Google Scholar]
- 21.Kelley, J. A., and K. L. Knight. 1997. Allosteric regulation of RecA protein function is mediated by Gln194. J. Biol. Chem. 27225778-25782. [DOI] [PubMed] [Google Scholar]
- 22.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 2241-15. [DOI] [PubMed] [Google Scholar]
- 23.Massey, T. H., C. P. Mercogliano, J. Yates, D. J. Sherratt, and J. Lowe. 2006. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell 23457-469. [DOI] [PubMed] [Google Scholar]
- 24.Mattson, G., E. Conklin, S. Desai, G. Nielander, M. D. Savage, and S. Morgensen. 1993. A practical approach to crosslinking. Mol. Biol. Rep. 17167-183. [DOI] [PubMed] [Google Scholar]
- 25.Middleton, R., K. Sjolander, N. Krishnamurthy, J. Foley, and P. Zambryski. 2005. Predicted hexameric structure of the Agrobacterium VirB4 C terminus suggests VirB4 acts as a docking site during type IV secretion. Proc. Natl. Acad. Sci. USA 1021685-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Miyamoto, K., D. J. Fisher, J. Li, S. Sayeed, S. Akimoto, and B. A. McClane. 2006. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J. Bacteriol. 1881585-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogura, T., S. W. Whiteheart, and A. J. Wilkinson. 2004. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 146106-112. [DOI] [PubMed] [Google Scholar]
- 29.Parsons, J. A., T. L. Bannam, R. J. Devenish, and J. I. Rood. 2007. TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens. J. Bacteriol. 1897782-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rood, J. I., E. A. Maher, E. B. Somers, E. Campos, and C. L. Duncan. 1978. Isolation and characterization of multiply antibiotic-resistant Clostridium perfringens strains of porcine origin. Antimicrob. Agents Chemother. 13871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rood, J. I., V. N. Scott, and C. L. Duncan. 1978. Identification of a transferable tetracycline resistance plasmid (pCW3) from Clostridium perfringens. Plasmid 1563-570. [DOI] [PubMed] [Google Scholar]
- 32.Savvides, S. N., H.-J. Yeo, M. R. Beck, F. Blaesing, R. Lurz, E. Lanka, R. Buhrdorf, W. Fischer, R. Haas, and G. Waksman. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 221969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroder, G., and E. Lanka. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J. Bacteriol. 1854371-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Story, R. M., I. T. Weber, and T. A. Steitz. 1992. The structure of the E. coli recA protein monomer and polymer. Nature 355318-325. [DOI] [PubMed] [Google Scholar]
- 35.Tato, I., I. Matilla, I. Arechaga, S. Zunzunegui, F. de la Cruz, and E. Cabezon. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J. Biol. Chem. 28225569-25576. [DOI] [PubMed] [Google Scholar]
- 36.Tato, I., S. Zunzunegui, F. de la Cruz, and E. Cabezon. 2005. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc. Natl. Acad. Sci. USA 1028156-8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng, W. L., T. L. Bannam, J. A. Parsons, and J. I. Rood. 2008. Functional characterization and localization of the TcpH conjugation protein from Clostridium perfringens. J. Bacteriol. 1905075-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 9911493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.