Abstract
While Eukarya, Bacteria, and Archaea are all capable of protein N glycosylation, the archaeal version of this posttranslational modification is the least understood. To redress this imbalance, recent studies of the halophilic archaeon Haloferax volcanii have identified a gene cluster encoding the Agl proteins involved in the assembly and attachment of a pentasaccharide to select Asn residues of the surface layer glycoprotein in this species. However, because the automated tools used for rapid annotation of genome sequences, including that of H. volcanii, are not always accurate, a reannotation of the agl cluster was undertaken in order to discover genes not previously recognized. In the present report, reanalysis of the gene cluster that includes aglB, aglE, aglF, aglG, aglI, and aglJ, which are known components of the H. volcanii protein N-glycosylation machinery, was undertaken. Using computer-based tools or visual inspection, together with transcriptional analysis and protein expression approaches, genes encoding AglP, AglQ, and AglR are now described.
Although the ability of Archaea to N glycosylate selected proteins has long been known (16), genes implicated in the archaeal version of this posttranslational modification have only recently been described (22). In the halophilic archaeon Haloferax volcanii, agl (archaeal glycosylation) genes, i.e., aglB, aglD, aglE, aglF, aglI, and aglJ, which were first identified through the homologies of their products to known elements of the eukaryal or bacterial N-glycosylation pathways, have been shown to participate in the assembly and attachment of a pentasaccharide decorating at least two Asn residues of the surface layer (S-layer) glycoprotein (1-3, 23; M. Abu-Qarn et al., unpublished data). However, except for the oligosaccharide transferase, AglB (2), little is known about the functions of the Agl proteins identified. Indeed, it is likely that not all components of the H. volcanii N-glycosylation pathway have been identified.
Recently, an annotated version of the H. volcanii genome has become available (20; http://archaea.ucsc.edu/cgi-bin/hgGateway?db=haloVolc1). Accordingly, in an effort to identify novel components of the H. volcanii N-glycosylation process not identified through earlier homology-based searches, open reading frames (ORFs) found adjacent to those genes known to participate in this posttranslational modification were investigated. Such an approach had earlier served to identify aglG, which is situated between the oligosaccharide transferase-encoding aglB and the aglI genes and is a homologue of Campylobacter jejuni pglI, the product of which adds a glucose branch to the lipid-linked polysaccharide structure that is ultimately attached to protein targets in this bacterium (15). Encouraged by this finding, in the present study we have examined the region of the H. volcanii genome between aglJ (HVO_1517) and aglB (HVO_1530). This region also includes aglG (HVO_1529), aglI (HVO_1528), and aglF (HVO_1527). Analysis of the genome region between aglJ and aglF in the hope of identifying novel N-glycosylation genes is hampered by the fact that the automated annotation of the H. volcanii genome currently available inadvertently includes at least one error, which became evident only upon subsequent manual annotation efforts. In those studies, it was shown that aglE, encoding a protein involved in adding the fourth subunit of the pentasaccharide decorating the H. volcanii S-layer glycoprotein, corresponds to the 5′ portion of HVO_1523 as well as most of the nonannotated region between HVO_1523 and HVO_1524 (3).
In the present report, computer-based approaches and visual inspection were employed to reannotate ORFs in the stretch of the genome between nucleotide 1382311, i.e., the start of aglJ, and nucleotide 1398898, i.e., the start of aglB (2), with the aim of identifying novel H. volcanii N-glycosylation pathway genes, including those not previously identified as a result of misannoation. Such reassessment of the H. volcanii agl gene cluster, followed by studies conducted at the RNA and protein levels, has served to reveal two novel gene sequences, annotated as aglQ and aglR, and to confirm that aglP is a true gene, in agreement with earlier results (12). The proximity and cotranscription of these genes with sequences known to be involved in N glycosylation point to aglP, aglQ, and aglR as also participating in this posttranslational modification.
MATERIALS AND METHODS
Cell growth.
H. volcanii cells were grown in complete medium containing 3.4 M NaCl, 0.15 M MgSO4·7H2O, 1 mM MnCl2, 4 mM KCl, 3 mM CaCl2, 0.3% (wt/vol) yeast extract, 0.5% (wt/vol) tryptone, and 50 mM Tris-HCl (pH 7.2) at 40°C (17).
RT-PCR.
Reverse transcriptase PCR (RT-PCR) was performed as described previously (1). Briefly, specific forward and reverse oligonucleotide primers were designed for each H. volcanii gene under consideration (see Table S1 in the supplemental material). RNA isolation was carried out using an Easy-spin RNA extraction kit (Intron Biotechnology, Kyungki-Do, Korea) according to the manufacturer's instructions. The RNA concentration was determined spectrophotometrically. After contaminating DNA was eliminated with a DNAFree kit (Ambion, Austin, TX), single-stranded cDNA was prepared for each sequence from the corresponding RNA (2 μg) using random hexamers (150 ng) in a SuperScript III first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA). The cDNA was then used for PCR amplification, together with appropriate forward and reverse primer pairs. cDNA amplification was monitored by electrophoresis in 1% agarose gels. The sequences of the PCR products were determined to confirm their identity. In control experiments designed to exclude any contribution from contaminating DNA, PCR amplification was performed on total RNA prior to cDNA preparation.
Generation and detection of GFP fusion proteins.
To generate constructs encoding the putative protein products of the ORF of interest fused to green fluorescent protein (GFP), DNA sequences corresponding to HVO_1518, HVO_1521, HVO_1522, HVO_1526, ORF 2, ORF 3, ORF 5, ORF 6, ORF 9, ManHv1, or ManHv2, together with the 200-bp region located directly preceding the 5′ end of each sequence, were PCR amplified using forward and reverse primers (see Table S1 in the supplemental material) designed to introduce XbaI and BglII restriction sites at the 5′ and 3′ ends of the fragments, respectively. The amplified fragments were digested with XbaI and BglII, purified by electrophoresis in 1% agarose gels, and ligated into plasmid pJAM1020 (19), which was predigested with the appropriate restriction enzymes to yield plasmids encoding the various fusion proteins, with the promoter normally found in the plasmid being replaced by the 200-bp region preceding the 5′ end of each ORF being considered.
To detect the expression of GFP fusion proteins, proteins were electrotransferred from sodium dodecyl sulfate-polyacrylamide gels to nitrocellulose membranes (0.45 μm; Schleicher & Schuell, Dassel, Germany) and incubated with anti-GFP antibodies (1:1,000; Roche) and then with horseradish peroxidase-conjugated goat anti-mouse antibodies (1:2,500; KPL, Gaithersburg, MD). Detection of antibody binding was achieved using ECL Western blotting detection reagent (Amersham, Bucks., United Kingdom).
Nucleotide sequence accession numbers.
The sequences of H. volcanii aglP, aglQ, and aglR have been deposited in the EMBL/GenBank/DDBJ databases and assigned accession numbers FM955369, FM955370, and FM955371, respectively.
RESULTS
Redefinition of ORFs contained within the H. volcanii genome region studied.
The version of the H. volcanii genome currently available at the USCS Archaeal Genome Browser (20; http://archaea.ucsc.edu/) includes sequence annotations based largely on in silico predictions using the Glimmer (Gene Locator and Interpolated Markov ModelER) system, as performed at The Institute for Genome Research (TIGR). However, given our earlier finding that such computer-based annotation had failed to recognize a sequence subsequently identified through visual inspection and experimentally verified as participating in N glycosylation, i.e., aglE (3), we chose to reannotate that region of the genome between nucleotide 1382311, i.e., the start of aglJ, and nucleotide 1398898, i.e., the start of aglB. Accordingly, in the hope of finding novel agl genes involved in N glycosylation, annotation of this segment of the H. volcanii genome was repeated, this time using the ORF Finder analysis tool available at NCBI (http://www.ncbi.nlm.nih.gov/projects/gorf/).
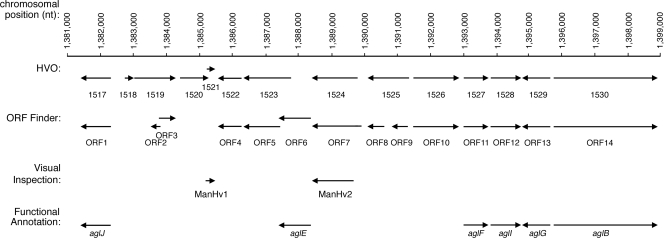
In several instances, as reflected in Fig. 1 and Table 1, the two algorithms identified identical ORFs. Specifically, the originally annotated HVO_1517, HVO_1522, HVO_1527, HVO_1528, HVO_1529, and HVO_1530 sequences correspond to the ORF 1, ORF 4, ORF 11, ORF 12, ORF 13, and ORF 14 sequences identified by ORF Finder, respectively. Of these sequences, HVO_1517 (ORF 1) corresponds to aglJ (M. Abu-Qarn et al., unpublished data); HVO_1527 (ORF 11), HVO_1528 (ORF 12), and HVO_1529 (ORF 13) correspond to aglF, aglI, and aglG, respectively (23). HVO_1530 (ORF 14) corresponds to aglB (2). In contrast, although each algorithm reported the presence of nine ORFs between nucleotides 1382311, i.e., the start of aglJ, and 1393000, i.e., the start of aglF, the two algorithms did not concur on the boundaries of the ORFs in this region, apart from the stretch between nucleotides 1385553 and 1386272, simultaneously annotated as HVO_1522 and ORF 4. In addition, HVO_1526 and ORF 10 can be distinguished by the presence of an additional ATG codon at the start of ORF 10 not found in HVO_1526, while HVO_1524 and ORF 7 overlap apart from an additional 51 nucleotides found at the start of ORF 7. Moreover, ORF 3 and ORF 8 differ by only 12 nucleotides, 11 of which do translate into differences at the deduced amino acid level. The remaining sequences of the original annotation, i.e., HVO_1518, HVO_1519, HVO_1520, HVO_1521, HVO_1523, and HVO_1525 are not repeated in the set comprising ORF 2, ORF 3, ORF 5, ORF 6, ORF 8, and ORF 9. Indeed, the two sets of ORFs can be further distinguished by the differential assignment of set members to the positive and negative strands of the genome (Table 1). Finally, ORF 6 has been shown to correspond to aglE (3).
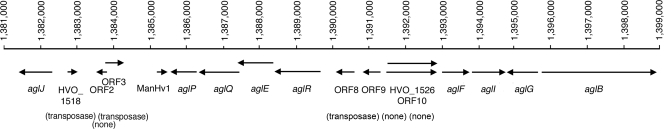
FIG. 1.
Schematic depiction of the agl cluster of the annotated H. volcanii genome. Shown are annotations automatically generated by the Glimmer (http://archaea.ucsc.edu/cgi-bin/hgGateway?db=haloVolc1; listed as HVO numbers) or ORF Finder (listed as ORF numbers) algorithm, as well as by visual inspection. Functional annotations are reflected as agl gene names.
TABLE 1.
ORFs recognized in the H. volcanii agl cluster
| Algorithm | Chromosomal position (strand) | ORFa | agl gene |
|---|---|---|---|
| USCS Archaeal | 1381400-1382311 (−) | HVO_1517 | aglJ |
| Genome | 1382719-1383006 (+) | HVO_1518 | |
| Browser | 1383007-1384200 (+) | HVO_1519 | |
| 1384455-1385342 (+) | HVO_1520 | ||
| 1385335-1385466 (+) | HVO_1521 | ||
| 1385553-1386272 (−) | HVO_1522 | ||
| 1386375-1387784 (−) | HVO_1523 | ||
| 1388454-1389884 (−) | HVO_1524 | ||
| 1390102-1391292 (−) | HVO_1525 | ||
| 1391458-1392831 (+) | HVO_1526 | ||
| 1393000-1393731 (+) | HVO_1527 | aglF | |
| 1393780-1394667 (+) | HVO_1528 | aglI | |
| 1394676-1395617 (+) | HVO_1529 | aglG | |
| 1395734-1398898 (+) | HVO_1530 | aglB | |
| NCBI ORF | 1381400-1382311 (−) | ORF 1 | aglJ |
| Finder | 1383421-1383831 (+) | ORF 2 | |
| 1383667-1384200 (+) | ORF 3 | ||
| 1385553-1386272 (−) | ORF 4 | ||
| 1386375-1387490 (−) | ORF 5 | ||
| 1387487-1388401 (−) | ORF 6 | aglE | |
| 1388454-1389935 (−) | ORF 7 | ||
| 1390102-1390635 (−) | ORF 8 | ||
| 1390821-1391270 (−) | ORF 9 | ||
| 1391455-1392831 (+) | ORF 10 | ||
| 1393000-1393731 (+) | ORF 11 | aglF | |
| 1393780-1394667 (+) | ORF 12 | aglI | |
| 1394676-1395617 (−) | ORF 13 | aglG | |
| 1395734-1398898 (+) | ORF 14 | aglB | |
| Manual | 1385200-1385463 (+) | ManHv1 | |
| annotation | 1388454-1389689 (−) | ManHv2 |
HVO_1517 = ORF 1, HVO_1522 = ORF 4, HVO_1527 = ORF 11, HVO_1528 = ORF 12, HVO_1529 = ORF 13, and HVO_1530 = ORF 14.
Transcriptional analysis of identified ORFs.
Experiments were next undertaken to determine which of those ORFs not previously experimentally verified as encoding Agl proteins (i.e., HVO_1518-HVO_1526, ORFs 2 to 5, and ORFs 7 to 10) indeed correspond to true genes. Since transcription of a given sequence is indicative of that sequence encoding a true protein, RT-PCR was performed using primers directed at the start and end of the each of the ORFs under consideration, in addition to HVO_1527 (ORF 11, i.e., aglF), which served as a positive control. In such experiments, cDNA generated from RNA collected from cells grown to mid-exponential phase in complete medium served as the PCR template. As reflected in Fig. 2, PCR products were obtained for HVO_1518, HVO_1521, HVO_1522 (ORF 4), HVO_1526, ORFs 2 to 6, and ORFs 8 to 10, as well as for HVO_1527 (ORF 11 [aglF]). Indeed, although the sequences of ORF 5 (nucleotides 1386375 to 1387490) and ORF 6 (nucleotides 1387487 to 1388401) overlap by 3 nucleotides, RT-PCR confirmed that both are transcribed. In contrast, no PCR products were obtained for HVO_1519, HVO_1520, HVO_1523 to HVO_1525, or ORF 7.
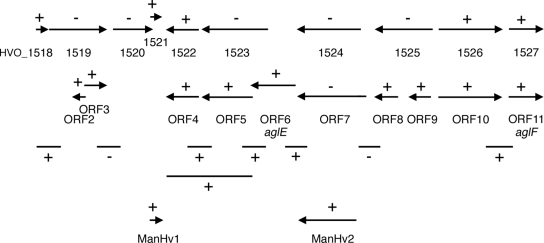
FIG. 2.
Schematic depiction of the transcripts generated by RT-PCR. +, sequences that generated transcripts; −, sequences that failed to generate transcripts. Primers raised against either the proposed 5′ and 3′ regions of the gene of interest or sequences spanning the 3′ region of a sequence and the 5′ region of the downstream sequence, designed to address cotranscription of the sequences of interest and listed in Table S1 in the supplemental material, were employed.
Next, RT-PCR was performed using primers designed to amplify sequences encompassing the region between the various ORFs under consideration, together with at least 100 bp of each of the flanking ORFs. PCR products were obtained from the region linking HVO_1526 (or ORF 10) and HVO_1527 (ORF 11, i.e., aglF) and from the region spanning the space between HVO_1522 (ORF 4) and ORF 5. In fact, a single PCR product spanning the complete HVO_1522 (ORF 4) and ORF 5 sequences could be obtained using the appropriate primer pair, pointing to the cotranscription of these ORFs. The regions between HVO_1518 and HVO_1519 and between ORF 6 (aglE) and HVO_1524 (or the similar ORF 7) also yielded PCR products, despite the fact that no amplification of either HVO_1519 or HVO_1524 (or the similar ORF 7) was achieved. This implies that the HVO_1518 transcript includes a region found at the start of HVO_1519, while the ORF 6 transcript includes a region found at the end of ORF 7. Finally, no PCR products could be amplified when the region between HVO_1519 and HVO_1520 or the common region between HVO_1524 and HVO_1525 (essentially corresponding to the common region between ORF 7 and ORF 8) served as the template (Fig. 2).
Visual inspection reveals novel ORFs in the agl cluster.
As a PCR product was obtained when DNA spanning the end of HVO_1524 (a region shared by ORF 7) and the beginning of ORF 6 (algE) served as the template whereas no product was generated when either HVO_1524 or ORF 7 served as the PCR template, the possibility exists that the automated annotation algorithms described above failed to recognize the true start site of the gene erroneously annotated as HVO_1524 or ORF 7 within this section of the genome. Moreover, the finding that a PCR product only 131 bp long was generated when HVO_1521 served as the template raised the question of whether here too the automated annotation had correctly identified the start site of the ORF in question. Therefore, the region downstream of the predicted start site of HVO_1524 and ORF 7 (corresponding to largely overlapping sequences, both found on the reverse strand) and the region upstream of HVO_1521 (found on the forward strand) were manually analyzed to identify alternative potential in-frame start sites. To then confirm whether any of these potential start sites are indeed employed, RT-PCR was performed using appropriate primers.
To manually analyze that region of the H. volcanii genome automatically annotated as either HVO_1524 (predicted to begin at position 1389884) or ORF 7 (predicted to begin at position 1389935), RT-PCR was performed using a forward primer encompassing those nucleotides surrounding position 1389689, together with the same reverse primer used in unsuccessful attempts to amplify HVO_1524 and ORF 7. The use of this new primer pair now yielded a PCR product. Hence, the amplified sequence, spanning the region between positions 1388454 and 1389689 identified through visual inspection, was annotated as ManHv2 (Table 1).
The sequence automatically annotated as HVO_1521 is proposed to begin at position 1385335. When RT-PCR was performed with a forward primer that encompassed those nucleotides surrounding position 1385200 (containing the only other start codon in the region between HVO_1520 and HVO_1521 and which could yield an in-frame product) together with the same reverse primer as used in the earlier amplification of the shorter sequence, a PCR product was generated. Therefore, the region spanning positions 1385200 to 1385463, a 264-bp sequence putatively encoding an 88-residue polypeptide, also now identified through visual inspection, was annotated as ManHv1 (Table 1).
Bioinformatics analysis of deduced ORF protein products.
As a next step toward identifying ORFs that encode N-glycosylation-related proteins in that portion of the agl gene cluster between aglJ and aglF, protein expression was considered. Initially, the deduced amino acid sequences of the products of HVO_1518-HVO_1526, ORFs 2 to 5, ORFs 7 to 10, and ManHv1 and -2 were analyzed. Apart from ORF 9, all of the ORFs under consideration can be translated into amino acid sequences expected for true proteins. In the case of ORF 9, 50% of the protein is deduced as corresponding to Ser and Thr residues, casting doubt as to whether this DNA sequence indeed represents a real gene.
The isoelectric points of the amino acid sequences deduced from the various ORFs under study were next considered. Haloarchaeal proteins are generally characterized by an enhanced acidic amino acid content, a property related to the ability of such proteins to remain properly folded in hypersaline surroundings (10, 13). In contrast to the acidic pI values predicted for many of the sequences examined (Table 2), the highly basic pI values of HVO_1524 (pH 9.0), ORF 3 (pH 10.2), ORF 7 (pH 8.7), ORF 8 (pH 9.9), and ORF 9 (12.1) raise the question of whether these sequences indeed encode true H. volcanii proteins.
TABLE 2.
Bioinformatics analysis of nonannotated ORFs in the H. volcanii agl cluster
| ORF | Homologue determined by:
|
BLAT result | Predicted protein
|
||
|---|---|---|---|---|---|
| Conserved-domain BLAST | BLAST | No. of amino acids | pI | ||
| HVO_1518 | None | Transposase (ISH5) | 15 | 95 | 5.2 |
| HVO_1519 | Transposase | Transposase (ISH3) | 39 | 397 | 6.0 |
| HVO_1520 | Transposase | Transposase (ISH5) | 14 | 295 | 7.1 |
| HVO_1521 | None | Hypothetical | 14 | 43 | 4.6 |
| HVO_1522a | None | Methyltransferase | 0 | 239 | 4.8 |
| HVO_1523 | None | Hypothetical | 0 | 469 | 5.5 |
| HVO_1524 | Involved in O-antigen export | Polysaccharide biosynthesis/transport | 0 | 476 | 9.0 |
| HVO_1525 | Transposase | Transposase (ISH3) | 37 | 396 | 5.6 |
| HVO_1526 | None | Hypothetical | 0 | 457 | 6.1 |
| ORF 2 | None | None | 38 | 136 | 5.5 |
| ORF 3 | Transposase | Transposase | 38 | 177 | 10.2 |
| ORF 5 | None | Hypothetical | 0 | 371 | 5.6 |
| ORF 7 | Involved in O-antigen export | Polysaccharide biosynthesis/transport | 0 | 493 | 8.7 |
| ORF 8 | None | Transposase | 37 | 177 | 9.9 |
| ORF 9 | None | Hypothetical | 37 | 149 | 12.1 |
| ORF 10 | Transposase | None | 0 | 457 | 6.1 |
HVO_1522 = ORF 4.
While deduced amino acid content and pI values can provide some indication of which of the sequences being investigated encode true proteins, the existence of homologues to a given sequence in other organisms offers stronger support than do those criteria considered above. Therefore, the H. volcanii ORFs under examination were used as bait in BLAST and conserved-domain BLAST searches. As shown in Table 2, only the deduced products of HVO_1522 (ORF 4) and of the overlapping HVO_1524, ORF 7, and ManHv2 sequences were revealed as being homologous to proteins putatively involved in glycosylation elsewhere. Specifically, these sequences were shown to be homologues of a methyltransferase (HVO_1522 [ORF 4]) or of proteins involved in polysaccharide biosynthesis/transport (HVO_1524, ORF 7, or ManHv2). HVO_1518, HVO_1519, HVO_1520, HVO_1525, ORF 3, ORF 8, and ORF 9 were reported as being homologous to members of the ISH3 and ISH5 transposase families. Given that BLAT analysis, a homology-based search tool available at the H. volcanii genome site (http://archaea.ucsc.edu/cgi-bin/hgGateway?db=haloVolc1), reveals the presence of numerous copies of these ISH3 and ISH5 transposase family sequences scattered throughout the H. volcanii genome, it is reasonable to assume that HVO_1518, HVO_1519, HVO_1520, HVO_1525, ORF 3, ORF 8, and ORF 9 serve some function in the cell, albeit one distinct from glycosylation. No homologues were detected for HVO_1521, HVO_1523, HVO_1526, ORF 2, ORF 5, ORF 10, or ManHv1 within H. volcanii or any other organism.
Confirmation of ORF identification at the protein expression level.
While offering some information on the H. volcanii sequences under investigation, the various computer-based approaches enlisted and discussed in the previous section are not sufficient to determine whether a given ORF encodes a true protein or not. To this end, a more direct strategy designed to assess protein translation was adopted. In this approach, H. volcanii cells were transformed to express constructs in which the ORF under study and a region containing the preceding 200 bp upstream (possibly containing the promoter of that ORF) were fused in frame (at the 3′ end of the ORF) to DNA encoding a version of GFP designed to work in the hypersaline environment of this haloarchaeon (19). Expression of fusion proteins containing the C-terminal GFP tag was then revealed by immunoblotting using anti-GFP antibodies.
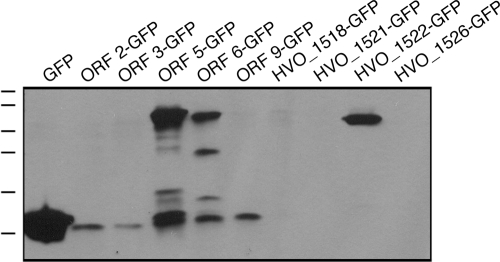
When cells were transformed to express either GFP-tagged ORF 5 or ORF 6 (aglE), which overlap by 3 nucleotides, protein bands of 66 and 58 kDa, respectively, in addition to GFP-sized bands (26 kDa) and intermediate-sized breakdown products, were labeled by the anti-GFP antibodies (Fig. 3). As reported above, both of these H. volcanii ORFs generated transcripts, with the ORF 6 product having been previously shown to correspond to AglE, a known component of the H. volcanii N-glycosylation apparatus (3). A construct encoding a GFP-tagged version of HVO_1522 (ORF 4) also generated a protein band migrating at a slightly higher mass than the combined predicted molecular masses of the H. volcanii ORF product and the GFP moiety (approximately 24 and 26 kDa, respectively), in keeping with the detection of a transcript for this H. volcanii sequence (see above). In this case, no intermediate breakdown products or GFP-sized bands were detected. Indeed, a previous proteomics-based study had also reported that HVO_1522 (ORF 4) encodes a true protein (12). That study did not, however, consider either ORF 5 or ORF 6 (aglE).
FIG. 3.
The appearance of GFP-containing fusion proteins reveals genes that are expressed. As described in Materials and Methods, H. volcanii cells were transformed with plasmids carrying a sequence of interest fused at its 3′ end to DNA encoding GFP. This construct was preceded by the 200-bp region immediately preceding the sequence of interest. In a control experiment, plasmid pJAM1020 (19), directing the expression of GFP alone, was employed. Expression of the GFP-containing chimeras was then assessed by immunoblot analysis using antibodies against GFP. The positions of molecular mass markers (100, 70, 55, 40, 35, and 25 kDa) are shown on the left.
When GFP-ManHv1 and GFP-ManHv2 expression was considered, only protein bands migrating slightly faster than GFP were antibody labeled (not shown). Similarly, ORF 2, ORF 3, and ORF 9, all of which also generated transcripts in the RT-PCR studies described above, failed to be expressed as GFP-bearing fusion proteins, with only GFP-sized or slightly heavier protein bands being detected. Finally, no GFP-incorporating protein products or GFP-sized bands were detected in cells transformed to express GFP fused to HVO_1518, HVO_1521, or HVO_1526 (or the highly similar ORF 10 sequence), despite these sequences also being transcribed. Finally, HVO_1519, HVO_1520, HVO_1523, HVO_1524, HVO_1525, and ORF 7, all of which failed to generate PCR products in the RT-PCR-based transcription studies described above, were not tested for their abilities to generate the GFP fusion proteins.
Based on these studies, it can be concluded that the H. volcanii portions of the full-length, C-terminally GFP-tagged fusion proteins detected, i.e., HVO_1522 (ORF 4), ORF 5, and ORF 6 (aglE), indeed encode expressed proteins. An earlier proteomic study also confirmed the translation of HVO_1522 (ORF 4) (12). In the cases of ManHv1, ManHv2, ORF 2, ORF 3, and ORF 9, sequences for which RNA was transcribed, it is not clear whether only the GFP portion of each fusion protein was expressed or whether degradation of the expressed full-length chimera had occurred. Similarly, in the case of the transcribed HVO_1518, HVO_1521, and HVO_1526 (or the highly similar ORF 10) sequences, one cannot distinguish between failure of expression (possibly in response to the growth conditions employed) and instability of the translated polypeptides. Thus, one cannot discount the possibility that these latter two groups of genes also encode protein products.
In other halophilic archaea, agl gene homologues are also found in a gene island.
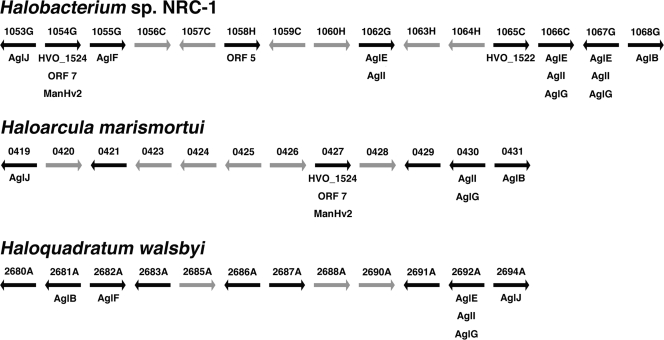
In addition to that of H. volcanii, the genomes of four other halophilic archaea, namely, Halobacterium sp. strain NRC-1 (Halobacterium salinarum) (18), Haloarcula marismortui (4), Haloquadratum walsbyi (5), and Natronomonas pharaonis (9), are publicly available. The detection of agl gene homologues and the proven or likely presence of S-layer glycoproteins in these species suggest that they too perform N glycosylation. Accordingly, efforts were directed at addressing the distribution of H. volcanii agl gene homologues in these other haloarchaea. As schematically depicted in Fig. 4, gene islands that include homologues of H. volcanii agl genes, in addition to other ORFs annotated as encoding products involved in glycosylation, are found in other haloarchaeal genomes, with the homologue of H. volcanii aglB, encoding the sole component of the archaeal oligosaccharide transferase (1, 6, 11), serving as the island-defining component. In Halobacterium sp. strain NRC-1, the gene region between VNG1053G and VNG1069C includes homologues of H. volcanii aglB, aglE, aglF, aglG, aglI, and aglJ, as well as of H. volcanii HVO_1522 (ORF 4) and HVO_1524, ORF 7, or ManHv2. Like H. volcanii, Halobacterium sp. strain NRC-1 has also been experimentally confirmed as performing N glycosylation (8, 14). The Halobacterium sp. strain NRC-1 genome also includes VNG0318G, a homologue of H. volcanii aglD, the product of which is involved in adding the final subunit to the pentasaccharide decorating the H. volcanii S-layer glycoprotein. In both haloarchaea, aglD is found outside the agl cluster described here. The Haloarcula marismortui genome also contains a gene cluster that includes homologues of H. volcanii aglB, aglF, aglI, and aglJ, together with homologues of H. volcanii HVO_1522 (ORF 4) and HVO_1524, ORF 7, or ManHv2. The Haloquadratum walsbyi gene island that includes a homologue of H. volcanii aglB also includes homologues of H. volcanii aglE, aglF, aglG, and aglJ. The same region also includes other ORFs annotated as a glycosyltransferase and nucleoside diphosphate sugar epimerases. In contrast, the N. pharaonis homologue of H. volcanii aglB (i.e., NP3720) is quite distant from the cluster situated between NP4654A and NP4680A that includes homologues of H. volcanii aglF and aglJ, as well as other sequences annotated as UDP-glucose dehydrogenases, phosphohexomutases, and nucleoside diphosphate epimerases (not shown).
FIG. 4.
Schematic representation showing the presence of gene clusters containing H. volcanii agl gene homologues in other haloarchaea. Regions of the genomes of Halobacterium sp. strain NRC-1, Haloarcula marismortui, and Haloquadratum walsbyi that include H. volcanii agl gene homologues, homologues to other previously annotated sequences found within the H. volcanii agl gene-containing island, homologues to H. volcanii ORF sequences defined in this study, or homologues to other sequences automatically annotated as participating in protein glycosylation are shown (black arrows). The gray arrows in each gene cluster represent sequences automatically annotated as hypothetical proteins or proteins assigned functions apparently unrelated to protein glycosylation. The arrows representing each ORF do not reflect the relative length of a given sequence; instead, they are of arbitrary but equal length. The number above each arrow corresponds to the identification number of that ORF in the genome sequence of that haloarcheaon.
DISCUSSION
To fully exploit the wealth of information contained within a completed genome sequence, correct sequence annotation is essential. While gene-finding algorithms offer relatively rapid delineation of gene sequences within a genome, the accuracy of such automated efforts are often questionable. Indeed, as revealed in this study (and in a previous effort [3]), the same shortcoming holds true for the available annotation of the H. volcanii genome. The gene encoding AglE, which has been experimentally proven to participate in addition of the fourth subunit of the pentasaccharide decorating the S-layer glycoprotein, corresponds to the 5′ portion of HVO_1523 as well as most of the nonannotated region between HVO_1523 and HVO_1524 (3) and, accordingly, was not detected by the automated annotation used to generate the available H. volcanii genome sequence. The same holds true for ORF 5, which has been confirmed to correspond to a true gene as revealed here by studies performed at the DNA, RNA, and protein levels. In the current annotation of the H. volcanii genome, ORF 5 corresponds to the last 1,115 bp of the predicted 1,409-bp HVO_1523 sequence. On the other hand, the other sequence verified as generating mRNA and a protein product in this study (and in a previous report [12]), i.e., ORF 4, was correctly annotated as HVO_1522.
Of the other H. volcanii sequences considered in this study, no transcripts were detected in the cases of HVO_1519, HVO_1520, HVO_1524, HVO_1525, and ORF 7. The reannotation efforts of the present study explain, in some instances, why this was the case. HVO_1519 was reannotated to encompass ORFs 2 and 3, while HVO_1525 was reannotated to encompass ORFs 8 and 9. All of these novel ORFs were transcribed. Visual inspection revealed that HVO_1524 and ORF 7 in fact contain ManHv2, which was transcribed. No GFP-ManHv2 protein product was detected, however. Similarly, despite the reannotation of HVO_1521 as the longer ManHv1 (both of which were transcribed), no GFP-tagged ManHv1 protein could be generated. Moreover, no protein products were translated from HVO_1518, HVO_1521, ORF 2, ORF 3 (or the highly similar ORF 8), ORF 9, or HVO_1526, despite the observed transcription of these sequences. It is thus possible that appropriate conditions for the transcription and/or expression of at least some of the sequences listed above were not realized in the present study. Therefore, gene products for some of the sequences could appear under suitable growth conditions. Indeed, differential expression of proteins involved in mediating H. volcanii N glycosylation could be related to the proposed adaptive role of this posttranslational modification in this organism (1, 23). Of course, the possibility remains that the set of sequences for which no transcription was obtained also includes ORFs that do not correspond to true genes.
Within the H. volcanii agl gene cluster, several sequences are cotranscribed. In addition to a previous report showing aglF and aglI to be simultaneously transcribed (22), the present study reveals cotranscription of at least two additional pairs of genes. A single transcript encoding both HVO_1522 (ORF 4) and ORF 5 was observed, while in the case of aglE and ManHv2, a single transcript corresponding to regions extending some 100 bp into each partner sequence, together with the stretch linking these sequences, could be generated. Hence, given their proximities to genes known to participate in N glycosylation, BLAST-predicted glycosylation-related roles in the case of HVO_122 (ORF 4) and ManHv2 (Table 2), and, in the case of ManHv2, cotranscription with aglE, a known N-glycosylation component pathway (3), it is reasonable to assume that HVO_1522 (ORF 4), ORF 5, and ManHv2 also participate in protein N glycosylation. Accordingly, HVO_1522 (ORF 4) is now renamed aglP, ORF 5 is now renamed aglQ, and ManHv2 is now renamed aglR (Fig. 5). In keeping with this reannotation, mass spectrometry studies have verified the participation of AglP and AglQ in S-layer glycoprotein N glycosylation (S. Yurist-Doutsch et al., unpublished data).
FIG. 5.
Revised version of the H. volcanii agl gene cluster. Numbers along the horizontal bar denote nucleotide positions in the genome. Proven or putative N-glycosylation genes are termed agl (archaeal glycosylation) sequences. For genes not assigned roles in N glycosylation, proposed functions are given below each sequence.
Although the precise chemical composition of the pentasaccharide decorating the H. volcanii S-layer glycoprotein remains to be described, earlier efforts have identified agl gene products that participate in the assembly or attachment of this oligosaccharide (1-3, 23; Abu-Qarn et al., unpublished data). Of these, all but aglD reside within the agl gene cluster now shown to also include the confirmed (12) HVO_1522 (ORF 4 [i.e., aglP]) and the newly delineated ORF 5 (aglQ), ManHv1, and ManHv2 (aglR) genes. In contrast to what is observed in H. volcanii, where the majority of agl sequences identified exist in a cluster (as is largely true for homologous sequences in Halobacterium sp. strain NRC-1, Haloarcula marismortui, and Haloquadratum walsbyi as well), the same does not hold true for N-glycosylation genes in the other model archaeal system where this posttranslational modification has been studied in detail, namely, the methanogen Methanococcus voltae. Instead, the M. voltae N-glycosylation genes identified thus far, namely, aglA (Mv151), aglB (Mv1749), aglC (Mv990), aglH (Mv1751), and aglK (Mv991) (6, 7, 21), are more widely distributed throughout the genome. Such differential distribution of genes involved in similar roles in the H. volcanii and M. voltae genomes may provide insight into the evolution of this protein-processing event.
In conclusion, genes involved in most of the steps of H. volcanii N glycosylation have now been identified. Future efforts can now determine the precise roles of products of these genes, including the newly delineated sequences reported here, in H. volcanii N glycosylation.
Supplementary Material
Acknowledgments
We thank Moshe Mevarech, Tel Aviv University, for suggesting the GFP fusion protein experiments.
J.E. is supported by grants from the Israel Science Foundation (grant 30/07) and the U.S. Air Force Office for Scientific Research (grant FA9550-07-10057). S.Y.-D. is the recipient of a Negev-Faran Associates Scholarship.
Footnotes
Published ahead of print on 27 February 2009.
Supplemental material for this article is available at http://jb.asm.org/.
REFERENCES
- 1.Abu-Qarn, M., and J. Eichler. 2006. Protein N-glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol. Microbiol. 61511-525. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Qarn, M., S. Yurist-Doutsch, A. Giordano, A. Trauner, H. R. Morris, P. Hitchen, O. Medalia, A. Dell, and J. Eichler. 2007. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 3741224-1236. [DOI] [PubMed] [Google Scholar]
- 3.Abu-Qarn, M., A. Giordano, F. Battaglia, A. Trauner, P. Hitchen, H. R. Morris, A. Dell, and J. Eichler. 2008. Identification of AglE, a second glycosyltransferase involved in N glycosylation of the Haloferax volcanii S-layer glycoprotein. J. Bacteriol. 1903140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baliga, N. S., R. Bonneau, M. T. Tacciotti, M. Pan, G. Glusman, E. W. Deutsch, P. Shannon, Y. Chui, R. S. Weng, R. R. Gan, P. Hung, S. V. Date, E. Marcotte, L. Hood, and W. V. Ng. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 142221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolhuis, H., P. Palm, A. Wende, M. Falb, M. Rampp, F. Rodriguez-Valera, F. Pfeiffer, and D. Oesterhelt. 2006. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaban, B., S. Voisin, J. Kelly, S. M. Logan, and K. F. Jarrell. 2006. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol. Microbiol. 61259-268. [DOI] [PubMed] [Google Scholar]
- 7.Chaban, B., S. M. Logan, J. F. Kelly, and K. F. Jarrell. 2009. AglC and AglK are involved in biosynthesis and attachment of diacetylated glucuronic acid to the N-glycan in Methanococcus voltae. J. Bacteriol. 191187-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichler, J., and M. W. W. Adams. 2005. Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69393-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falb, M., F. Pfeiffer, P. Palm, K. Rodewald, V. Hickmann, J. Tittor, and D. Oesterhelt. 2005. Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis, Genome Res. 151336-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuchi, S., K. Yoshimune, M. Wakayama, M. Moriguchi, and K. Nishikawa. 2003. Unique amino acid composition of proteins in halophilic bacteria. J. Mol. Biol. 327347-357. [DOI] [PubMed] [Google Scholar]
- 11.Igura, M., N. Maita, J. Kamishikiryo, M. Yamada, T. Obita, K. Maenaka, and D. Kohda. 2008. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 27234-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkland, P. A., M. A. Humbard, C. J. Daniels, and J. A. Maupin-Furlow. 2008. Shotgun proteomics of the haloarchaeon Haloferax volcanii. J. Proteome Res. 75033-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanyi, J. K. 1974. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol. Rev. 38272-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lechner, J., and F. Wieland. 1989. Structure and biosynthesis of prokaryotic glycoproteins. Annu. Rev. Biochem. 58173-194. [DOI] [PubMed] [Google Scholar]
- 15.Linton, D., N. Dorell, P. G. Hitchen, S. Amber, A. V. Karlyshev, H. R. Morris, A. Dell, M. A. Valvano, M. Aebi, and B. W. Wren. 2005. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 551695-1703. [DOI] [PubMed] [Google Scholar]
- 16.Mescher, M. F., and J. L. Strominger. 1976. Purification and characterization of a prokaryotic glycoprotein from the cell envelope of Halobacterium salinarium. J. Biol. Chem. 2512005-2014. [PubMed] [Google Scholar]
- 17.Mevarech, M., and R. Werczberger. 1985. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162461-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 9712176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuter, C. J., and J. A. Maupin-Furlow. 2004. Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins, Appl. Environ. Microbiol. 707530-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider, K. L., K. S. Pollard, R. Baertsch, A. Pohl, and T. M. Lowe. 2006. The UCSC Archaeal Genome Browser. Nucleic Acids Res. 34D407-D410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shams-Eldin, H., B. Chaban, S. Niehus, R. T. Schwarz, and K. F. Jarrell. 2008. Identification of the archaeal alg7 gene homolog (encoding N-acetylglucosamine-1-phosphate transferase) of the N-linked glycosylation system by cross-domain complementation in Saccharomyces cerevisiae. J. Bacteriol. 1902217-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yurist-Doutsch, S., B. Chaban, D. VanDyke, K. F. Jarrell, and J. Eichler. 2008. Sweet to the extreme: protein glycosylation in Archaea. Mol. Microbiol. 681079-1084. [DOI] [PubMed] [Google Scholar]
- 23.Yurist-Doutsch, S., M. Abu-Qarn, F. Battaglia, H. R. Morris, P. G. Hitchen, A. Dell, and J. Eichler. 2008. aglF, aglG and aglI, novel members of a gene cluster involved in the N-glycosylation of the Haloferax volcanii S-layer glycoprotein. Mol. Microbiol. 691234-1245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.